Abstract
Cells rely on diffusion to move metabolites and biomolecules. Diffusion is highly efficient but only over short distances. Although eukaryotic cells have broken free of diffusion-dictated constraints on cell size, most bacteria and archaea are forced to remain small. Exceptions to this rule are found among the bacterial symbionts of surgeonfish; Epulopiscium spp. are cigar-shaped cells that reach lengths in excess of 600 μm. A large Epulopiscium contains thousands of times more DNA than a bacterium such as Escherichia coli, but the composition of this DNA is not well understood. Here, we present evidence that Epulopiscium contains tens of thousands of copies of its genome. Using quantitative, single-cell PCR assays targeting single-copy genes, we have determined that copy number is positively correlated with Epulopiscium cell size. Although other bacteria are known to possess multiple genomes, polyploidy of the magnitude observed in Epulopiscium is unprecedented. The arrangement of genomes around the cell periphery may permit regional responses to local stimuli, thus allowing Epulopiscium to maintain its unusually large size. Surveys of the sequences of single-copy genes (dnaA, recA, and ftsZ) revealed genetic homogeneity within a cell consistent with only a small amount (≈1%) of the parental DNA being transferred to the next generation. The results also suggest that the abundance of genome copies in Epulopiscium may allow for an unstable genetic feature, a long mononucleotide tract, in an essential gene. With the evolution of extreme polyploidy and large cell size, Epulopiscium has acquired some of the advantages of eukaryotic cells.
Keywords: dnaA, Epulopiscium, large bacteria, mononucleotide repeat, polyploid
It is well appreciated that many eukaryotes are orders of magnitude larger than all known members of the Bacterial and Archaeal domains. Eukaryotes have broken free of constraints on cell size by the development of sophisticated nutrient uptake systems, subcellular compartmentalization, and the use of a cytoskeleton and motor proteins to transport vesicles. With the further advance of multicellularity, cell and tissue specialization have allowed eukaryotes to attain tremendous dimensions (1, 2). Until recently (3, 4), bacterial (and archaeal) cells were considered simple, displaying little subcellular organization. Although we now know that bacterial cells are also highly organized, possessing motor and cytoskeletal proteins, and even extensive intracellular membranes in some instances (5), these cells are believed to rely on diffusion to access nutrients and other metabolically important chemicals. Diffusion coefficients of small molecules may impose time constraints on metabolite flux (6) that require bacterial cells to maintain very high surface-to-volume ratios. As a result, no part of the cytoplasm is very far from the external environment, and so exchange is rapid enough to sustain metabolic processes. Most large bacteria fit this paradigm and maintain a very thin cytoplasm; they are extremely long and slender, or if spherical they contain an intracellular vacuole to press the cytoplasm into a thin layer just under the cytoplasmic membrane (7–10). In addition, many large bacteria contain intracellular mineral inclusions, which further reduce the volume of active cytoplasm (11). Possible exceptions to this standard are found within the Firmicutes (12).
Our model for studying the cell biology of large bacteria is Epulopiscium sp. type B, which occurs in the intestinal tract of the unicornfish Naso tonganus (13, 14). These type B cells attain lengths of 200–300 μm and widths of 50–60 μm (15) and reproduce solely by the formation of multiple internal offspring (Fig. 1). This reproductive strategy likely evolved from endospore formation (16). A large Epulopiscium contains a substantial amount of DNA arranged around the periphery of the cytoplasm (17). This unusual feature may be key in the ability of these large cells to maintain an active metabolism despite having a low surface-to-volume ratio. To characterize the size and conformation of the Epulopiscium sp. type B genome, we used quantitative PCR to enumerate the copy number of genes in individuals and in DNA extracted from populations of cells. The results of these surveys suggest that Epulopiscium is highly polyploid throughout its life cycle, and an individual contains tens of thousands of copies of its genome.
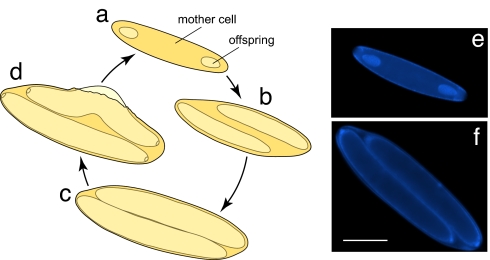
Fig. 1.
Epulopiscium sp. type B life cycle. Offspring production follows a circadian cycle. (a) Early in the day, a mother cell possesses small, internal offspring. (b and c) Offspring size increases throughout the day (b) until they fill the mother-cell cytoplasm (c). (d) Finally, “mature” offspring cells emerge from the mother-cell envelope. Note that before emergence, these cells begin to develop the next generation of offspring. (e and f) Images of DAPI-stained cells representing the populations of small (e) and large (f) Epulopiscium cells used in these studies. (Scale bar: 50 μm.)
Results and Discussion
DNA Content of Large and Small Epulopiscium Cells.
Currently, no Epulopiscium sp. is available in culture, which prevents the use of standard methods (18) for assessing the composition or conformation of the genome. Based on 16S rRNA gene sequence surveys, Epulopiscium type B populations are the most homogeneous of the characterized Epulopiscium morphotypes (13), and therefore they are well suited for the gene-based studies presented here. By quantifying the amount of DNA extracted from 5,000 purified large cells with mature offspring (Fig. 1f), and 5,000 cells with small, immature offspring (Fig. 1e), we found that large cells contain ≈250 pg of DNA, whereas small cells contain ≈85 pg of DNA. In contrast, a human diploid cell contains 6 pg of DNA. The tremendous amount of DNA in Epulopiscium is most likely in one of three conformations: (i) a few copies of an enormous genome (19), (ii) thousands of copies of a small genome (17, 20), or (iii) many copies of the complete genome but with portions that are highly amplified. Because only a small proportion of the mother-cell DNA is partitioned into newly formed offspring, it is unlikely that Epulopiscium cells possess only a few copies of an enormous genome (15). Instead, we hypothesized that Epulopiscium has a genome comparable to the size of other bacterial genomes (21), and that it is present in very high numbers.
Ploidy of Large and Small Epulopiscium Cells.
The composition of DNA in individual Epulopiscium cells was assessed by using real-time quantitative PCR. Four genes were assayed: ftsZ, dnaA, recA, and the 16S rRNA gene. The first three of these are generally unlinked, single-copy genes (22–24), and thus they were used to represent the unit genome of Epulopiscium. Large Epulopiscium cells on average possess 50,000 to 120,000 copies of each of these markers (Table 1). The genomes of many Firmicutes (low G+C Gram-positive bacteria) have multiple (as many as 15) rRNA operons (25). The ribosomal RNA operon is preferentially amplified in some eukaryotes (26–28). Ribosomal RNA synthesis is the rate-limiting step in assembly of ribosomes in many bacteria (29). For these reasons, we considered the rRNA operon a good candidate for gene amplification in Epulopiscium. Real-time PCR assays of the 16S rRNA gene showed that large Epulopiscium cells have 240,000 to 740,000 copies of this gene (Table 1). Single-cell PCR amplification of the internal transcribed spacer (ITS) between the 16S and 23S rRNA genes showed that these cells have at least four unique ITSs, indicative of multiple rRNA operons [supporting information (SI) Fig. S1]. These results suggest that Epulopiscium type B also has multiple rRNA operons per genome; however, the rRNA gene copy number in individuals is not large enough, relative to single-copy markers, to indicate substantial amplification of this operon.
Table 1.
Gene copy number in individual Epulopiscium
| Gene | Copy number* | Range† | Control‡ |
|---|---|---|---|
| ftsZ | 80,600 | 35,800 – 198,000 | 240 |
| dnaA | 48,700 | 29,800 – 153,000 | 240 |
| recA | 120,000 | 60,300 – 205,000 | 70 |
| 16S | 368,000 | 241,000 – 737,000 | 310 |
*Median gene copy number, n = 10.
†Copy number range (minimum – maximum).
‡Mean negative controls, surgeonfish gut contents without Epulopiscium cells, n = 3.
It is difficult to determine from single-cell data whether all single-copy markers are equally present in Epulopiscium type B because cell size and gene copy number varied greatly in individuals from a given population. Additionally, using cells instead of purified genomic DNA in these real-time PCR assays could introduce factors that may alter amplification efficiency. We therefore assayed relative gene copy numbers by using purified Epulopiscium genomic DNA. This approach also allowed for a rough estimation of genome size. Gene copy numbers of the three single-copy markers were statistically similar in genomic DNA extracted from large cells (one-way ANOVA, F = 1.25, P = 0.301), with markers averaging 40,900 copies in 156 pg of DNA (Table 2). These results support the idea that large Epulopiscium cells contain tens of thousands of copies of a fully replicated, ≈3.8 Mb genome. However, the marker numbers obtained from small-cell genomic DNA varied (F = 11.82, P = 0.000). These small cells were taken at an early stage of their growth cycle, which is presumably a time of active DNA replication. This supposition is supported by the observation that the replication origin-linked marker dnaA (30) was more numerous than ftsZ or recA.
Table 2.
Gene copy number in 156 pg of Epulopiscium genomic DNA
| Gene | Large-cell DNA* | Small-cell DNA* | Control DNA† |
|---|---|---|---|
| ftsZ | 42,600 ± 5,820 | 31,900 ± 1,640 | 290 |
| dnaA | 38,200 ± 7,940 | 36,500 ± 7,000 | 120 |
| recA | 41,900 ± 8,330 | 26,300 ± 4,300 | 30 |
| 16S | 285,000 ± 36,000 | 177,000 ± 11,200 | 40 |
*Mean ± SD, n = 10.
†Mean negative controls, 150 pg of DNA extracted from surgeonfish gut contents with no Epulopiscium cells, n = 3.
Cell Volume per Genome.
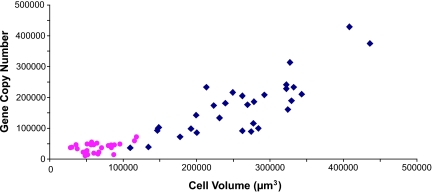
Bacillus subtilis (also a member of the Firmicutes) maintains a fairly constant cell-volume-to-genomic-DNA ratio over a variety of exponential growth rates (31). We took advantage of the natural variation in cell size in Epulopiscium populations to investigate whether genome copy number varied with respect to cytoplasmic volume, and whether this ratio was similar to that of B. subtilis. The genome copy number proxy ftsZ was assayed in individuals taken from two natural Epulopiscium type B populations that represent the extremes of offspring development and cell size (examples shown in Fig. 1 e and f). A linear relationship between cell size and copy number was observed in the two populations (Fig. 2). Cells maintained an average ratio of one genome for every 1.9 μm3 of cytoplasm over a range of volumes from 28,200 to 436,000 μm3. When analyzed separately, the means of ratios for cells of the two Epulopiscium populations were not significantly different (t test, P = 0.481). In comparison, B. subtilis in exponential growth in rich media harbors one chromosome per 0.7 μm3 of cytoplasm (based on data from Sharpe et al. in ref. 31). Epulopiscium cells appear to follow the same rules as other bacteria that link cell growth and DNA replication (31–33); however, at least in the populations we compared, Epulopiscium maintains a larger cytoplasmic-volume-to-genome ratio than B. subtilis. Further studies are needed to determine whether all cytoplasmic regions of these large bacterial cells are functionally equivalent.
Fig. 2.
The relationship between gene copy number and cytoplasmic volume in Epulopiscium sp. type B cells. The pink circles represent data from individuals of the small-cell population (n = 26), and the blue diamonds represent data from the large-cell population (n = 31).
Factors Leading to Extreme Polyploidy.
The significance of the substantial polyploidy and genetic expansion recorded for Epulopiscium may be considered with respect to the two dominant attributes of this bacterium: its nutritional biology as a symbiont and its extraordinarily large size. Genetic amplification is seen in diverse organisms (bacteria, archaea, protists, fungi, plants, and animals). Amplification provides resources to support rapid growth and division, cell specialization and adaptation, and may enhance repair of genetic lesions (34–40). For some bacteria, polyploidy is allied with metabolic adaptation and symbiont differentiation. Buchnera aphidicola, the proteobacterial symbiont of aphids, contains on average 120 copies of its chromosome, and expansion of genetic resources correlates with host development and presumably increased host demand for the essential biomolecules provided by Buchnera (41). During differentiation to symbiotic bacteroids, some species of rhizobia undergo modest genome proliferation (42). Therefore, one factor that may have led to polyploidy in Epulopiscium is selection pressure for the evolution of a symbiotic relationship that contributes to host metabolism.
Alternatively, the extreme polyploidy in Epulopiscium may be more tightly linked with cell size. In insect and plant cells, an expansion of genomic resources is accompanied with an increase in size (42–45), although size is not always proportionate to ploidy (46). Although the advantage of large cell size in Epulopiscium spp. has yet to be determined, two features of the symbiont's environment should be considered. The host feeding behavior varies substantially on a daily basis, with concentrations of food occurring at different points in the relatively long alimentary tract. Populations of large Epulopiscium cells migrate to distinct regions of the intestinal tract at different times of day (47), presumably in response to host digestive processes. We speculate that large size allows these motile cells better control over their position along the length of the intestinal tract of the host. In addition, the alimentary microbial community of the host is complex and supports very high numbers of ciliate bacterial predators (48). Epulopiscium type B cells appear to avoid predation by all but the largest ciliates that inhabit the N. tonganus intestinal tract (49).
The biased distribution of DNA within the cytoplasm of Epulopiscium permits functional compartmentalization and regional specialization within these large cells. A similar peripheral arrangement of nucleoids has been reported in another large bacterium, Thiomargarita namibiensis (50), although the composition of these nucleoids has yet to be determined. Single spherical cells of Thiomargarita are generally 100–300 μm in diameter but cells as large as 800 μm occur. Each Thiomargarita cell contains a large, fluid-filled vacuole, which takes up ≈98% of the cell volume. This central vacuole confines the active cytoplasm to a shallow, 0.5– to 2-μm layer just under the cytoplasmic membrane. As with all bacteria, the close association of DNA with the cell membrane accommodates transertion (51), the linked processes of transcription, translation, and insertion of proteins into the cytoplasmic membrane. In large bacteria, genomic copies arrayed around the cellular periphery would permit transcription of any gene at disparate locations within the cell, thus reducing transit time of proteins and metabolites from site of synthesis or entry to site of action. In this way, a large bacterium could function like a microcolony, with different regions of the cell independently responding to local stimuli, which would alleviate some of the pressure to remain small for the sake of rapid intracellular diffusive transport. In stark contrast to Thiomargarita, the central cytoplasm of Epulopiscium is relatively free of DNA but apparently active. This subcellular arrangement allows for the rapid growth of internal offspring, as seen in the related, but smaller (12–35 μm long), endospore-forming bacterium Metabacterium polyspora (16). It still remains to be seen whether other mechanisms in large Epulopiscium cells enhance movement of molecules throughout the bulk of the central cytoplasm.
Polyploidy and Intracellular Genetic Diversity.
It is coming to light that genome duplication and subsequent divergence of orthologs has been an important driving force in genome evolution and the generation of morphological complexity (44, 52). Polyploidy in Epulopiscium could allow for diversification of genome copies while supporting an increase in cell size. Such a simple cellular modification may have been an important advance toward the development of the contemporary eukaryotic cell. In a previous study, we estimated that ≈1% of the DNA in an Epulopiscium sp. type B cell is inherited (15). For an average type B cell, this amount of DNA could comprise 230 genome equivalents. Although reproduction imposes a significant genomic population bottleneck, some genetic diversity could be passed on to the offspring.
To evaluate gene diversity in Epulopiscium, we followed an approach used by Mark Welch and Meselson (53, 54) to study bdelloid rotifers. There is mounting evidence for functional divergence of genes that were once alleles in these asexual, but highly successful, rotifers (55). For Epulopiscium gene surveys, we cloned the PCR products from single-cell amplification reactions targeting single-copy genes ftsZ, dnaA, or recA (SI Text). As mentioned above, multiple ITS sequences have been recovered from an Epulopiscium single-cell PCR amplification (Fig. S1), so this method should allow for the recovery of abundant variants in single-copy genes, if present. Alignments of single-copy gene clones revealed that a consensus gene sequence was predominant in each library; however, we did detect sequence variants (SI Text and Tables S1, S2, and S3). For ftsZ and recA, clones varied from the consensus by 1 or 2 nt, and all variants were unique (Tables S1 and S2). Single-nucleotide changes appeared to be transitions and the vast majority coded for nonsynonymous amino acid substitutions. The overall frequency of nucleotide differences was comparable to published TaqDNA polymerase PCR error rates (56–58). For dnaA, we observed similar trends (Table S3). During our analysis of dnaA, however, we were surprised to find a common, single-nucleotide deletion in 12 of the 30 dnaA clones (SI Text and Table S3). The deletion occurred within a mononucleotide tract of 10 adenines. Further investigations (SI Text and Fig. S2) indicate that at least some of these deletion variants are produced during amplification with thermostable polymerases. A high frequency of slippage in mononucleotide tracts of this length during PCR amplification has been observed by others (59, 60). Long, mononucleotide repeats are common in eukaryotic genomes, but rare in bacterial or archaeal genomes (61–63). In bacteria, these highly mutable motifs tend to be found within genes that encode variable surface proteins. We have found a unique, long (10 bp) mononucleotide repeat within an essential bacterial gene. The functional significance of this dnaA adenine-deletion variant and frequency of its expression have yet to be determined.
Evolutionary Implications of Extreme Polyploidy in Bacteria.
Together, the findings support the idea that Epulopiscium sp. type B cells are highly polyploid throughout their life cycle. Gene sequence surveys suggest that the bottleneck imposed on Epulopiscium genomes during reproduction may restrict diversification of orthologs in an individual. Nevertheless, extreme polyploidy may allow Epulopiscium to harbor unstable genetic features, such as mononucleotide tracts, within essential genes, without detriment.
The functional dichotomy of “somatic” and “germ-line” genomes within an enormous and highly polyploid bacterium represents a unique intermediate between the “typical” asexual, single-celled microorganism and a multicellular organism. The genetic material of a successful, solitary bacterium is replicated and faithfully passed on to its offspring. For multicellular organisms, only the germ line may be inherited; the waste of genetic resources in somatic cells is offset by the diversification of cellular function, which commonly leads to increased size, enhanced access to resources, and improved metabolic capacity. Compared with a solitary existence, colonial microbes (e.g., some actinobacteria) or populations of cooperative microorganisms (e.g., cellular slime molds, myxobacteria) benefit from improved metabolic potential and perhaps better dispersal at the cost of the genomes of cells that play a supporting role (64, 65). The enormous, polyploid Epulopiscium cell has converged on the advantages of social microbes but with additional benefits (exceptional motility, enhanced resistance to predation) normally found in large eukaryotic microbes or multicellular organisms.
Methods
Epulopiscium Collection.
Epulopiscium sp. morphotype B cells were obtained from N. tonganus, collected by spearfishing on reefs around Lizard Island, Australia. Sections of the gut were removed, and intestinal contents were fixed in 80% ethanol and stored at −20°C. Individual Epulopiscium cells were collected from gut contents by using a standard Gilson micropipettor and a dissecting microscope (Nikon SMZ-U). Cells were transferred five times through sterile ethanol wash buffer [80% ethanol, 145 mM NaCl, 50 mM Tris·HCl (pH 8.0), 0.05% Igepal] and rinsed in sterile deionized water.
DNA Extraction and Quantification.
DNA was extracted from 5,000 handpicked, washed cells. DNA extraction and quantification protocols were based on standard procedures (66). Epulopiscium cells were lysed by incubation in proteinase K (100 μg/ml in 10 mM Tris, pH 8.0) at 50°C for 1 h followed by six rounds of freeze–thaw. Cell lysate was extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1), and the nucleic acids were precipitated with 0.3 M sodium acetate and ethanol. The pellet was rinsed with 70% ethanol and air-dried. The pellet was dissolved in sterile water and treated with RNase A (10 μg/ml) at 37°C for 30 min.
PicoGreen assays to quantify genomic DNA were performed as follows. DNA from bacteriophage lambda was diluted in TE [10 mM Tris (pH 8.0), 1 mM EDTA] to generate a dilution series ranging from 500 to 10 ng/ml. Genomic DNA from Epulopiscium was diluted 1:5,000 in TE. Aliquots (50 μl) of the standards and genomic DNA were dispensed in triplicate into wells of a microtiter plate. PicoGreen (Molecular Probes) was prepared according to the manufacturer's instructions, and 50-μl aliquots were mixed with the DNA solutions in each microtiter plate well. Relative fluorescence was determined by using a Perkin–Elmer LS50B fluorometer. Genomic DNA was also quantified by using an ethidium bromide spot test. For the spot test, lambda DNA was used to generate a serial dilution ranging from 50 to 1 μg/ml. Equal volumes of DNA (unknowns or standards) and a 2 μg/ml solution of ethidium bromide were mixed, and 10 μl of each mixture was spotted on a Petri dish. The spots were illuminated with UV light and photographed. The fluorescence intensity of each unknown was compared with the fluorescence intensities of the DNA standards.
Primer and Probe Design for Quantitative PCR.
Epulopiscium type B dnaA (GenBank accession no. EF127641) and recA (GenBank accession no. EF127642) genes were cloned by using an approach previously used to clone ftsZ (67). TaqMan probes and primer sequences for Epulopiscium dnaA (probe: 5′-TTCTTTCTTTTCCGGCGATAAATTGAATATCATCTATTAG; forward primer: 5′-GACCAACCTCCTGCCTTCAGAAATAAA; reverse primer: 5′-GTGTTAAATGTATGGAAAAATTCTTCTTGT), ftsZ (probe: 5′-CACAGGTGACTCGACACTTGCAATC; forward primer: 5′-ATTAAAGGTGCAGGTGGCGTACT; reverse primer: 5′-GCTAGCTCCCGCAT TCAACT), recA (probe: 5′-TTAAGAATAAAATTGCTCCTCCATTTAAACAAGCAG; forward primer: 5′-TGCGACAGAAATAATCGGCAGCAAAA; reverse primer: 5′-AGAAGAAATTCCTTCGCCATAGATAA), and the 16S rRNA gene (probe: 5′-CCATGCCGCCTACACACCCTTTACA; forward primer: 5′-ATTAAAGGTGCAGGTGGCGTACT; reverse primer: 5′-GCTAGCTCCCGCATTCAACT) used in real-time PCR were designed by using Primer Express (Applied Biosystems). TaqMan probes used in the real-time PCR assays were 5′ end-labeled with the reporter dye 6-carboxyfluorescein (FAM) and 3′-labeled with the quencher dye carboxytetramethylrhodamine (TAMRA).
TaqMan Quantitative PCR Assays.
Primer sets were tested in standard PCR amplifications using Epulopiscium cells, microbial community DNA extracted from surgeonfish intestinal contents, and genomic DNA from five other Firmicutes. A proteinase K solution (as above) was irradiated for 3 min by using a UV transilluminator (FisherBiotech) and then aliquoted into each tube in a 96-well PCR plate. One washed Epulopiscium was added to each tube and incubated at 50°C for 1 h. The plate was heated, 95°C for 15 min, to inactivate the proteinase K. TaqMan assay reaction mixtures were prepared on ice and contained 1× TaqMan Universal Master Mix (ABI), 900 nM of the appropriate forward and reverse primers, and 200 nM of the appropriate fluorogenic probe. To determine the copy number of genes in Epulopiscium genomic DNA, the DNA extracts from large- and small-cell populations were diluted 1:10 and 1:100 in TE. The concentrations of all dilutions were determined by using the PicoGreen assay described above, except 1 μl of diluted Epulopiscium DNA was used in the assays. The small-cell DNA concentration was adjusted to 156 pg/μl to match the concentration of the large-cell DNA sample. For gene quantification, 1 μl of DNA was added to PCR plate well containing the TaqMan reaction mixture. Each genomic DNA sample was run in triplicate for each gene assay. Quantification standards were generated from serial dilutions of plasmid DNA (2.0 × 107 copies to 2.0 × 102 copies per μl) containing dnaA, ftsZ, recA, or the 16S rRNA gene cloned from Epulopiscium type B. Standards were run in duplicate, and no template controls were run in triplicate for each assay. Thermal cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Data were compiled and analyzed by using Sequence Detection Software, version 1.3 (ABI).
Cell Volume and Gene Copy Number Comparisons.
Images of Epulopiscium cells were acquired with a Cooke SensiCam CCD camera and an Olympus BX61 microscope equipped with a LCPlanF1 ×40 objective. Using Slidebook software (calibrated with a stage micrometer), the cell length and width were determined. The cell volume was calculated by using the formula for a prolate ellipsoid. After image acquisition, each cell was transferred into a well of a 96-well plate and processed for quantitative PCR as described above. Statistical analyses (t test and ANOVA) were performed with SAS software, version 9.1 of the SAS system for Windows.
Supplementary Material
Acknowledgments.
We thank the staff and directors of the Lizard Island Research Station for support; Will Robbins and David Raubenheimer for help in the field; Rebekah Ward, David Miller, John Helmann, and two anonymous reviewers for comments on the manuscript; and Simona Despa for statistical consultation. Funding was provided by National Science Foundation Molecular and Cellular Biology Program Grant MCB 0721583.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF127641, EF127642, EU500234, EU500235, EU500236, and EU500237).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707522105/DCSupplemental.
References
- 1.Bonner JT. Perspective: The size-complexity rule. Evol Int J Org Evol. 2004;58:1883–1890. doi: 10.1111/j.0014-3820.2004.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 2.West GB, Brown JH. The origin of allometric scaling laws in biology from genomes to ecosystems: Toward a quantitative unifying theory of biological structure and organization. J Exp Biol. 2005;208:1575–1592. doi: 10.1242/jeb.01589. [DOI] [PubMed] [Google Scholar]
- 3.Losick R, Shapiro L. Changing views on the nature of the bacterial cell: From biochemistry to cytology. J Bacteriol. 1999;181:4143–4145. doi: 10.1128/jb.181.14.4143-4145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro L, Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 5.Shively JM. Prokaryote complex intracellular structures: Descriptions and discoveries. In: Shively JM, editor. Microbiology Monographs. Vol 2. Berlin: Springer; 2006. pp. 3–22. [Google Scholar]
- 6.Berg HC. Random Walks in Biology. Princeton: Princeton Univ Press; 1993. [Google Scholar]
- 7.Blakemore RP, Canale-Parola E. Morphological and ecological characteristics of Spirochaeta plicatilis. Arch Microbiol. 1973;89:273–289. doi: 10.1007/BF00408895. [DOI] [PubMed] [Google Scholar]
- 8.Delaporta B. Descriptive study of bacteria of very large size. Ann Institut Pasteur. 1964;107:845–862. [PubMed] [Google Scholar]
- 9.Gundersen JK, Jorgensen BB, Larsen E, Jannasch HW. Mats of giant sulphur bacteria on deep-sea sediments due to fluctuating hydrothermal flow. Nature. 1992;360:454–456. [Google Scholar]
- 10.Schulz HN, et al. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science. 1999;284:493–495. doi: 10.1126/science.284.5413.493. [DOI] [PubMed] [Google Scholar]
- 11.Head IM, Gray ND, Babenzien H, Glöckner FO. Uncultured giant sulfur bacteria of the genus Achromatium. FEMS Microbiol Ecol. 2000;33:171–180. doi: 10.1111/j.1574-6941.2000.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 12.Angert ER. The enigmatic cytoarchitechture of Epulopiscium spp. In: Shively JM, editor. Microbiology Monographs. Vol 2. Berlin: Springer; 2006. pp. 285–301. [Google Scholar]
- 13.Angert ER, Clements KD, Pace NR. The largest bacterium. Nature. 1993;362:239–241. doi: 10.1038/362239a0. [DOI] [PubMed] [Google Scholar]
- 14.Clements KD, Sutton DC, Choat JH. Occurrence and characteristics of unusual protistan symbionts from surgeonfishes Acanthuridae of the Great Barrier Reef Australia. Marine Biol. 1989;102:403–412. [Google Scholar]
- 15.Angert ER, Clements KD. Initiation of intracellular offspring in Epulopiscium. Mol Microbiol. 2004;51:827–835. doi: 10.1046/j.1365-2958.2003.03869.x. [DOI] [PubMed] [Google Scholar]
- 16.Angert ER. Alternatives to binary fission in bacteria. Nat Rev Microbiol. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- 17.Robinow C, Angert ER. Nucleoids and coated vesicles of “Epulopiscium” spp. Arch Microbiol. 1998;170:227–235. doi: 10.1007/s002030050637. [DOI] [PubMed] [Google Scholar]
- 18.Herschleb J, Ananiev G, Schwartz DC. Pulsed-field gel electrophoresis. Nat Protoc. 2007;2:677–684. doi: 10.1038/nprot.2007.94. [DOI] [PubMed] [Google Scholar]
- 19.Bresler V, Montgomery WL, Fishelson L, Pollak PE. Gigantism in a bacterium, Epulopiscium fishelsoni, correlates with complex patterns in arrangement, quantity, and segregation of DNA. J Bacteriol. 1998;180:5601–5611. doi: 10.1128/jb.180.21.5601-5611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bresler V, Fishelson L. Polyploidy and polyteny in the gigantic eubacterium Epulopiscium fishelsoni. Mar Biol. 2003;143:17–21. [Google Scholar]
- 21.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides NC. The Genomes On Line Database (GOLD) version 2: A monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:332–334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- 23.Eisen JA. The RecA protein as a model molecule for molecular systematic studies of bacteria: Comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlin S, Weinstock GM, Brendel V. Bacterial classifications derived from recA protein sequence comparisons. J Bacteriol. 1995;177:6881–6893. doi: 10.1128/jb.177.23.6881-6893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rainey FA, Ward-Rainey NL, Janssen PH, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 26.McGrath CL, Katz LA. Genome diversity in microbial eukaryotes. Trends Ecol Evol. 2004;19:32–38. doi: 10.1016/j.tree.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Tower J. Developmental gene amplification and origin regulation. Annu Rev Genet. 2004;38:273–304. doi: 10.1146/annurev.genet.37.110801.143851. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Macalpine DM, Kapler GM. Developmental regulation of DNA replication: Replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol Cell Biol. 1997;17:6147–6156. doi: 10.1128/mcb.17.10.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider DA, Ross W, Gourse RL. Control of rRNA expression in Escherichia coli. Curr Opin Microbiol. 2003;6:151–156. doi: 10.1016/s1369-5274(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 30.Ogasawara N, Yoshikawa H. Genes and their organization in the replication origin region of the bacterial chromosome. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- 31.Sharpe ME, Hauser PM, Sharpe RG, Errington J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: Constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol. 1998;180:547–555. doi: 10.1128/jb.180.3.547-555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 33.Donachie WD, Blakely GW. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol. 2003;6:146–150. doi: 10.1016/s1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- 34.Brown DD. Gene expression in eukaryotes. Science. 1981;211:667–674. doi: 10.1126/science.6256857. [DOI] [PubMed] [Google Scholar]
- 35.Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster PL. Adaptive mutation in Escherichia coli. J Bacteriol. 2004;186:4846–4852. doi: 10.1128/JB.186.15.4846-4852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova KS, et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reams AB, Neidle EL. Genome plasticity in Acinetobacter: New degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol Microbiol. 2003;47:1291–1304. doi: 10.1046/j.1365-2958.2003.03342.x. [DOI] [PubMed] [Google Scholar]
- 39.Romero D, Palacios R. Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet. 1997;31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg SM, Hastings PJ. Adaptive point mutation and adaptive amplification pathways in the Escherichia coli Lac system: Stress responses producing genetic change. J Bacteriol. 2004;186:4838–4843. doi: 10.1128/JB.186.15.4838-4843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komaki K, Ishikawa H. Genomic copy number of intracellular bacterial symbionts of aphids varies in response to developmental stage and morph of their host. Insect Biochem Mol Biol. 2000;30:253–258. doi: 10.1016/s0965-1748(99)00125-3. [DOI] [PubMed] [Google Scholar]
- 42.Mergaert P, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson KK, Kelley AC, Bienz M. Loss of Drosophila borealin causes polyploidy, delayed apoptosis, and abnormal tissue development. Development. 2005;132:4777–4787. doi: 10.1242/dev.02057. [DOI] [PubMed] [Google Scholar]
- 44.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saucedo LJ, Edgar BA. Why size matters: Altering cell size. Curr Opin Genet Dev. 2002;12:565–571. doi: 10.1016/s0959-437x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- 46.Sugimoto-Shirasu K, Roberts K. Big it up: Endoreduplication and cell-size control in plants. Curr Opin Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Fishelson L, Montgomery WL, Myrberg AA. A unique symbiosis in the gut of a tropical herbivorous surgeonfish (Acanthuridae: Teleostei) from the Red Sea. Science. 1985;229:49–51. doi: 10.1126/science.229.4708.49. [DOI] [PubMed] [Google Scholar]
- 48.Grim JN. Description of somatic kineties and vestibular organization of Balantidium jocularum sp. n., and possible implications for the class Litostomatea and genus Balantidium. Acta Protozool. 1993;32:37–45. [Google Scholar]
- 49.Grim JN. Food vacuole contents in the ciliate, Balantidium jocularum (Balantididae), a symbiont in the intestine of the surgeonfish, Naso tonganus (Acanthuridae) J Eukaryotic Microbiol. 2006;53:269–274. doi: 10.1111/j.1550-7408.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- 50.Schulz HN. The genus Thiomargarita. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. Vol 6. New York: Springer; 2006. pp. 1156–1163. [Google Scholar]
- 51.Norris V, Turnock G, Sigee D. The Escherichia coli enzoskeleton. Mol Microbiol. 1996;19:197–204. doi: 10.1046/j.1365-2958.1996.373899.x. [DOI] [PubMed] [Google Scholar]
- 52.Infante JJ, Dombek KM, Rebordinos L, Cantoral JM, Young ET. Genomewide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics. 2003;165:1745–1759. doi: 10.1093/genetics/165.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mark Welch D, Meselson M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 54.Mark Welch D, Meselson MS. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc Natl Acad Sci USA. 2001;98:6720–6724. doi: 10.1073/pnas.111144598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pouchkina-Stantcheva NN, et al. Functional divergence of former alleles in an ancient asexual invertebrate. Science. 2007;318:268–271. doi: 10.1126/science.1144363. [DOI] [PubMed] [Google Scholar]
- 56.Eckert KA, Kunkel TA. DNA polymerase fidelity and the polymerase chain reaction. PCR Methods Appl. 1991;1:17–24. doi: 10.1101/gr.1.1.17. [DOI] [PubMed] [Google Scholar]
- 57.Ennis PD, Zemmour J, Salter RD, Parham P. Rapid cloning of HLA-A,B cDNA by using the polymerase chain reaction: Frequency and nature of errors produced in amplification. Proc Natl Acad Sci USA. 1990;87:2833–2837. doi: 10.1073/pnas.87.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunning AM, Talmud P, Humphries SE. Errors in the polymerase chain reaction. Nucleic Acids Res. 1988;16:10393. doi: 10.1093/nar/16.21.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke LA, Rebelo CS, Goncalves J, Boavida MG, Jordan P. PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. Mol Pathol. 2001;54:351–353. doi: 10.1136/mp.54.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shinde D, Lai Y, Sun F, Arnheim N. TaqDNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 2003;31:974–980. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ackermann M, Chao L. DNA sequences shaped by selection for stability. PLoS Genet. 2006;2:e22. doi: 10.1371/journal.pgen.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moxon ER, Rainey PB, Nowak MA, Lenski RE. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 63.Mrázek J, Guo X, Shah A. Simple sequence repeats in prokaryotic genomes. Proc Natl Acad Sci USA. 2007;104:8472–8477. doi: 10.1073/pnas.0702412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 65.Keller L, Surette MG. Communication in bacteria: An ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 67.Angert ER, Losick RM. Propagation by sporulation in the guinea pig symbiont Metabacterium polyspora. Proc Natl Acad Sci USA. 1998;95:10218–10223. doi: 10.1073/pnas.95.17.10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.