Abstract
Recent work by several groups has significantly expanded our knowledge of the structure, regulation of assembly, and function of components of the extracellular portion of the type III secretion system (T3SS) of Gram-negative bacteria. This perspective presents a structure-informed analysis of functional data and discusses three nonmutually exclusive models of how a key aspect of T3SS biology, the sensing of host cells, may be performed.
Keywords: flagella, structure
Type III secretion systems (T3SS) are protein transport apparatuses required for the interaction of many Gram-negative bacteria with eukaryotic hosts. Their major purpose is to inject, directly into host cells, effector proteins that modulate eukaryotic cell function to aid infection (1, 2). T3SS are genetically, structurally, and functionally related to bacterial flagella (3), which are motility devices that also contain an export apparatus (4). Expression and self-assembly of both flagella and T3SS are regulated in response to external stimuli on reaching the correct physiological niche (e.g., temperature, chemical environment). Furthermore, stepwise assembly and activation is hierarchical, involving the tightly coordinated interplay of >25 proteins; including structural elements, intracellular chaperones, regulatory components, and secreted effectors.
Structurally T3SS consist of a basal body that spans both bacterial membranes (5), thereby allowing secretion of substrates without a periplasmic intermediate, and an external needle, which is regulated in length to bridge the LPS on the bacterial surface and the distance to the host cell, ensuring delivery of effectors to the host cell cytoplasm (6, 7). Key to the assembly and subsequent function of the T3SS is the ≈25-Å channel that runs through the center of the entire structure (8). First to be assembled is the basal body in a process that is still poorly characterized at a structural level and therefore not further discussed here. Subsequently, secretion of the major needle subunit through the basal body leads to assembly of the extracellular needle, whose basic structure was recently revealed to be highly conserved (described below). Needle assembly is under tight control by regulatory mechanisms that determine its length. This issue is complex and has recently been reviewed (1, 2), we therefore do not touch on this issue here. After needle completion other proteins are secreted through the needle and assemble at the tip, where they are thought to perform roles in host sensing (9–13). Activation of the T3SS by host cells results in the secretion of two hydrophobic proteins, which, probably from the tip of the needle, insert directly into the eukaryotic membrane to form an apparatus known as the translocon (13). The translocon forms an ≈25-Å pore in the host membrane that allows translocation of partially unfolded effectors into the host cytoplasm (14–16). These latter steps of translocon insertion and effector secretion also lack the required structural information to underpin a structure-informed synthesis and hence are also not discussed herein. Recent structural and functional work in a number of the important mammalian pathogen T3SS families has revealed important information regarding the assembly and activation of the extracellular portion of the T3SS. By bringing together the structural and functional data, our aim is to interpret earlier hypotheses concerning the mechanisms underlying host–cell sensing at a molecular level.
The Needle
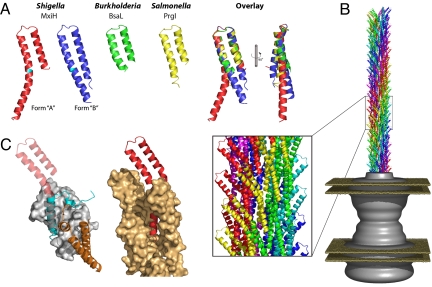
The major extracellular component of the T3SS is a needle (17–19) that extends from the outer-membrane portion of the apparatus and through which runs an ≈25-Å channel forming the secretion conduit (8). The needle is formed by a helical assembly of multiple copies (on the order of 100–150) of a single, small (≈9 kDa) protein, which is highly conserved between T3SS from animal pathogens (20). The helical parameters of this needle (≈5.5 subunits per turn; ≈4.6-Å axial rise per subunit) are very similar to those defining the assembly of the components of the extracellular portion of bacterial flagella, although the subunits show no detectable sequence similarity (21). The inherent ability of these proteins to polymerize renders them unsuitable for high-resolution structural studies. However, analogies to the proteins of the flagellar filament and hook, which have unstable N and C termini known to be required for polymerization, suggested that truncations of these termini could yield structurally manipulable forms of the proteins (22, 23). There are now structures for three needle subunits (Fig. 1A) generated by x-ray crystallography and NMR: MxiH (Shigella flexneri) (24), BsaL (Burkholderia pseudomallei) (25), and PrgI (Salmonella typhimurium) (26). All of these structures are of proteins that have been rendered monodisperse by expression of constructs lacking five C-terminal residues (≈6% of the sequence). The structures (Fig. 1A) reveal a consistent picture of the central half of the T3SS needle subunit as a coiled-coil with a short (≈6 residue) connecting loop (rmsd = 1.1 ± 0.3 Å over ≈40 residues). Outside of this central portion, all of these structures reveal a highly mobile N terminus and most structures are also disordered at the C terminus. However, both NMR structures (BsaL and PrgI) show that these regions maintain some degree of helical structure in solution. These structures together suggest that the termini are connected to the central portion of the molecule via highly flexible hinges. Indeed, one of the MxiH structures (form A shown in red in Fig. 1A) contains an ordered C terminus, which continues the helix in the central portion but is bent around Tyr-60. Based on the conservation of the structure of the central portion and relative mobility of the termini we have termed the central portion the “head,” with the termini forming a “tail” flexibly hinged to the head. The low level of sequence diversity among T3SS needle subunits of mammalian pathogens, in combination with these structures, now allow us to predict that they will all share this common fold.
Fig. 1.
Structures of needle proteins and their complexes. (A) Cartoons of monomeric needle proteins: the crystal structures of two conformations of MxiH from S. flexneri (form A, red; form B, blue) with the position of Tyr-60 at the bend highlighted in cyan [ref. 24 and Protein Data Bank (PDB) ID code 2CA5], the NMR structures (well ordered regions only) of BsaL from Burkholderia pseudomallei (green) (ref. 25 and PDB ID code 2G0U) and PrgI from Salmonella typhimurium (yellow) (ref. 26 and PDB ID code 2JOW) and an overlay using the head region. (B) The Shigella T3SS needle (120 copies of the MxiH monomer as docked (PDB ID code 2V6l) into the EM reconstruction, EMD-1416) shown protruding from the EM reconstruction of the Shigella basal apparatus [gray surface (8) EMD-1422] with the inner and outer membranes illustrated (planes in sand). (Inset) A magnified region of the needle assembly shows the arrangement of needle subunits. (C) (Left) The crystal structure (ref. 27 and PDB ID code 2UWJ) of a fragment of PscF from Pseudomonas aeruginosa (cyan) bound to the chaperones PscE (orange) and PscG (surface in light gray) overlaid with MxiH (transparent red) via the C terminus (residues 65–80 of MxiH) is shown. (Right) A portion of the Shigella needle showing a MxiH monomer (red) surrounded by adjacent MxiH molecules in the assembly (surface in light orange) is shown.
The problems seen in generating nonpolymerizing forms of the subunits for structural work highlight a problem for the bacteria, which must also prevent inappropriate, intracellular, polymerization. Overlay of the MxiH A subunit onto the portion of the Pseudomonas aeruginosa needle subunit seen in complex with its specific intracellular chaperone (27) (Fig. 1C) suggests a solution to the problem. The face of the C-terminal helix that forms the majority of the subunit–chaperone contacts is also the face of the helix that forms the majority of the subunit–subunit contacts in the assembled needle. By masking this face, premature polymerization is prevented. Additionally, the bulk of the chaperone prevents the N terminus packing against the C terminus as seen in the model of the intact needle. As specific intracellular chaperones are a feature of many T3SS secreted proteins (reviewed in ref. 28), this mechanism would seem to provide a general method for chaperoning of all T3SS needle subunits. However, instead it highlights a further problem: no direct homologues of this chaperone are found in the Shigella/Salmonella T3SS family (29).
Although monomeric forms of the needle subunit were required to reveal their atomic structures, they could not yield insight into the assembly of the intact needle. However, for the Shigella system we had earlier determined a 16-Å resolution electron microscopy reconstruction of the polymerized needle (21). Combination of both the helical parameters that define the assembly and the shape of the reconstructed volume led to a unique solution for fitting of the MxiH A structure into the EM density, thus yielding an atomic model for the assembled needle (24). The MxiH structures lacked the N-terminal 20 residues, and examination of the needle assembly in the reconstruction revealed an unmodeled piece of density, running alongside the C-terminal helix, of the appropriate size for a 20-residue helix at the N terminus of the truncated model. The model for the intact needle assembly is shown in Fig. 1B in the context of the earlier reconstruction for the Shigella basal body (8) and shows that the MxiH head groups decorate the needle surface. The N-terminal helix lines the inner channel, while the C-terminal helix is almost completely buried in forming the assembly, explaining why truncations in this region led to nonpolymerizing forms of the protein. Because T3SS needle subunits share a common subunit structure, it is likely they will also share a common needle architecture. Therefore, our assembled needle provides a reasonable paradigm for all needles of T3SS from animal pathogens.
There are, however, some caveats to bear in mind. Principally, the moderate resolution of the EM reconstruction means that the accuracy of the pseudoatomic model is limited, particularly when the 20 N-terminal residues are modeled rather than taken from a high-resolution structure. This means that while the backbone and perhaps also the side chains of exposed residues (where their environment is likely to be similar to that seen in the atomic structures) are likely to be accurate, we cannot be confident of side-chain orientations and specific interactions in the core of the assembly. Therefore, we cannot yet analyze side chain-dependent properties (such as electrostatic potential) of the assembly. Higher-resolution reconstructions of the intact needle or atomic structures of subcomplexes are needed to allow meaningful consideration of such properties.
The needle model displays clear structural homology to the packing of the flagellar D0 domains within the filament (30), raising the question of whether the mechanism of assembly is also conserved. Construction of the flagellum absolutely requires a distal cap protein to rotationally catalyze the correct insertion of each subunit at the tip of the growing filament (31). However, no such cap protein has been identified in any T3SS. The smaller size of the needle subunits, relative to the flagellin subunit, may mean that the termini of the unfolded subunit remain in sufficiently close proximity to the top of the assembling needle that they correctly associate, without needing to be trapped at the assembly site by a separate capping system. This argument assumes that the needle assembles from the tip as demonstrated for other related systems (32, 33).
Proteins Assembled at the Needle Tip: The Adaptor Proteins
The needle culminates in a tip complex that contains multiple copies of an adaptor protein (9, 10, 12, 13, 34). The adaptor protein is required in several species for regulation of secretion activity and insertion of the translocation pore (e.g., refs. 35–38). Additionally, the Shigella adaptor protein (IpaD) was recently found to be required for association of the most highly conserved translocon component within the tip complex (11, 13, 39). Thus, the adaptor appears to mediate the interaction between the needle and the translocon and may also be involved in sensing the host cell (discussed below).
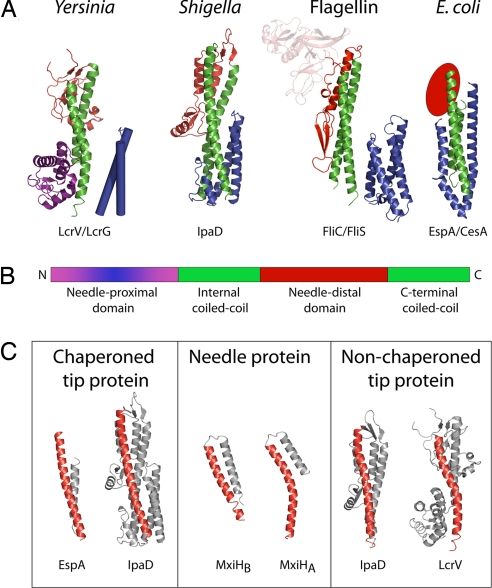
Despite the high degree of conservation among the needle subunits, there is low sequence identity between the adaptor proteins. However, recent structures have revealed significant structural similarities (22, 39–42), as originally predicted based on functional homologies (3). Representative structures for members of the different families are shown in Fig. 2A and reveal that the proteins share a common topology consisting of a long, central coiled-coil formed between the extreme C-terminal helix and a helix earlier in the sequence, an inserted domain between these two helices and, in some cases, a domain at the extreme N terminus (Fig. 2B). Presence of a coiled-coil region in these proteins and the needle subunits suggest a common mechanism of assembly. The wide variation in the type of domains found at either end of the coiled-coil suggests that different functionalities can be bolted on to the basic structure required for assembly at the needle tip. This variation is presumably to accomplish species-specific biological activities and may explain the low level of overall sequence identity among the adaptor proteins. However, the high degree of structural homology between the Shigella family of adaptor proteins (IpaD in Fig. 2A) and the flagellin family (FliC in Fig. 2A) and even closer similarities to FlgL, one of the flagellar hook/junction proteins (K. Imada, H. Matsunami, and K. Namba, personal communication) may give insight into the structure of the evolutionary ancestor of the adaptors.
Fig. 2.
Needle tip-associated adaptor protein structures. (A) Cartoon representation of the four classes of adaptor proteins thus solved: Yersinia family (LcrV; PDB ID code 1R6F) (41); Shigella family (IpaD; PDB ID code 2J0O) (39); Flagellin family (FliC; PDB ID code 1IO1) (22); E. coli family (EspA; PDB ID code 1XOU) (42). The structures demonstrate a common architecture, consisting of a central coiled-coil (green), a needle-distal domain (red), and a needle-proximal domain (purple/blue). The needle-proximal portion of the coiled-coil, which is postulated to be involved in assembly at the needle-tip, is chaperoned (shown in blue). This chaperoning may be via an intramolecular mechanism (IpaD: IpaD N-terminal domain) or separate chaperones [EspA:CesA, PDB ID code 1XOU (42), FliC:FliS, PDB ID codes 2IO1/1ORJ (22, 40), LcrV:LcrG, LcrV; PDB ID code 1R6F; no structure available for LcrG, modeled on IpaD N-terminal domain structure]. (B) Schematic representation of the common architecture. (C) The C-terminal helix of the central coiled-coil is straight in chaperoned adaptor proteins (IpaD chaperoned, PDB ID zj0o and bent in the nonchaperoned structure (IpaD nonchaperoned, PBD ID 2j0n). This bend is comparable to the kink in the C-terminal helix of the Shigella needle protein MxiH (PDB ID code 2CA5) (24).
As these adaptor proteins are structurally designed to polymerize atop the needle, it is unsurprising that they are chaperoned and that the chaperoning strategies are structurally analogous to those used in chaperoning needle subunits, i.e., masking of the area of the coiled-coil that needs to be buried on assembly. What is surprising is the range of strategies used (Fig. 2A). They vary from use of the N-terminal domain of the same polypeptide chain [e.g., IpaD (39)], to association of a separate chaperone [e.g., CesA chaperones EspA (43), FliS chaperones FliC (40, 44)]. Interestingly, although all of the adaptor proteins contain a long coiled-coil, comparison of their structures reveals that there is significant conformational variation and flexibility in the coiled-coil, which seems to correlate with chaperoning (Fig. 2C). Chaperoned adaptor proteins contain more linear forms of the coiled-coil, whileas those of unchaperoned forms are bent, producing structures reminiscent of both the straight B-form and bent A-form of the MxiH needle subunit. This inherent flexibility in the regions involved in assembly may be relevant to the process of assembly or another biological role, such as transmission of a signal through the apparatus.
In addition to variation in chaperoning strategies, there is significant diversity in the proposed stoichiometries of these proteins, varying from ≈5 copies (≈1 helical turn) of LcrV per Yersinia needle (10, 20, 24) to ≈11 copies (≈2 helical turns) of FlgL per flagellum (45, 46) to thousands of copies of EspA per enteropathogenic Escherichia coli needle (42). This diversity raises questions about how different systems regulate assembly of the adaptors. Such differences in regulation may be partly explained by the wide variation in the nature and size of the domains bolted on to the coiled-coil. For example, the relatively bulky domains of the Yersinia LcrV protein sterically prevent assembly of more than five copies, whereas the flexible C-terminal domain of EspA, combined with the lack of an N-terminal domain, imposes no such constraints.
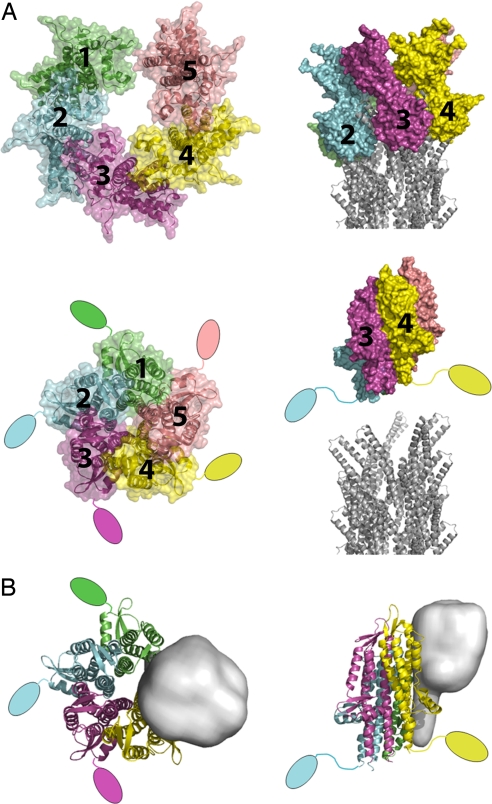
Given the structural conservation of the coiled-coil between the needle and adaptor proteins, the simplest structural model for tip-complex assembly is that it will be driven by similar interactions to those used to drive needle subunit assembly. Consistent with this idea is the finding that the C-terminal residues of the Shigella family adaptor proteins are critical for assembly on the tip (9, 13, 20). This knowledge, combined with the structural homology between the bent C-terminal helices of LcrV and the A form of MxiH, allowed us to construct an atomic model for the Yersinia tip complex on the Shigella needle (Fig. 3A and ref. 24). Our LcrV tip-complex model bears a central channel that matches the diameter of the needle channel (≈25 Å). The full assembly of the MxiH needle with the LcrV tip complex therefore provides us with a model for the needle/tip complex that is in an “open” state. Others (10) have modeled a tip-complex structure that has rotational rather than helical symmetry based on averages of electron microscopy of negatively stained structures. Although the detail of the molecular interactions is quite different between the models, there are many overall similarities (e.g., proposed copy number, assembly driven by the coiled-coil region). High-resolution 3D reconstructions of a needle-tip complex are required to unambiguously distinguish between the two hypotheses. In their absence we discuss the most structurally conservative proposal, the assembly of a helical tip complex on the helical needle structure.
Fig. 3.
Models for tip-associated protein assembly. (A) Adaptor protein homopentamers as suggested by the LcrV EM data (10) (Upper) and the IpaD crystal structure (39) (Lower) suggest open and closed forms of the tip complex, respectively. The position of the N-terminal chaperoning domain of IpaD is unknown in the pentamer and so is shown in a more extended conformation projecting away from the tip/needle (only two are shown in the side view for clarity). The Shigella needle is shown as cartoons (PDB ID code 2V6L) (24). (B) Ribbon representation of a proposed hetero-pentamer tip complex, consisting of four copies of IpaD (colored as in A) and one copy of IpaB (shown in gray as a blurred surface and modeled on the structure of the pore-forming domain of the Colicin family of pore-forming toxins).
By contrast, crystals of the unchaperoned form of the Shigella adaptor (39) contained an IpaD–IpaD homodimer where the two molecules were related to one another by rotation and translation parameters similar to those that define the helical arrangement of the needle subunits. Expansion of this relationship to assemble additional IpaDs allowed insertion of no more than five subunits to reveal a tight homopentamer (Fig. 3A and ref. 39). This alternate model of a tip complex is clearly closed to protein export, with a central pore that would only allow passage of water or other small molecules. Mutation of residues involved in IpaD–IpaD interactions in this closed tip led to reduced translocon-insertion capacity (39), supporting the functional relevance of this complex. The packing of the IpaD C termini in this complex is significantly tighter than the packing of the MxiH C termini in the needle assembly, therefore this model for a closed tip cannot be easily positioned on the top of our present needle model.
The Shigella Needle Point: From Sensor to Translocon?
Although details of the needle and adaptor protein structures are now emerging from several species, allowing us to understand which features are conserved and which are species-specific, there is still relatively little information about the molecular basis by which these systems sense the host cell. In the last year, however, significant advances have been made in understanding the molecular basis of host cell sensing for Shigella species. The structural conservation between all needle and adaptor subunits and sequence conservation between the first translocon components implies conservation at a mechanistic level. It is clear that there may be differences between the specific molecules involved between species. However, because all T3SS must detect and transmit a signal from the extracellular needle tip to the secretion regulators at the base of the basal body, it is likely that the overall molecular strategies associated with host-cell sensing and signaling will be conserved.
Recent biochemical evidence has conclusively demonstrated that one of the translocon components, IpaB, is also present in the tip complex of Shigella (11, 13, 39), although in ≈5-fold lower amounts than IpaD (13). IpaB is the most conserved translocon component in most T3SS families but, to date, none of the homologs have been localized to needle tips. Therefore, it is presently uncertain whether these findings are applicable to all animal pathogen T3SS, but we find their mechanistic implications relevant enough to the sensing process to discuss them here. IpaB (and its homologs from other T3SS families) is currently structurally uncharacterized, but bioinformatic approaches (39) suggest that its topology will be consistent with that of IpaD and other needle-assembled proteins, i.e., a coiled-coil with two bolted-on domains. Using this information it was possible to build a heteropentamer consisting of four molecules of IpaD and one molecule of IpaB. Taking the fact that IpaD is required for assembly of IpaB (13, 39) at the tip to mean that IpaD assembles first, the first copy of IpaD would assemble onto the needle via interactions with MxiH subunit below. The second, third, and fourth copies of IpaD would then all assemble by using the same interactions with MxiH below and a neighboring IpaD on one side. The fifth site for assembly, however, would differ from all previous sites in that it would be lined on both sides by IpaD. We therefore assume that the unique nature of this site would favor insertion of IpaB. This logic allowed us to generate the cartoon shown in Fig. 3B for the Shigella tip complex that represents our current state of knowledge. One consequence of this model is that IpaB must interact with the needle-distal domains of IpaD, which would explain the observation that deletion of this domain does not prevent IpaD assembly at the tip, but does prevent assembly of IpaB (39).
The second component of the Shigella translocon, IpaC, has been shown in a needle mutant to specifically and functionally associate with the tip complex before activation by host-cell contact (13). IpaC cannot obviously be assigned to the same predicted topology as IpaD and IpaB. However, the last 50 residues of IpaC are strongly predicted to contain a C-terminal coiled-coil-forming helix, suggesting some commonalities in architecture with IpaD and IpaB. This C-terminal region has also been demonstrated to be involved in biological activities inside the host-cell cytoplasm (47). However, it is not known whether this role depends on membrane-inserted IpaC attached at the needle tip or on further molecules of IpaC subsequently translocated into the cytoplasm. Evidently, therefore, further experiments will be needed to ascertain whether IpaC assembles atop IpaB and IpaD as they do atop the needle, to complete the translocon within the host membrane.
Models for T3SS Activation by Host-Cell Sensing
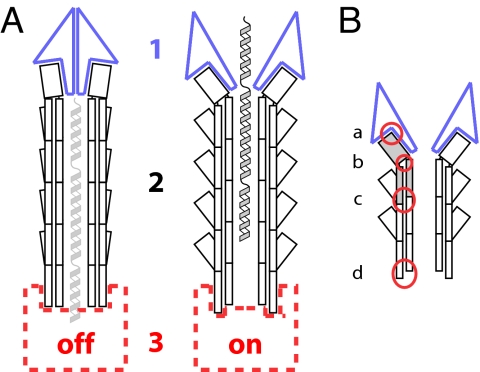
Over the years, various models have been proposed for how the signal of host-cell contact is relayed through the T3SS to activate secretion within the bacterial cytoplasm. All of the models, described below, involve structural and functional changes at some combination of three different sites within the T3SS (Fig. 4A). Site 1 corresponds to the tip complex, and site 2 is the needle. The composition and structure of both are now well understood. Site 3, the base of the needle, is poorly defined in terms of either composition or structure. The first model proposed (site 1, Fig. 4A), long before the existence of the needle and the tip complex were recognized, was that of an extracellular plug clogging the exit of the T3SS (37). The logical extension of this model is that the plug would have to be structurally altered, i.e., opened or removed, by host-cell contact. This idea developed from the discovery that mutants in IpaD and IpaB, or several of their homologs in other species, were incapable of translocon insertion and constitutively secreted effectors (48). Thus, this model was the first to suggest that a change at the extracellular top of the apparatus might be sufficient to allow activation of the secretion machinery (site 3, Fig. 4A). Testing of this model has been complicated by the fact that these proteins also seem to be involved in regulation of secretion internally (38). Nevertheless, the extracellular plug model has been remarkably borne out by the findings that both IpaD and IpaB localize to the needle tip complex. Furthermore, experimental interest in it can only be stimulated by the structural evidence presented in Fig. 3, which suggests that adaptor proteins can form both closed and open structures, which could act to physically block or release the secretion channel of the needle.
Fig. 4.
Schematic illustrations of possible different states of the T3SS needle complex and sites of action for needle mutants. (A) Changes in three distinct regions of the T3SS may be involved in the regulation of secretion after host-cell sensing: the tip complex (1, blue), the needle itself (2, black), and the basal apparatus (3, red). Secretion substrates are shown in the needle channel in a partially unfolded state (light gray and gray). (B) Mutations of the needle protein can alter secretion patterns by disruption of four different interfaces (red circles): needle subunit to tip complex (a), within the needle subunit, i.e., intramolecular (b), between the needle subunits, i.e., intermolecular (c), and needle subunit to base (d).
Other experimental evidence suggests that there are gating mechanisms located in the bacterial cytoplasm or on the cytoplasmic face of the inner membrane (site 3, Fig. 4A). Evidence exists for multiple mechanisms regulating secretion at the base of the apparatus, such as the YopN/TyeA family (e.g., refs. 49 and 50); briefly reviewed in ref. 51. These proteins are not essential for activation of secretion per se, but rather seem to act as selective repressors or activators of the secretion of certain classes of substrates, serving to generate a hierarchy of secretion. Absence of members of this family has been linked to reduced/abolished secretion of translocators and enhanced secretion of effectors (e.g., ref. 52). It is therefore possible that some of these proteins, many of which become secreted upon T3SS activation, are also involved in coupling this gating mechanism with exogenous activation signals (e.g., ref. 53). The most crucial, but also most difficult, part of this model to investigate experimentally is the mode of transmission of exogenous activation signals to the cytoplasmic or inner-membrane T3SS apparatus. If one excludes the needle as a means of transmission, the only direct physical connection between tip and base would have to be hypothetical polypeptide(s) in the process of being secreted (light gray helices in Fig. 4A). Evidence is mounting that during T3SS/flagellar assembly sensing and termination of needle/hook growth occurs through just such a mechanism and involves a molecular “ruler”/“tape measure” protein, which is secreted (54, 55). It is hypothesized that while in the process of secretion these extended proteins have their N terminus interacting with the needle/hook top, while the C terminus is still within the bacterial cytoplasm, abutting the inner-membrane export apparatus and so ideally placed to flip an activation switch (54, 56). Although no such data exist for activation of secretion by host-cell contact this model proposes that needle assembly, tip assembly, and effector secretion activation all are controlled by variations on a common signaling mechanism.
Our final model proposes that the needle (site 2) plays an active role in transmitting the host-cell contact signal from the tip to the base of the T3SS (3). Contact of the proteins bound at the top of the needle with the host cell would generate a signal that is relayed downward via structural changes in each needle subunit in turn to the base. We do not know the nature of any such structural change, but current evidence suggests that it would have to be relatively subtle, because a change in the helical parameters of the needle has been excluded (57). Therefore, it may relate to inherent flexibility of the subunit or simply to rearrangements of side chains (represented in Fig. 4 as an entirely hypothetical change in the relative orientation of the head and tail of the needle protein). However, the atomic model of the Shigella needle indicates that such a signal cannot be transmitted via the most conceptually simple route, through the shallowest helix of the needle. The small size of the needle protein monomer, combined with its fold and the helical parameters, means that there are no contacts between the molecules of the one-start helix. Should such a signal be transmitted through the needle, therefore, it must involve a different mechanism, such as one involving the protofilament.
These models are supported by the identification of numerous single amino acid changes within the Yersinia and Shigella needles that lead to altered secretion phenotypes (58–60). The different locations of the mutations leading to such deregulation of T3SS activation are indicated in Fig. 4B, where the four different types of interactions that can be affected are circled: (a) needle subunit to tip complex; (b) within the needle subunit; (c) between the needle subunits; and (d) needle subunit to base. Mutations at any of these sites can, via different mechanisms, lead to phenotypes such as the inability to correctly assemble the needle/tip complex or to detect the host cell and insert the transolocon. It is clear that the same surfaces on the needle subunit are used to make different contacts (e.g., needle protein/needle protein, needle protein/tip complex, and needle protein/base), meaning that it is very difficult to dissect precisely which is the major step affected by each mutant. In addition, rapid signal transduction systems are notoriously difficult to dissect. For this reason most needle mutagenesis studies can be reinterpreted to support any of the individual models above (or combinations thereof). However, mutagenizing the needle is sufficient to affect the ability of the T3SS to regulate its activity in response to host-cell contact (59, 60). Therefore, mapping of the needle mutants onto the needle structure currently provides the best clues to the path of transduction of the host-cell contact signal from tip to base (24).
Outlook
In addition to an informed mutational and functional reanalysis of tip-complex components guided by the new atomic structures available, what is now most urgently required to help positively distinguish between the different mechanistic models is moderate to high-resolution structural data for different parts of this supramolecular assembly separately and, crucially, together. Given the present state of our knowledge, initial efforts will likely center on obtaining reconstructions of needle/tip-complex assemblies in different functional states. Such studies are likely to require development of novel methodologies, including using tip structures to aid alignment of needle EM images and use of methods such as FRET or EPR capable of investigating dynamic changes in needle structure, to overcome the technical difficulties associated with study of small helical assemblies such as the needle. It is likely, however, that the mechanistic concepts that emerge from new high-resolution information about how T3SSs sense host cells will in time help to understand functionally and structurally analogous properties of a broad variety of microbial systems (61–65).
Acknowledgments.
We thank Tohru Minamino for comments on the manuscript. S.J. and P.R. are funded by Medical Research Council Grants G0400389 and G0400775 (to S.M.L.). J.E.D. is funded by Wellcome Trust Grant 077082 (to S.M.L. and P.R.). AK.J.V. is funded by European Community Marie Curie Postdoctoral Fellowship MEIF-CT-2005-023694. J.L.H. is supported by Medical Research Council Grant G0401595 (to A.J.B.). A.J.B. is supported by a Guy G. F. Newton Senior Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 3.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proc Natl Acad Sci USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minamino T, Namba K. Self-assembly and type III protein export of the bacterial flagellum. J Mol Microbiol Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- 5.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 6.Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR. Bacterial injectisomes: Needle length does matter. Science. 2005;307:1278. doi: 10.1126/science.1107679. [DOI] [PubMed] [Google Scholar]
- 7.West NP, et al. Optimization of virulence functions through glucosylation of Shigella LPS. Science. 2005;307:1313–1317. doi: 10.1126/science.1108472. [DOI] [PubMed] [Google Scholar]
- 8.Blocker A, et al. Structure and composition of the Shigella flexneri“needle complex,” a part of its type III secreton. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 9.Espina M, et al. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller CA, et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 11.Olive AJ, et al. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun. 2007;75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sani M, et al. IpaD is localized at the tip of the Shigella flexneri type III secretion apparatus. Biochim Biophys Acta. 2007;1770:307–311. doi: 10.1016/j.bbagen.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Veenendaal AK, et al. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol. 2007;63:1719–1730. doi: 10.1111/j.1365-2958.2007.05620.x. [DOI] [PubMed] [Google Scholar]
- 14.Blocker A, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakansson S, et al. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 16.Neyt C, Cornelis GR. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: Requirement for translocators YopB and YopD, but not LcrG. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 17.Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamano K, et al. Supramolecular structure of the Shigella type III secretion machinery: The needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broz P, et al. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol. 2007;65:1311–1320. doi: 10.1111/j.1365-2958.2007.05871.x. [DOI] [PubMed] [Google Scholar]
- 21.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 22.Samatey FA, et al. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- 23.Deane JE, et al. Expression, purification, crystallization, and preliminary crystallographic analysis of MxiH, a subunit of the Shigella flexneri type III secretion system needle. Acta Crystallogr. 2006;62:302–305. doi: 10.1107/S1744309106006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deane JE, et al. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci USA. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J Mol Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinaud M, et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci USA. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsot C, Hamiaux C, Page AL. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 29.Troisfontaines P, Cornelis GR. Type III secretion: More systems than you think. Physiology (Bethesda) 2005;20:326–339. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- 30.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 31.Yonekura K, et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 32.Crepin VF, Shaw R, Abe CM, Knutton S, Frankel G. Polarity of enteropathogenic Escherichia coli EspA filament assembly and protein secretion. J Bacteriol. 2005;187:2881–2889. doi: 10.1128/JB.187.8.2881-2889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Q, He SY. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science. 2001;294:2556–2558. doi: 10.1126/science.1066397. [DOI] [PubMed] [Google Scholar]
- 34.Wilson RK, Shaw RK, Daniell S, Knutton S, Frankel G. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:753–762. doi: 10.1046/j.1462-5822.2001.00159.x. [DOI] [PubMed] [Google Scholar]
- 35.Bergman T, et al. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: Evidence for a regulatory role of LcrH and LcrV. J Bacteriol. 1991;173:1607–1616. doi: 10.1128/jb.173.5.1607-1616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersson J, et al. The V-antigen of Yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol Microbiol. 1999;32:961–976. doi: 10.1046/j.1365-2958.1999.01408.x. [DOI] [PubMed] [Google Scholar]
- 37.Menard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picking WL, et al. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson S, et al. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evdokimov AG, et al. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Mol Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- 41.Derewenda U, et al. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure. 2004;12:301–306. doi: 10.1016/j.str.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Yip CK, Finlay BB, Strynadka NC. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 43.Creasey EA, et al. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol Microbiol. 2003;47:209–221. doi: 10.1046/j.1365-2958.2003.03290.x. [DOI] [PubMed] [Google Scholar]
- 44.Auvray F, Thomas J, Frase GM, Hughes C. Flagellin polymerization control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda T, Homma M, Iino T, Asakura S, Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987;169:1168–1173. doi: 10.1128/jb.169.3.1168-1173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones CJ, Macnab RM, Okino H, Aizawa S. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 47.Tran Van Nhieu G, Caron E, Hall A, Sansonetti PJ. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 1999;18:3249–3262. doi: 10.1093/emboj/18.12.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsot C, Menard R, Gounon P, Sansonetti PJ. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 49.Ferracci F, Schubot FD, Waugh DS, Plano GV. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol Microbiol. 2005;57:970–987. doi: 10.1111/j.1365-2958.2005.04738.x. [DOI] [PubMed] [Google Scholar]
- 50.Schubot FD, et al. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J Mol Biol. 2005;346:1147–1161. doi: 10.1016/j.jmb.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 51.Pallen MJ, Beatson SA, Bailey CM. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type III secretion. BMC Microbiol. 2005;5:9. doi: 10.1186/1471-2180-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng W, et al. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun. 2005;73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, et al. Regulatory role of PopN and its interacting partners in type III secretion of Pseudomonas aeruginosa. J Bacteriol. 2007;189:2599–2609. doi: 10.1128/JB.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 55.Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J Mol Biol. 2006;359:466–477. doi: 10.1016/j.jmb.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 56.Minamino T, et al. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J Mol Biol. 2004;341:491–502. doi: 10.1016/j.jmb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 57.Cordes FS, et al. Helical packing of needles from functionally altered Shigella type III secretion systems. J Mol Biol. 2005;354:206–211. doi: 10.1016/j.jmb.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 58.Davis AJ, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J Bacteriol. 2007;189:83–97. doi: 10.1128/JB.01396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenjale R, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 60.Torruellas J, Jackson MW, Pennock JW, Plano GV. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 61.Gerlach RG, Hensel M. Protein secretion systems and adhesins: The molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 62.Burrows LL. Weapons of mass retraction. Mol Microbiol. 2005;57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- 63.Deghmane AE, Giorgini D, Maigre L, Taha MK. Analysis in vitro and in vivo of the transcriptional regulator CrgA of Neisseria meningitidis upon contact with target cells. Mol Microbiol. 2004;53:917–927. doi: 10.1111/j.1365-2958.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 64.Zahrl D, Wagner M, Bischof K, Koraimann G. Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. J Bacteriol. 2006;188:6611–6621. doi: 10.1128/JB.00632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kostyuchenko VA, et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]