Abstract
The biological functions of individual members of the large family of chitinase-like proteins from the red flour beetle, Tribolium castaneum (Tc), were examined by using gene-specific RNAi. One chitinase, TcCHT5, was found to be required for pupal–adult molting only. A lethal phenotype was observed when the transcript level of TcCHT5 was down-regulated by injection of TcCHT5-specific dsRNA into larvae. The larvae had metamorphosed into pupae and then to pharate adults but did not complete adult eclosion. Specific knockdown of transcripts for another chitinase, TcCHT10, which has multiple catalytic domains, prevented embryo hatch, larval molting, pupation, and adult metamorphosis, indicating a vital role for TcCHT10 during each of these processes. A third chitinase-like protein, TcCHT7, was required for abdominal contraction and wing/elytra extension immediately after pupation but was dispensable for larval–larval molting, pupation, and adult eclosion. The wing/elytra abnormalities found in TcCHT7-silenced pupae were also manifest in the ensuing adults. A fourth chitinase-like protein, TcIDGF4, exhibited no chitinolytic activity but contributed to adult eclosion. No phenotypic effects were observed after knockdown of transcripts for several other chitinase-like proteins, including imaginal disk growth factor IDGF2. These data indicate functional specialization among insect chitinase family genes, primarily during the molting process, and provide a biological rationale for the presence of a large assortment of chitinase-like proteins.
Keywords: functional genomics, gene family, molting defects, wing abnormalities
Chitinases (β-1,4-poly-N-acetylglucosaminidase; EC 3.2.1.14) are members of the O-glycoside hydrolase superfamily and are found in many species, including microbes, plants, insects, and mammals (1). Insect chitinases, which belong to family 18 glycosylhydrolases, have been detected in molting fluid and gut tissues and are predicted to mediate the digestion of chitin present in the exoskeleton and peritrophic membrane (PM) in the gut to chitooligosaccharides (2, 3). Genes and cDNAs encoding insect chitinases have been identified and characterized from several lepidopteran, dipteran, and coleopteran insects (4–6). Even though only one (or occasionally two) chitinase gene had been previously identified in studies involving many insect species, database searches of fully sequenced genomes from Drosophila, Anopheles, and, more recently, Tribolium, have revealed that each of these insects has a rather large family of genes encoding chitinase and chitinase-like proteins with 16–23 members, depending on the species (refs. 7 and 8 and unpublished data). Based on amino acid sequence similarity and phylogenetic analysis, insect chitinase family proteins have been classified into five or possibly more groups (7, 8). From currently available data on a large number of biochemically characterized insect chitinases, we have hypothesized that many and perhaps all group I insect chitinases are involved in chitinolysis associated with insect molting. However, the functions of proteins belonging to the other four groups, some of which lack chitinase activity, have not been explored in detail. In this study, we have used RNA interference (RNAi) to gain insight into the functions of several Tribolium chitinase-like genes. This coleopteran is sensitive to systemic RNAi by single injections of 50- to 200-ng amounts of dsRNA at any developmental stage (9, 10). We chose the following genes representing each of the five groups as targets for RNAi: the group I gene TcCHT5; group II gene, TcCHT10; group III gene, TcCHT7; for group IV (which has many members), TcCHT2, -6, -8, -14, and -16; and group V genes, TcIDGF2 and TcIDGF4. Transcript levels of specific chitinase genes were shown to be substantially down-regulated by injection of gene-specific dsRNAs. By monitoring these dsRNA-injected animals and observing the developmental abnormalities that ensue, distinct functions for some of these genes have been identified. Both TcCHT5 and TcCHT10 are involved in molting. TcCHT5 is required for adult eclosion only, whereas TcCHT10 contributes to every molt and also to egg hatch. TcCHT7 is required for abdominal contraction and wing expansion after ecdysis. There were no detectable abnormalities in TcCHT2, -6, -8, -14, -16, and TcIDGF2 dsRNA-treated insects. Although Tribolium castaneum (Tc)IDGF4 has no chitinolytic activity, it is required for adult eclosion. Injection of dsRNA for this gene had no effect on pupation, but the pupal–adult molt was incomplete and pharate adults that were trapped in the pupal cuticle subsequently died.
Results
Expression Profiles of Genes for Tribolium Chitinase-Like Family.
To help determine the optimal time(s) for dsRNA injections, the expression profiles of Tribolium chitinase-like genes were determined by RT-PCR by using cDNAs prepared from RNAs isolated at different stages of Tribolium development [supporting information (SI) Fig. S1]. All group IV genes (TcCHT2, -4, -6, -8, -9, -11, -12, -13, -14, -15, and -16) were expressed mainly in the feeding stages (larvae and adults). TcCHT4, TcCHT9, and TcCHT16 were also expressed in the pharate pupal and pupal stages. The group I gene TcCHT5, group II gene TcCHT10, group III gene TcCHT7, and group V genes TcIDGF2 and TcIDGF4 were expressed throughout all stages of development, from young larvae to adults. TcCHT5 and TcCHT7 were mainly expressed in the pupal and adult stages. TcCHT10 and TcIDGF4 transcripts were also detected in the embryonic stages.
Specificity of Knockdown of Chitinase Transcripts by dsRNA.
To confirm the specificity of RNAi, dsRNAs for TcCHT2, TcCHT5, TcCHT6, TcCHT10, and TcCHT16 were injected into insects in the penultimate larval instar (200 ng per insect, n = 20), at which time all of these genes are expressed. RT-PCR analyses of RNA isolated 3 days after injection showed that each dsRNA suppressed transcript levels of its target gene only, without reducing transcript levels of nontarget chitinase family genes (Fig. S2).
TcCHT5 Is Required for the Pupal–Adult Molt.
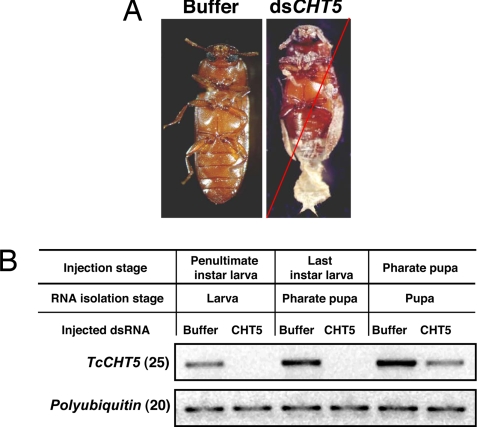
All animals injected with TcCHT5 dsRNA (n = 20) exhibited a lethal phenotype only at the pharate adult stage, no matter when the dsRNA was injected (larval, pharate pupal, or pupal stages). The insects died ≈5–6 days after pupation. The larval–larval and larval–pupal molts were normal. At the time of death, the adult cuticle was visible under the old pupal cuticle. The old pronotal cuticle was split open, but the animals failed to shed their old cuticles, which remained attached to their abdomens (Fig. 1A). Buffer-injected animals showed no phenotypic abnormalities and developed into healthy adults. RT-PCR analyses carried out by using cDNA templates prepared from dsRNA-injected insects indicated that there were no detectable transcripts for TcCHT5 when dsRNA for TcCHT5 was injected into either penultimate instar or last instar larvae (Fig. 1B). The transcript knockdown efficiency in pharate pupae was lower when compared with larvae, but, nonetheless, a pupal–adult lethal phenotype was observed in all dsRNA-injected animals. Injection of dsRNA for TcCHT5 into young adult females had no effect on their morphology, survival, or number of eggs laid.
Fig. 1.
Effect of dsRNA for TcCHT5 on larval, pupal, and adult development in Tribolium. (A) Typical terminal phenotype produced by injection of dsRNA for TcCHT5 (Right). Injection of dsTcCHT5 (200 ng per insect) into penultimate and last instar larvae, as well as pharate pupae, prevented only the pupal–adult molt. Larvae (n = 20) injected with buffer developed normally into pupae and then into adults (Left). The red slash across the image indicates the phenotype of the insect at the time of death. (B) Effect of dsRNA for TcCHT5 on transcript levels. Three to 4 days after injections, four insects from each treatment at the indicated stage of development were collected for total RNA preparation, followed by RT-PCR. Another reaction with a pair of primers for polyubiquitin gene was used as the internal loading control.
TcCHT10 Contributes to Larval–Larval, Larval–Pupal, and Pupal–Adult Molting and Egg Hatch.
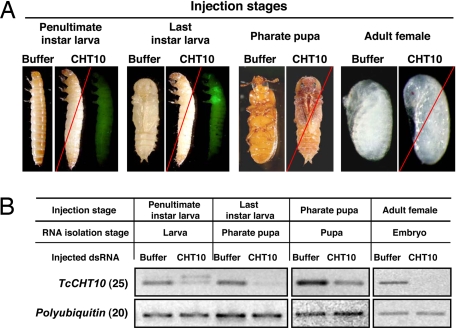
Preliminary experiments with Tribolium GA-1 strain larvae indicated that, unlike dsRNA for TcCHT5, injection of dsRNA for TcCHT10 resulted in two different terminal phenotypes, presumably because of differences in developmental stage at the time of injection (data not shown). For subsequent larval injections, the Pig-23 strain of Tribolium was used, which allows more precise discrimination between penultimate and last instar larvae. This strain carries an EGFP transgene under the control of an eye-specific promoter. The transgene is inserted near an imaginal disk enhancer, which leads to expression of EGFP in the imaginal discs only in the last instar, which is easily detectable under fluorescence microscopy. When dsRNA for TcCHT10 was injected into penultimate instar larvae, they failed to molt to the next larval instar. Apolysis and slippage of the old cuticle was visible, but these insects failed to complete ecdysis. The beetles died, apparently when they tried to molt to the last larval stadium, because there was no EGFP expression in the wing imaginal discs (Fig. 2A, “penultimate instar larvae”). When these insects were dissected and examined, a newly synthesized larval cuticle was observed under the old cuticle (data not shown). When TcCHT10 dsRNA was injected into last instar larvae, pupation was affected (Fig. 2A, “last instar larvae”). The pupal cuticle was fully developed under the loosened larval cuticle that remained attached to the lower abdomen (data not shown). EGFP expression was observed in the wing imaginal discs, confirming that developmental arrest had occurred at the last larval instar. Injection of TcCHT10 dsRNA also prevented pupal–adult molting. These insects developed into pharate adults when dsRNA for TcCHT10 was injected into pharate pupae, but they failed to shed their old pupal cuticle (Fig. 2A, “pharate pupae”). When dsRNA for TcCHT10 was injected into female adults, there was no observable effect on adult morphology or survival. The adults oviposited a similar number of eggs compared with control insects. Fully developed embryos were observed inside these eggs (Fig. 2A, “adult females”). However, none of the eggs hatched into larvae, suggesting a role for this chitinase in egg hatching.
Fig. 2.
Effect of dsRNA for TcCHT10 on embryonic, larval, pupal, and adult development and egg hatch of Tribolium. The Pig-23 strain that has an EGFP tag to identify last instar larvae was used in this experiment. (A) dsRNA for TcCHT10 (200 ng per insect, n = 20) was injected into penultimate- or last-instar larvae, pharate pupae, or adult females as indicated above each image. All TcCHT10 dsRNA-injected animals died at the ensuing molt. Terminal phenotypes are shown. The left two sets of images (injections into penultimate and last instar larvae) show the same two individuals viewed microscopically under visible light (specimen on the left within the image labeled CHT10) and after excitation at 480 nm and monitoring emission at 510 nm to view the EGFP fluorescence (specimen on the right within the image labeled CHT10). When last instar larvae were injected with dsRNA for TcCHT10 (note EGFP fluorescence in wing imaginal discs), death occurred at the pupal stage with the larval cuticle still loosely attached to the posterior abdomen. The pharate pupa specimen labeled CHT10 (third image from the right) shows a typical terminal phenotype of insects injected at the pharate pupal stage. The far right image labeled CHT10 shows the embryos inside eggs laid by adult females injected with dsRNA for TcCHT10. These embryos are fully developed but failed to hatch. (B) Effect of buffer or dsRNA for TcCHT10 on transcript levels. Three to 4 days after injection, four insects from each treatment were collected for total RNA preparation and cDNAs were prepared and used as templates for RT-PCR. The number of RT-PCR cycles is indicated in parentheses except for egg samples (Right), in which RT-PCR was carried for 25 cycles with polyubiquitin primers and 30 cycles with TcCHT10 primers. The appearance of a larger band in some samples is attributable to contamination of RNA preparations with pre-mRNA for the target gene. The increase in size of the PCR product is consistent with the presence and size of a known intron between the two primer-binding sites.
RT-PCR analyses revealed that the levels of transcripts for TcCHT10 were significantly lower in dsRNA-injected animals compared with buffer-injected controls (Fig. 2B). Even though the knockdown of TcCHT10 transcripts was incomplete in some cases, all dsRNA-injected animals (n = 20) exhibited the same lethal phenotype and none survived to adulthood.
TcCHT7 Is Required for Abdominal Contraction and Wing Extension.
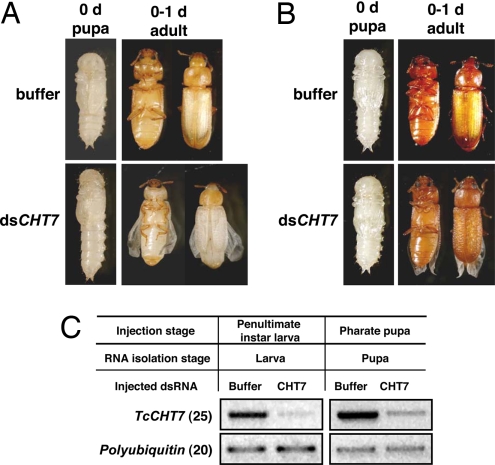
Injection of dsRNA for TcCHT7 did not interfere with larval or pupal development. When dsRNA for TcCHT7 was injected into penultimate instar larvae, the larvae completed the larval–larval and larval–pupal molts. However, there were some noticeable differences during the molts. The resulting pupae failed to contract their abdomens to the same extent as control insects and also failed to fully expand their elytra. They, thus, retained a more larval-like appearance than buffer-injected pupae. Other aspects of pupal maturation also were interrupted, including extension and folding of legs and antennae and expansion of the pronotum (Fig. 3A). Nonetheless, these pupae survived and developed into adults. However, the elytra were very short, approximately two-thirds of the normal size, and the surface of the elytra was rumpled instead of smooth. The hind wings did not fold or unfold properly. When injection of dsRNA was carried out at the pharate pupal stage, pupation occurred without any delay relative to buffer-injected animals (Fig. 3B). Adult eclosion also occurred as in the control insects, but the elytra did not expand to their full length and had a rough upper surface. RT-PCR results indicated that the level of the transcript for TcCHT7 was significantly down-regulated by TcCHT7 dsRNA in both larval and pupal stages compared with buffer-injected controls (Fig. 3C).
Fig. 3.
Effect of dsRNA for TcCHT7 on pupal or adult development of Tribolium. dsRNA for TcCHT7 (200 ng of dsRNA per insect; n = 20) was injected into last instar larvae (A) or into pharate pupae (B), and morphologies were recorded as shown. Normal phenotypes were observed in the buffer-injected control insects (top images of each set). Note that TcCHT7 dsRNA-injected larvae pupated on schedule, but the resulting pupae failed to contract their abdomens and to expand their wings/elytra. After adult eclosion, the hind wings also did not fold or unfold properly. The surface of the elytra was very uneven, and the elytra were substantially shorter than normal. When dsRNA for TcCHT7 was injected into pharate pupae, near-normal phenotypes were observed in the pupal stage. However, unlike buffer-injected controls, TcCHT7 dsRNA-treated insects failed to fully expand their adult wings/elytra, and their wings did not fold or unfold properly. (C) Effect of dsRNA on TcCHT7 transcript levels. Four insects were collected from each treatment group for RNA extraction. RT-PCR was used to detect the transcription level of TcCHT7.
TcIDGF4 Is Necessary for Adult Eclosion.
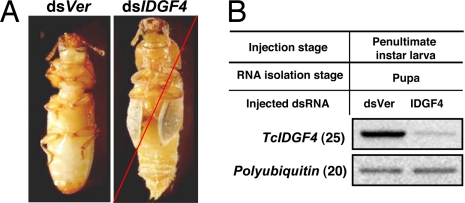
dsRNAs for the two imaginal disk growth factors (IDGFs) were injected into penultimate instar and last instar larvae as well as female adults. dsRNA for TcIDGF2 had no effect on pupation or adult eclosion (data not shown). When dsRNA for TcIDGF4 was injected into penultimate or last instar larvae, pupation was normal, but the insects died during adult eclosion (Fig. 4; data for penultimate instar injection not shown). Injection of dsRNA for TcIDGF2 or TcIDGF4 into young adult females had no effect on their fecundity or fertility.
Fig. 4.
Effect of dsRNA for TcIDGF4 on Tribolium pupal development and pupal–adult molting. (A) dsRNA for TcIDGF4 or vermillion (200 ng of dsRNA per insect; n = 20) was injected into penultimate instar larvae. Normal larval–larval molt and pupation were observed in both dsVer-injected control insects and TcIDGF4 dsRNA-injected insects. Approximately 14 days after injection, the control pupae became normal adults (Left), but the pupae treated with dsRNA for TcIDGF4 were unable to molt and died entrapped in their pupal cuticles (Right). (B) Four insects from each treatment were collected for total RNA preparation. RT-PCR product with a pair of primers for the polyubiquitin gene was used as the internal loading control.
No Phenotypes Observed After Knockdown of Other Chitinase Gene Transcripts.
Injection of dsRNAs corresponding to TcCHT2, TcCHT6, TcCHT8, TcCHT14, and TcCHT16 did not result in any observable abnormalities at any developmental stage (data not shown). Injection of a mixture of dsRNAs for three group IV genes (TcCHT2, TcCHT6, and TcCHT16) also failed to result in any observable abnormalities.
Discussion
We have identified 23, 17, and 16 chitinase family genes in Tribolium, Drosophila, and Anopheles, respectively (refs. 7 and 8 and unpublished data on seven additional chitinase-like genes belonging to group IV which have not been fully characterized). Some of the chitinases present in molting fluid or gut are presumably involved in the turnover of chitin in the exoskeleton and PM during the molting process. However, the rationale for the presence of such a large family of chitinases and related proteins has not been analyzed in detail to date in any insect species. It is possible that differences in the types of chitin (α, β, or γ) or their modification (such as partial deacetylation) may require different chitinases for effective turnover of chitin in different extracellular structures. The chitinases belonging to different groups have distinctly different developmental patterns of expression and tissue specificity, suggestive of distinct biological functions (Fig. S1 and Fig. S3). We also have found substantial differences in the biochemical properties of chitinase-like proteins belonging to different groups (11, 12). In this paper, we investigated the biological roles of individual insect chitinase-like genes by using T. castaneum, a species that is sensitive to RNAi at all developmental stages and in different cell types (9, 10). Even though the members of different groups of the Tribolium chitinase family of proteins share substantial sequence similarity in the catalytic domain, they differ in the linker and chitin-binding domains, allowing us to design gene-specific dsRNAs for use in RNAi experiments. We were able to specifically knock down the transcript level of the target gene without affecting transcript levels of related chitinase-like genes by injecting gene-specific dsRNAs. Unique lethal phenotypes were observed in beetles treated with dsRNAs for TcCHT5, -7, and -10, as well as dsRNA for TcIDGF4 at one or more developmental stages. Although the TcCHT5 gene is highly expressed in the late developmental stages, including larval and pharate pupal stages predominantly in the carcass (whole body minus head, gut, and posterior tip; see Fig. S3), it was essential only for the pupal–adult molt and not for earlier molts, because the terminal phenotype was the same irrespective of the developmental stage at which the dsRNA was injected (i.e., larval, pharate pupal, or pupal stage). All insects died as pharate adults entrapped in the old pupal cuticle. Apparently, the role of this chitinase can be fulfilled by other chitinases at all stages of development except at the pharate adult stage. In contrast, TcCHT10 (which is also expressed predominantly in the carcass) appears to be required for successful completion of all molts including larval–larval, larval–pupal, and pupal–adult. Injections of dsRNA for this gene led to death of the treated insects at the time of the first molt following the injection. TcCHT5 apparently cannot fulfill this function even though it is also expressed at all developmental stages. A interesting finding is that there is a unique requirement for TcCHT10 during egg hatch. The target for this chitinase is unknown. However, one possibility is that this chitinase is required for embryonic molting, as has been reported to occur in some lepidopteran species. To our knowledge, there is no evidence reported to date for an embryonic molt occurring in Tribolium. We propose that, in addition to a role in cuticle turnover during molting, this chitinase also may be needed to digest chitin or chitin-like material in the eggshell. Even though the Tribolium eggshell has not been directly analyzed for chitin, we speculate that it may contain chitin or a related material. Consistent with this notion is the recent report that a chitin-like substance is present in mosquito eggshells, eggs, and ovaries (13).
TcCHT7 dsRNA produced a unique and unexpected phenotype. When larvae were injected with this dsRNA, larval–larval, larval–pupal, and even pupal–adult molts were completed on schedule without mortality, but there were noticeable abnormalities in the abdomen, elytra, and wings during morphogenesis. These results indicated that the primary role of TcCHT7 is probably not in global chitinolysis and general cuticle turnover. Unlike other Tribolium chitinases, this enzyme is a most likely a membrane-bound protein (Y.A., unpublished data). The precise metabolic target of TcCHT7 is unknown, but we speculate that it may act on an extracellular or membrane-bound chitin-like polysaccharide or glycoprotein with NAG–NAG linkages, which is required for abdominal contraction. The failure of the abdomen to contract after molting may have an indirect effect on hemolymph pressure, leading to a failure to expand the wings, elytra, and pronotum and also to unfold the legs and antennae.
An important finding of this work is that down-regulation of transcripts for chitinase genes belonging to different groups yielded quite different phenotypes. TcCHT5 belongs to the group I family of chitinase genes. Group I chitinase cDNAs have been characterized from Manduca sexta (4), Bombyx mori (14), Hyphantria cunea (14), Spodoptera litura (15), Choristoneura fumiferana (16), and Helicoverpa armigera (17) and the corresponding proteins have been isolated from molting fluid. The group I chitinase mRNAs/proteins reached their maximal levels during the molting periods of larval and/or pupal stages, and the levels decreased immediately after molting. Based on the time course of expression, group I genes have been predicted to encode molting-associated chitinases, although only one or two larval/pupal stages were analyzed in most cases. Only TcCHT5 and the C. fumiferana chitinase have been studied throughout insect development and shown to be expressed at every molt (16). We anticipated molting defects in Tribolium at every developmental stage with dsRNA for this gene. It is unclear why injection of dsRNA for TcCHT5 into young larvae prevented only the pupal–adult molt. During this interval, the gene is indeed expressed, and treated larvae underwent one to two successful larval–larval molts, followed by a successful larval–pupal molt. Furthermore, there was no unusual phenotype observed in the larval or pupal stages. The insects developed normally until a lethal pharate adult phenotype ensued. One possibility is that during larval molting, the role of the missing TcCHT5 chitinase could be compensated by other chitinases. However, the TcCHT5 enzyme appears to be indispensable during adult metamorphosis and could not be substituted by other chitinases. It is interesting to point out that at this stage TcCHT5 is maximally transcribed, whereas transcripts for many chitinases that belong to group IV are barely detectable.
Unlike group I genes, relatively little information is available about the expression or the role of group II chitinases. Group II chitinases contain at least four catalytic domains and four putative chitin-binding domains, three of which are clustered between two catalytic domains (8). The domain organization of TcCHT10 is similar to that of the T. molitor ortholog, which contains five catalytic domains, of which the first and second were predicted to lack enzymatic activity (6). The presence of an enzyme with a similar arrangement of catalytically competent and inactive domains in all lepidopteran and coleopteran insects analyzed to date suggests an important function for this unusual enzyme with multiple catalytic and predicted chitin-binding domains (ChBDs). A detailed study of the kinetics of accumulation of transcripts for this group II T. molitor chitinase indicated that this mRNA appeared hours before apolysis in both pharate pupal and pharate adult stages and decreased substantially after apolysis (6). A careful analysis of the expression profile of another molt-associated chitinase belonging to group I from C. fumiferana during the larval and pupal stages indicated that this chitinase is secreted after apolysis. This group I chitinase is required only for adult ecdysis when the thick pupal cuticle is being digested and recycled, probably after digestion by molting-associated proteases (18). This hypothesis is consistent with our finding that injection of dsRNA for TcCHT10 prevented every larval molt, pupation, adult metamorphosis, and egg hatch, whereas only adult metamorphosis was disrupted after injection of TcCHT5 dsRNA. The multiple predicted ChBDs and the catalytically inactive chitinase domains of group II chitinases may have a role in separating or loosening the individual chitin chains in the chitin microfibril, causing disaggregation of the crystalline α-chitin. We propose that these accessible chitin chains are then bound and hydrolyzed by the multiple catalytic chitinase domains of this large enzyme, leading to separation of the old procuticle from the waxy outer layer referred to as “cuticulin” or more recently as “envelope” (19) of the newly synthesized cuticle, thus initiating apolysis. In support of this hypothesis is the finding that the chitin-binding protein CBP21 from S. marcescens, which contains an inactive chitinase-like domain, promotes hydrolysis of crystalline β-chitin by other active chitinases (20).
The group III chitinase, TcCHT7, may function in tissue differentiation rather than in molting, because dsRNA for this gene interferes with wing expansion without preventing the molt. It is interesting that the expression of the Drosophila ortholog DmCht7 (CG1869), which encodes a group III chitinase, increased substantially in the wing discs from 24 to 40 h during the pupal stage (21). The only member of this group studied at the protein level is a group III chitinase from the tick, Haemaphysalis longicornis, which, unlike TcCht7 and DmCht7 chitinases, is a secreted protein and localized between the old and new cuticle in the epidermis, as well as in the midgut of molting nymphs. This protein was expressed and purified from the culture medium of Sf9 cells infected with a recombinant baculovirus containing this cDNA. The purified H. longicornis chitinase had a size of 116 kDa, which was ≈12 kDa larger than the theoretical value, glycosylated and enzymatically active (22). These observations suggest that at least some of the larger-sized chitinases are indeed catalytically active and do not require proteolytic processing to yield enzymatically active proteins. In the tick, a similar large protein was also identified in vivo by 2D immunoblot analysis. The protein contains two “ELR” motifs and appears to be a chitinase of the chemokine family, which is involved in angiogenesis (22).
Although the group V protein TcIDGF4 has no chitinolytic activity (11), it is required during adult eclosion. This is quite surprising because IDGFs are thought to be involved in cell proliferation and/or differentiation (23) but not in ecdysis. It is possible that this chitinase-like protein is involved in tracheal proliferation, which is known to occur at this stage (24). Of particular interest is that knockdown of Tribolium IDGF2 does not result in any altered phenotype compared with the control. Injection of dsRNA for either IDGF into adult females also did not reveal any abnormalities in the developing embryos or in egg hatch. Additional work is required to elucidate the precise role of IDGF genes in Tribolium development.
To date, we have not observed any phenotypic abnormalities after injection of dsRNAs for any of the group IV chitinase genes. For some unknown reason, the knockdown efficiency of dsRNAs for TcCHT2 and TcCHT6 was low, and there was no observable adverse effect in larvae that were treated with dsRNA for TcCHT2, -6, -8, -14, and -16 (Fig. S2 and data not shown). Attempts to suppress transcript levels of these chitinase-like genes to even lower levels with larger amounts of dsRNA and/or combinations of dsRNAs have not resulted in any observable phenotypes. The finding that these genes are expressed in the gut suggested the possibility that they may be involved in the turnover of PM-associated chitin or in digestion of dietary chitin. Interestingly, the expression patterns of individual chitinases of group IV were different along the length of the midgut. Some of them are expressed predominantly in the posterior midgut, whereas others predominate in the anterior and middle parts of the midgut (Fig. S3). It is also possible that these proteins have functions unrelated to chitin turnover. Shi and Paskewitz (25) have identified two group V chitinase-like proteins, AgBR1 and AgBR2, in the hemolymph of Anopheles gambiae. AgBR1 and AgBR2 (orthologs of TcIDGF4 and TcIDGF2) do not have chitinolytic activity and were induced specifically in response to bacterial infection. Interestingly, in adults AgBR1 is expressed only in the fat body, whereas AgBR2 is expressed in several tissues. It is possible that some of the Tribolium chitinases identified in this work may have similar immune-related functions.
In summary, a rather large number of chitinase-like genes are present and expressed in Tribolium. They differ in their developmental patterns and tissue specificities of expression. Phylogenetic analysis indicates that the appearance of these distinct groups of chitinases predates the separation of coleopteran and lepidopteran lineages of insects (8). Results from RNAi studies reported here support the hypothesis that genes in different groups have distinctly different biological functions. At least two of these groups, the group I and group II enzymes, are involved in molting by digesting cuticular chitin, whereas the group III enzymes have a morphogenetic role in development such as regulating abdominal contraction and wing expansion. Some IDGFs belonging to group V, which have been shown to affect cell proliferation in imaginal disks, also appear to affect molting even though these proteins lack catalytic activity.
Materials and Methods
Tribolium Strains.
Beetles were reared in whole wheat flour containing 5% brewer's yeast at 30°C under standard conditions (26). The strains used were T. castaneum strain GA-1 and an enhancer trap line, Pig-23, which expresses the inserted gene for an EGFP in the wing and elytral discs at the end of the last larval instar (27).
Expression Profiles of Tribolium Chitinase Family Genes.
Total RNA was prepared from pools of embryos, young larvae, last instar larvae, pharate pupae, pupae, or adults. cDNAs were synthesized from total RNA as described previously (9). RT-PCR was used to check the expression profile for each gene by using pairs of gene-specific primers (Table S1).
dsRNA Synthesis.
dsRNAs were prepared as described in Arakane et al. (9). Unique regions of each Tribolium chitinase-like gene were chosen as templates for the synthesis of gene-specific dsRNA (Table S2). In most cases, more than one region from the same gene was chosen to prepare dsRNAs. dsRNA was synthesized from linearized templates by using the AmpliScribe T7-Flash transcription kit (Epicentre).
Injection of dsRNA into Tribolium.
dsRNAs were injected into young larvae, last instar larvae, and pharate pupae by using a microinjection system and a dissection stereomicroscope (10). Approximately 200 ng of dsRNA dissolved in ≈0.2 μl of injection buffer (0.1 mM sodium phosphate, 5 mM KCl, pH 7) was injected into the dorsal abdomen of each beetle. After injection, beetles were kept under standard conditions, as described above, for visual monitoring of phenotypes and further analysis. Buffer alone or buffer containing dsRNA for the Tribolium tryptophan oxygenase gene (dsVer) was used as a negative control. Replications were carried out with at least 20 insects for each dsRNA.
dsRNAs of TcCHT5, TcCHT7, TcCHT10, TcIDGF2, and TcIDGF4 were also injected into adult females. Two days after injection, dsRNA-treated females were mated with equal numbers of untreated adult males. The insects were maintained under standard conditions. Every 3 days, eggs were collected and observations for any abnormal phenotypes were made on a daily basis.
Measurement of Transcripts for Tribolium Chitinase Family Genes After dsRNA Treatment.
Total RNA was prepared from pools of three or four insects after injection of dsRNA or buffer by using the RNeasy Mini Kit (Qiagen). cDNA was synthesized by using these RNA preparations and the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). RT-PCR was carried out to monitor the effect of dsRNA or buffer injections on levels of transcripts by using gene-specific primers. A pair of Tribolium polyubiquitin gene primers was used as an internal control to monitor equal loading of cDNA for analysis of transcript levels.
Supplementary Material
Acknowledgments.
This was a cooperative investigation between Kansas State University (Kansas Agricultural Experiment Station Contribution No. 08-222-J) and the Department of Agriculture. Support from National Science Foundation Grants IBN-0316963 and IOS-615818 is gratefully acknowledged.
Footnotes
This work was presented in part at the Seventh International Congress of Comparative Physiology and Biochemistry, Salvador, Brazil, August 12–16, 2007.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800739105/DCSupplemental.
References
- 1.Kramer KJ, Muthukrishnan S. Chitin metabolism in insects. In: Gilbert LI, Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science. Oxford, UK: Elsevier; 2005. pp. 111–144. [Google Scholar]
- 2.Kramer KJ, Muthukrishnan S. Insect chitinases: Molecular biology and potential use as biopesticides. Insect Biochem Mol Biol. 1997;27:887–900. doi: 10.1016/s0965-1748(97)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Kramer KJ, Koga D. Insect chitin: Physical state, synthesis, degradation and metabolic regulation. Insect Biochem. 1986;16:851–877. [Google Scholar]
- 4.Kramer KJ, Corpuz L, Choi HK, Muthukrishnan S. Sequence of a cDNA and expression of the gene encoding epidermal and gut chitinases of Manduca sexta. Insect Biochem Mol Biol. 1993;23:691–701. doi: 10.1016/0965-1748(93)90043-r. [DOI] [PubMed] [Google Scholar]
- 5.de la Vega H, Specht CA, Liu Y, Robbins PW. Chitinases are a multi-gene family in Aedes, Anopheles and Drosophila. Insect Mol Biol. 1998;7:233–239. doi: 10.1111/j.1365-2583.1998.00065.x. [DOI] [PubMed] [Google Scholar]
- 6.Royer V, Fraichard S, Bouhin H. A novel putative insect chitinase with multiple catalytic domains: Hormonal regulation during metamorphosis. Biochem J. 2002;366:921–928. doi: 10.1042/BJ20011764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Q, et al. Computational identification of novel chitinase-like proteins in the Drosophila melanogaster genome. Bioinformatics. 2004;20:161–169. doi: 10.1093/bioinformatics/bth020. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Q, et al. Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect Biochem Mol Biol 2008. 2007;38:452–466. doi: 10.1016/j.ibmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc Natl Acad Sci USA. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Q, Arakane Y, Kramer KJ, Beeman RW, Muthukrishnan S. Characterization of recombinant chitinase-like proteins of Drosophila melanogaster and Tribolium castaneum. Insect Biochem Mol Biol 2008. 2007;38:467–477. doi: 10.1016/j.ibmb.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Matsumiya M, Arakane Y, Haga A, Muthukrishnan S, Kramer KJ. Substrate specificity of chitinases from two species of fish, greenling, Hexagrammos otakii, and common mackerel, Scomber japonicus, and the insect, tobacco hornworm, Manduca sexta. Biosci Biotech Biochem. 2006;70:971–979. doi: 10.1271/bbb.70.971. [DOI] [PubMed] [Google Scholar]
- 13.Moreira MF, et al. A chitin-like component in Aedes aegypti eggshells, eggs and ovaries. Insect Biochem Mol Biol. 2007;37:1249–1262. doi: 10.1016/j.ibmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Kim MG, Shin SW, Bae KS, Kim SC, Park HY. Molecular cloning of chitinase cDNAs from the silkworm, Bombyx mori and the fall webworm, Hyphantria cunea. Insect Biochem Mol Biol. 1998;28:163–171. doi: 10.1016/s0965-1748(97)00112-4. [DOI] [PubMed] [Google Scholar]
- 15.Shinoda T, Kobayashi J, Matsui M, Chinzei Y. Cloning and functional expression of a chitinase cDNA from the common cutworm, Spodoptera litura, using a recombinant baculovirus lacking the virus-encoded chitinase gene. Insect Biochem Mol Biol. 2001;31:521–532. doi: 10.1016/s0965-1748(00)00133-8. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, et al. A molt-associated chitinase cDNA from the spruce budworm, Choristoneura fumiferana. Insect Biochem Mol Biol. 2002;32:1813–1823. doi: 10.1016/s0965-1748(02)00166-2. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad T, Rajagopal R, Bhatnagar RK. Molecular characterization of chitinase from polyphagous pest Helicoverpa armigera. Biochem Biophys Res Commun. 2003;310:188–195. doi: 10.1016/j.bbrc.2003.08.136. [DOI] [PubMed] [Google Scholar]
- 18.Samuels RI, Reynolds SE. Timing of appearance of proteolytic enzymes in the moulting fluid of Manduca sexta pharate adults. Arch Insect Biochem Physiol. 1993;24:33–44. doi: 10.1002/(SICI)1520-6327(200001)43:1<33::AID-ARCH5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 19.Locke M. The Wigglesworth lecture: Insects for studying fundamental problems in biology. J Insect Physiol. 2001;47:495–507. doi: 10.1016/s0022-1910(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 20.Vaaje-Kolstad G, Horn SJ, van Aalten DM, Synstad B, Eijsink VG. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem. 2005;280:28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- 21.Ren N, Zhu C, Lee H, Adler PN. Gene expression during Drosophila wing morphogenesis and differentiation. Genetics. 2005;171:625–638. doi: 10.1534/genetics.105.043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You M, et al. Identification and molecular characterization of a chitinase from the hard tick Haemaphysalis longicornis. J Biol Chem. 2003;278:8556–8563. doi: 10.1074/jbc.M206831200. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura K, Shibata T, Saget O, Peel D, Bryant PJ. A new family of growth factors produced by the fat body and active on Drosophila imaginal disc cells. Development. 1999;126:211–219. doi: 10.1242/dev.126.2.211. [DOI] [PubMed] [Google Scholar]
- 24.Zimoch L, Hogenkamp DG, Kramer KJ, Muthukrishnan S, Merzendorfer H. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochem Mol Biol. 2005;35:515–527. doi: 10.1016/j.ibmb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Paskewitz SM. Identification and molecular characterization of two immune-responsive chitinase-like proteins from Anopheles gambiae. Insect Mol Biol. 2004;13:387–398. doi: 10.1111/j.0962-1075.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- 26.Haliscak JP, Beeman RW. Status of malathion resistance in five genera of beetles infesting farm-stored corn, wheat and oats in the United States. J Econ Entomol. 1983;76:717–722. [Google Scholar]
- 27.Lorenzen MD, et al. piggyBac-mediated germline transformation in the beetle Tribolium castaneum. Insect Mol Biol. 2003;12:433–440. doi: 10.1046/j.1365-2583.2003.00427.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.