Abstract
Microfibrils are essential elements in elastic and nonelastic tissues contributing to homeostasis and growth factor regulation. Fibrillins form the core of these multicomponent assemblies. Various human genetic disorders, the fibrillinopathies, arise from mutations in fibrillins and are frequently associated with aberrant microfibril assembly. These disorders include Marfan syndrome, Weill–Marchesani syndrome, Beals syndrome, and others. Although homotypic and heterotypic fibrillin self-interactions are considered to provide critical initial steps, the detailed mechanisms for microfibril assembly are unknown. We show here that the C-terminal recombinant half of fibrillin-1 assembles into disulfide-bonded multimeric globular structures with peripheral arms and a dense core. These globules are similar to the beaded structures observed in microfibrils isolated from tissues. Only these C-terminal fibrillin-1 multimers interacted strongly with the fibrillin-1 N terminus, whereas the monomers showed very little self-interaction activity. The multimers strongly inhibited microfibril formation in cell culture, providing evidence that these recombinant assemblies can also interact with endogenous fibrillin-1. The C-terminal self-interaction site was fine-mapped to the last three calcium-binding EGF domains in fibrillin-1. These results suggest a new mechanism for microfibril formation where fibrillin-1 first oligomerizes via its C terminus before the partially or fully assembled bead-like structures can further interact with other beads via the fibrillin-1 N termini.
Keywords: extracellular matrix, fibrillinopathies, Marfan syndrome, protein assembly
Fibrillins, found in almost all vertebrate tissues as well as in invertebrates, are large cysteine-rich extracellular matrix glycoproteins. The three human fibrillins (fibrillin-1, -2, and -3) show a highly conserved domain organization that includes primarily arrays of calcium-binding EGF-like (cbEGF) domains interspersed with TGF-β binding protein domains (TB/8-Cys) and hybrid domains (1). Mutations in fibrillins lead to a number of heritable disorders, the fibrillinopathies. These include Marfan syndrome, autosomal dominant Weill–Marchesani syndrome (both fibrillin-1), and Beals syndrome (fibrillin-2) (2).
Fibrillins constitute the core components of large assemblies, termed microfibrils (3). Microfibrils are believed to be multicomponent structures in extracellular matrices, and numerous ligands including fibulins, latent TGF-β binding proteins, perlecan, versican, and others have been identified, although the functional relevance for most of these proteins remains to be elucidated (4). Besides fibrillins, the only other integral molecule identified in microfibrils in a periodic fashion is microfibril-associated glycoprotein 1 (MAGP-1) (5). Functionally, microfibrils play critical roles in the biogenesis of elastic fibers, as stress-bearing units in various organs including the kidney and the eye, in homeostasis of tissues such as the cardiovasculature, and as growth factor-regulating entities (6).
Microfibrils extracted from tissue or cell culture show a typical bead-on-the-string ultrastructure with periodicities of 50–55 nm when analyzed by electron microscopy after rotary shadowing (7, 8). Very little is known about the dynamic mechanisms required for fibrillin-1 to form the backbone of these structures. In fibrillinopathies, such as Marfan syndrome, aberrant microfibril assembly is frequently observed (9). To advance the understanding of disease pathogenesis, detailed information on microfibril assembly mechanisms is urgently needed. Although current models for the static organization of fibrillins in microfibrils agree on a head-to-tail alignment with approximately eight fibrillin monomers in cross section, it is not clear whether the molecules are arranged in an unstaggered or staggered orientation (10–18). The initial formation of microfibrils from monomeric fibrillins most likely occurs close to or at the cell surface and involves formation of disulfide bonds in early stages (19). One of the early steps is considered the self-interaction of fibrillin-1, where lateral (N-to-N and C-to-C) and head-to-tail (N-to-C) self-interaction sites have been identified in N- and C-terminal regions of the molecule (20–23). It is generally believed that fibrillin-1 monomers have the intrinsic ability to mediate elongation of the immature microfibril through its N-to-C self-interaction properties (22, 23). The respective epitopes have been mapped to a region between the unique N terminus and cbEGF2 and to a region between TB/8-Cys-7 and the unique furin-processed C terminus (23). On the other hand, N-to-N or C-to-C interactions are believed to provide a proper lateral alignment of fibrillin molecules in microfibrils (20, 21, 23).
We demonstrate here that the N-to-C self-interaction of fibrillin-1 depends on disulfide bond stabilized multimerization of the C-terminal region, resulting in globular particles similar to the bead portions in extracted microfibrils. Only these high-molecular-weight assemblies are able to interact with the fibrillin-1 N terminus in vitro and in cell culture. These results suggest a novel mechanism of microfibril assembly, where the fibrillin-1 C terminus initially forms bead-like structures that then align and interact via the N terminus to form the typical bead-on-the-string assemblies.
Results
Multimerization of C-Terminal Fibrillin-1 Constructs.
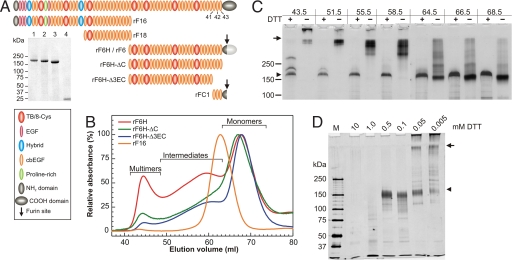
To fine-map and analyze the C-terminal head-to-tail fibrillin-1 self-interaction site, we generated recombinant fibrillin-1 constructs with deletions on their C termini based on published data (22, 23) (Fig. 1A). Although the affinity-purified constructs showed single bands under reducing conditions (Fig. 1A Inset), they were more heterogeneous under nonreducing conditions. Therefore, we size-fractionated the constructs further by gel filtration chromatography (Fig. 1B). Whereas the N-terminal control fragment rF16 primarily eluted as monomers, the C-terminal fragments demonstrated various degrees of monomeric, intermediate, and multimeric species (Fig. 1B and Table 1). For the nontruncated rF6H, ≈35% of the molecules were present in an intermediate and ≈11% in a high multimeric state, which did not migrate into the separating gel under nonreducing conditions (Fig. 1C, 43.5 ml). The rF6H intermediates are of distinct sizes, indicated by discrete bands in their nonreduced state. Both the intermediates and the multimers are reducible to a monomeric band of the expected size for rF6H, although in case of the multimers reduction without subsequent alkylation is typically incomplete (Fig. 1C, 43.5 ml). The required concentration of dithiothreitol (DTT) to reduce rF6H multimers was ≈0.05–0.1 mM, whereas 10- to 20-fold more (≈1 mM) was necessary to reduce intramolecular disulfide bonds, indicating that most of the intermolecular disulfide bonds are more readily accessible compared with the buried intramolecular disulfide bonds (Fig. 1D). The formation of disulfide-bonded rF6H occurred cell-associated, and purified monomers were not able to multimerize in vitro [supporting information (SI) Fig. S1]. Multimerization was not significantly different with fragment rF6, which is identical to rF6H but is expressed with its C-terminal propeptide, which becomes processed during or after secretion (Figs. S1 and S2).
Fig. 1.
Properties and multimerization of recombinant fibrillin-1 constructs. (A) Schematic representation of recombinant human fibrillin-1 constructs. The last three cbEGF domains are numbered. (Inset) Coomassie-stained purified and reduced recombinant fibrillin-1 fragments: lane 1, rF6H; lane 2, rF6H-ΔC; lane 3, rF6H-Δ3EC; lane 4, rFC1. Fragments rF6, rF16, and rF18 were described in detail previously (13, 31). rF6H is expressed without C-terminal propeptide (dark gray region of C-terminal domain), and rF6 is expressed with propeptide (additional light gray region of C-terminal domain) that becomes cleaved during or after secretion (arrow). (B) Gel filtration chromatography of recombinant fibrillin-1 fragments (as indicated) demonstrating various degrees of monomeric, intermediate, and multimeric fractions. The absorbance at 280 nm for each fragment was set to 100% for the monomeric peaks. Minor differences between monomeric rF6H and rF6H-ΔC elution volumes are statistically not significant. Note that rF6H-ΔC resulted in variable degrees of multimerization (see Table 1). (C) Individual fractions of the gel filtration chromatography of rF6H shown in B were subjected to SDS gel electrophoresis under reducing (+) or nonreducing (−) conditions and silver-stained. Elution volumes in milliliters on top correlate with elution volumes in B. The two additional bands above the rF6H monomer in the 43.5-ml fraction were determined to be rF6H by reaction with specific antibodies. (D) rF6H multimers were treated with various concentrations of DTT as indicated, alkylated, digested with trypsin, separated by SDS gel electrophoresis, and silver-stained. Note that reduction of the multimers to monomers requires ≈10- to 20-fold less DTT than reduction of intramolecular disulfide bonds as monitored by trypsin degradation of the monomers. In C and D, arrows denote the start of the separating gels and arrowheads indicate the position of monomeric and completely reduced rF6H.
Table 1.
Relative quantification of fibrillin construct multimerization (percentage of total)
| Recombinant construct | Monomers | Intermediates | Multimers |
|---|---|---|---|
| rF16 (n = 2) | 92.9 ± 6.3 | 2.9 ± 2.7 | 4.2 ± 3.6 |
| rF6H (n = 8) | 54.1 ± 2.3 | 34.6 ± 1.1 | 11.4 ± 1.2 |
| rF6H-ΔC (n = 3) | 54.5 ± 9.8 | 34.6 ± 10.7 | 10.9 ± 9.0 |
| rF6H-Δ3EC (n = 3) | 61.3 ± 4.8 | 36.2 ± 5.9 | 2.5 ± 1.3 |
Significant differences (Student's t test, P < 0.05) were found between the peak sizes of rF6H and rF6H-Δ3EC multimers and of rF6H and rF6H-Δ3EC monomers. The relative proportions of the three species varied significantly for rF6H-ΔC and were much more stable for all other constructs.
A similar distribution of monomers and multimers was found for the fibrillin-1 deletion construct rF6H-ΔC lacking the unique C-terminal domain, although this fragment showed significant variations in peak distribution (Table 1). However, the multimeric portion of rF6H-Δ3EC, lacking the last four domains of fibrillin-1, decreased significantly (≈5-fold) compared with the nontruncated rF6H (Table 1 and Fig. S3A). These data demonstrate that the last four C-terminal domains in rF6H influence its multimerization. To rule out potential effects of the hexahistidine tag on the multimerization process, rF6H was analyzed by gel filtration chromatography in the presence of 5 mM EDTA demonstrating virtually no differences in the ratio of multimeric to monomeric peak size or in SDS gel electrophoresis (data not shown). In addition, all deletion constructs are tagged in an identical fashion.
Structural Analysis.
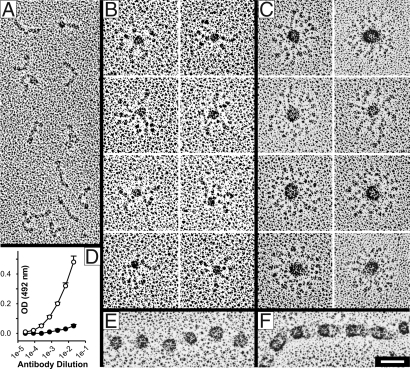
rF6H fractions from gel filtration chromatographies marked as “monomers,” “intermediates,” and “multimers” in Fig. 1B were analyzed by electron microscopy after rotary shadowing (Fig. 2). The monomeric fraction showed a highly homogenous population of rod-like molecules with a length of 60.5 ± 8.2 nm (n = 120) correlating well with previous observations (13) (Fig. 2A). One end of the molecules frequently showed a small globule. Particles from the intermediate fraction demonstrated an electron-dense globular core of 18.2 ± 4.6 nm (n = 203) with three to seven arms extending from the core (Fig. 2B). The multimeric particles, eluting in the void volume of the column, demonstrated a regular star-like globular shape with dense cores, a lucid center, and arms extending to the periphery (Fig. 2C). Because the extensions were apparently tangled, it was not possible to determine their precise number on each particle. However, we estimate that the particles contain approximately eight to maximal 12 peripheral extensions, indicating that steric hindrance determines the maximal particle size. These projecting arms frequently have a small globular end at the periphery, a feature that correlates with the appearance of the monomeric particles (Fig. 2 A and C). Antibodies against the hexahistidine tag located at the C terminus of rF6H reacted significantly less with the multimers than the monomers, indicating that this sequence is buried in the core of the particles (Fig. 2D). Quantification of the bead dimensions composed of rF6H revealed a diameter for the entire particle of 88.6 ± 14.0 nm with the inner electron dense core of 30.3 ± 5.2 nm (n = 75). Surprisingly, the diameter determined for the inner core is similar to the dimensions of the beads in microfibrils analyzed by rotary shadowing electron microscopy, which ranges between 22 and 29 nm (24–26). For comparison, a collagenase-extracted beaded microfibril demonstrates similar shapes and dimensions of the beads including the somewhat more lucent center (Fig. 2 E and F). Multimers of rF6H-ΔC and rF6H-Δ3EC resulted in similar globular shapes compared with rF6H, although for rF6H-Δ3EC, as mentioned above, the relative amounts were significantly lower than those for rF6H or rF6H-ΔC (Table 1 and Fig. S3B). This suggests that the last four fibrillin-1 domains help to regulate multimerization but do not contain a multimerization domain per se.
Fig. 2.
Structural analysis of rF6H by electron microscopy after rotary shadowing (A–C, E, and F) and by ELISA (D). Fragment rF6H was subjected to gel filtration chromatography (Fig. 1B), and representative fractions of the monomers (A), intermediates (B), and multimers (C) were analyzed. (D) ELISA analysis with an antibody against the C-terminal hexahistidine tag showed significant differences (P < 0.001) in the reactivity of monomers (open symbols) and multimers (filled symbols). A beaded microfibril isolated from skin fibroblasts after collagenase digestion (32) is shown in the open (E) or closed (F) form. Note the similar appearance of the beaded structures in C, E, and F. (Scale bar: 50 nm.)
The solution structure of rF6H was analyzed by dynamic light scattering (Fig. S4). The polydispersity for monomers (21.6 ± 1.1%), intermediates (19.8 ± 1.0%), and multimers (21.4 ± 1.2%) indicated monodisperse samples and thus the presence of one species of molecules. The hydrodynamic radius steadily increased from 10.4 ± 0.2 nm for the monomers to 17.6 ± 0.9 nm for intermediates to 40.0 ± 2.0 nm for the multimers. Calculation of the molecular mass for the monomers, based on a linear model, correlated well with values derived from the amino acid sequence. No mass calculation was possible for the intermediates and the multimers because of the lack of appropriate mathematical models for the shape of these assemblies. The hydrodynamic radius for rF6H multimers (≈40 nm) correlated very well with the radius of these particles after rotary shadowing (≈44 nm). We conclude from these data that the multimeric rF6H particles assume a globular shape in solution similar to that observed by electron microscopy after rotary shadowing of surface-coated particles.
Functional Consequences of Multimerization and C-Terminal Deletions.
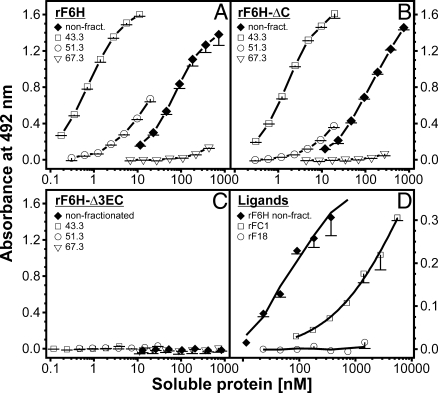
The functional consequences of C-terminal multimerization and deletions were tested in an established fibrillin-1 N-to-C self-interaction assay (22) with representative fractions from the gel filtration chromatographies (Fig. 3). The hypothesis was that the monomeric C-terminal fibrillin-1 fragment interacts with the monomeric N-terminal fragment, which represents an early mechanistic step in fibrillin-1 self-assembly. Unexpectedly, we found that the multimeric forms of rF6H interacted strongly with the N-terminal fragment rF16, whereas the monomers showed virtually no binding activity (Fig. 3A). Only when the monomers were used at very high concentrations (>3 μM) was some residual binding observed (data not shown), indicating the presence of a very-low-affinity self-interaction site present on monomers. Intermediate fractions showed size-dependent binding affinities. Deletion of the C-terminal domain (Ser2688-Lys2725) in rF6H-ΔC resulted in significant binding activity of the multimers, but oligomers with lower masses interacted much less with rF16 (Fig. 3B). Additional removal of cbEGF41–43 in rF6H-Δ3EC abolished binding to rF16 completely (Fig. 3C). In general, the binding strengths of the rF6H and rF6H-ΔC multimers were considerably stronger (≈100-fold) compared with the crude nonpurified mixtures, which explains why only a relatively small portion of multimers can account for the entire binding activity of a mixture (Table 1 and Fig. 3 A and B). Based on these results, we generated a small fragment comprising cbEGF41–43 and parts of the C-terminal domain (rFC1; Fig. 1A). This fragment interacted with rF16, albeit with a ≈20-fold lower affinity compared with the rF6H mixture and a >1,500-fold lower affinity compared with the rF6H multimers (Fig. 3 A and D). The control fragment rF18 did not interact. In summary, these data clearly demonstrate (i) that multimerization of the fibrillin-1 C terminus is required for high-affinity interaction with the N terminus and (ii) that the N-terminal binding site is located in cbEGF41–43 (Asp2567-Val2687).
Fig. 3.
N-to-C interaction of fibrillin-1 constructs. In this previously established solid-phase interaction assay (22), the N-terminal half of fibrillin-1 (rF16) was immobilized. Construct rF6H (A) and the deletion constructs rF6H-ΔC (B) and rF6H-Δ3EC (C) were size-fractionated by gel filtration chromatography (Fig. 1B), and individual fractions as indicated in each graph by their respective elution volumes in milliliters were used as soluble ligands. Mixtures of monomers and multimers not subjected to gel filtration were included as controls (filled diamonds). (D) The biotinylated small C-terminal fragment rFC1 bound to rF16 albeit with a significantly lower affinity compared with the rF6H mixture. A biotinylated control fragment rF18 did not interact. Data represent means of triplicates (A–C) or duplicates (D) including standard deviations.
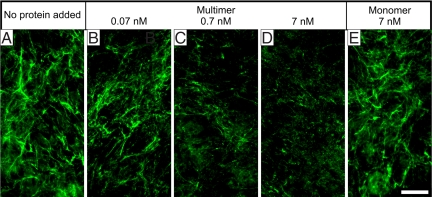
To test whether monomeric or multimeric rF6H interacts with fibrillin-1 in a cell culture system, the purified fractions were added to fibroblast cultures and fibrillin-1 assembly was analyzed by indirect immunofluorescence (Fig. 4). Purified rF6H multimers caused a dose-dependent reduction of the fibrillin-1 network in the range of 0.07–7 nM, whereas addition of 7 nM rF6H monomer resulted in no or only little inhibition of fibrillin-1 network formation. However, minor inhibition with the monomer was observed when the concentration was raised to >70 nM (data not shown). Addition of rF6H multimers and monomers did not change the endogenous fibrillin-1 mRNA and protein expression or the fibronectin or fibulin-2 network formation (Fig. S5). These results demonstrate that multimeric rF6H effectively interacts with endogenous fibrillin-1 produced by fibroblasts and thereby inhibits fibrillin-1 network formation.
Fig. 4.
Inhibition of the fibrillin-1 network produced by fibroblasts. At the time of seeding of human skin fibroblasts into chamber slides, no protein (A), rF6H multimers (B–D), or rF6H monomers (E) were added at the concentrations indicated. The cells were grown for 3 days, and network formation was monitored by indirect immunofluorescence with antibodies against fibrillin-1. Fibronectin and fibulin-2 network formation are shown as controls in Fig. S5C. (Scale bar: 50 μm.)
Discussion
Although many publications over the last years address the static spatial organization of fibrillins in microfibrils (11–18), very little information is available about the dynamic processes of microfibril formation. Revealing these mechanisms is critical for the understanding of the pathobiology of fibrillinopathies. Based on previously published data, the general, but not yet substantiated, belief was that fibrillin-1 monomers interact in a N-to-C fashion to generate higher-order elongated multimers, which lose the capacity to self-interact once the multimers have matured (22, 23). However, in this study, we surprisingly found (i) that the C-terminal half of fibrillin-1 alone has the intrinsic ability to multimerize into structures resembling the beads present in extracted microfibrils and (ii) that this multimerization is a prerequisite for high-affinity fibrillin-1 N-to-C self-interaction, mediated by cbEGF41–43, which is considered one of the early steps in microfibril assembly.
Recently, we and others have described high-affinity self-interaction properties between N- and C-terminal regions of fibrillin-1 (22, 23). Here we found that a C-terminal fibrillin-1 construct (rF6H), spanning one half of the molecule, showed significant amounts (≈46%) of disulfide bond-mediated multimerization. This is in line with a previous report demonstrating C-to-C self-interaction of a recombinant fragment contained in rF6H (23). Multimerization was observed in association with cells before the multimers were released into the culture medium, suggesting that cellular mechanisms are required for multimerization. This was further supported by the fact that the purified monomeric fraction did not show multimerization in vitro. In contrast to tissues, recombinant rF6H produced in cell culture is immediately exposed after secretion to culture medium allowing rapid free diffusion away from the cells, which may explain why half of the rF6H molecules are not multimerized. Similar observations were reported for endogenous fibrillin-1 produced by cultured cells (10). Alternatively, the cellular machinery for multimer formation might be overloaded by the high recombinant expression levels. Remarkably, construct rF6H-ΔC (missing the C-terminal domain) showed extremely variable amounts of intermediates and multimers, suggesting that the unique C-terminal domain (after furin processing) may play a regulatory role in stabilization of multimers. A deletion construct missing cbEGF41–43 and the unique C-terminal domain still multimerizes albeit at a significant lower level compared with the nontruncated construct, demonstrating that the multimerization domain is not directly localized in the deleted region of the molecule. However, this observation suggests that the multimerization domain is in close physical proximity to this region and is functionally affected by structural changes imposed by the deletion. The multimers of the recombinant C-terminal fragments were reducible, demonstrating the participation of intermolecular disulfide bonds. Based on high-resolution data available for the 8-Cys/TB and the cbEGF domains in fibrillin-1, it is predicted that cysteine residues in domains close to the deleted region are involved in intramolecular disulfide bonds. Thus, intermolecular disulfide bond formation would require appropriate disulfide bond reshuffling mechanisms, potentially mediated by members of the protein disulfide isomerases located on the cell surface (27). Intermolecular disulfide bonds were much more accessible to reduction compared with intramolecular disulfide bonds, suggesting that the former are solvent-exposed. This differential property could potentially be involved in the regulation of microfibril formation through redox mechanisms.
Analysis of the structural properties of C-terminal fibrillin-1 multimers by electron microscopy after rotary shadowing and dynamic light scattering unexpectedly revealed the rF6H multimers as unique, star-like globular structures with dense cores including a somewhat more lucid center and projecting arms on the periphery. The appearance and dimensions of these globular particles resembled the beads in extracted microfibrils. These data establish for the first time, to our knowledge, that fibrillin-1 alone, and in this case a fibrillin-1 C-terminal fragment, can form globular multimeric structures with a considerable degree of similarity to the beads in microfibrils, which provides a potential mechanism for microfibril formation. The “PF3” fragment produced by Maddox et al. (28) from pepsin-digested microfibrils extracted from human placenta showed somewhat similar properties to the rF6H multimers with (i) reducible high-molecular-weight assemblies and (ii) the globular bead structures, although the number of arms were lower. These PF3 particles reacted with a monoclonal antibody mAb69 directed against the region of fibrillin-1 between 8-Cys/TB6 and 8-Cys/TB7 (18), an epitope also contained in rF6H. It was further reported that attempts to prepare monoclonal antibodies against PF3 resulted only in antibodies against fibrillin-1 but not against other proteins. Along these lines, our data suggest that fibrillin-1 may have an intrinsic property to form the beaded structures in microfibrils in the absence of other microfibril-associated proteins. Other globular structures similar to the rF6H multimers have been observed in preparations of microfibrils isolated from chick vitreous humor without the use of proteolytic enzymes (25). In addition, Kuo et al. (18) recently described that cleavage of guanidine-extracted microfibrils from cell culture with crude collagenase, which cleaves fibrillin-1 at various sites almost exclusively in the N-terminal half, led to clumps of beads with peripheral extensions, again similar to the rF6H multimers. Based on our data, we propose that the globular parts of microfibrils can be formed by C-terminal portions of fibrillin-1 molecules. The number of arms in the microfibril preparations described above from placenta and vitreous humor appeared somewhat lower than what we observe with the rF6H multimers. This could potentially be attributed to back-folding of the rF6H arms into the globule, or alternatively to missing microfibril-associated proteins that regulate the number of arms. Antibodies specific for the C-terminal region of rF6H reacted less with the multimers than the monomers, indicating that the rF6H C terminus primarily contributes to the core (and is thus less available for the antibodies) and the N-terminal region of rF6H constitutes the peripheral arms. However, cbEGF41–43 must be present in a topological orientation, likely close to the surface of the beads, which allows this region to interact with the N terminus. Attempts to produce an entire bead-on-the-string microfibril in vitro from purified full-length fibrillin-1 is currently hampered by the propensity of recombinant full-length fibrillin-1 to precipitate (22) or by the extremely low yields of endogenous fibrillin-1 preparations from cell culture (10).
Surprisingly, after separation of monomers, intermediates, and multimers by gel filtration chromatography, we observed that only the highly multimeric fractions of rF6H bound to the N-terminal fibrillin-1 fragment with high affinity. The intermediates bound with moderate affinity, and the monomers bound only marginally and at very high concentrations. These data demonstrate that a relatively low-affinity self-interaction site, present on the monomers, dramatically increases its avidity for the N terminus (>1,500-fold) as a consequence of multimerization. Whereas deletion of the C-terminal domain had virtually no effect on the ability of the rF6H-ΔC multimers to interact with the N-terminal region of fibrillin-1, further deletion of cbEGF41–43 abrogated self-interaction completely. The data clearly localize the N-to-C fibrillin-1 self-interaction site to cbEGF41–43. These results were further substantiated with a small deletion construct, rFC1, comprising cbEGF41–43 and the unique C-terminal domain after furin processing. This construct interacted with the N-terminal portion albeit with a significant lower affinity compared with the multimerized large C-terminal constructs, again highlighting the presence of very-low-affinity binding sites in this region before multimerization.
Further evidence for the importance of C-terminal fibrillin-1 multimerization came from our cell culture model of microfibril assembly. In low concentrations (<10 nM), the multimers of the fibrillin-1 C-terminal half were able to inhibit fibrillin-1 network formation in fibroblasts, whereas the monomeric fraction had only a small effect at much higher concentrations. The inhibitory effect is specific because network formation of other extracellular matrix proteins (fibronectin, fibulin-2) was not affected. Inhibition must have occurred by interference with the in vivo assembly process by blocking formation of microfibrils due to the missing N-terminal region.
In summary, our data suggest the following new model for fibrillin-1 assembly. The C terminus of fibrillin-1 multimerizes via a disulfide bond-mediated mechanism into bead-like structures. This multimerization enhances the avidity of the N-to-C self-interaction site contained within cbEGF41–43 for binding to the N-terminal region of fibrillin-1. When the avidity of a nascent bead has sufficiently increased, the peripheral N-terminal sites begin to bind to the C-terminal sites in appropriate adjacent beads. Such a universal mechanism is compatible with all parallel head-to-tail models of fibrillin-1 organization in microfibrils, independent of whether the molecules were staggered or unstaggered or whether the interbead arms were spanning one, two, or even three bead-to-bead distances (10–18). Parallel staggered or unstaggered fibrillin-1 organization requires unidirectionality of the peripheral arms. This feature could be conferred to a nascent bead by increasing avidities of the next appropriate downstream bead causing the N termini to bind. Alternatively, microfibril-associated proteins such as MAGP-1 could potentially act to confer unidirectionality to the fibrillin molecules.
Materials and Methods
Recombinant Expression of Fibrillin-1 Fragments.
Production of rF16 (N-terminal half) and rF6H or rF6 (C-terminal halves) were described previously (13, 29). rF6H is designed as a processed molecule; i.e., it has a stop codon at the furin cleavage site within the unique C terminus, whereas rF6 is designed with the propeptide (Fig. 1A). The C-terminal deletion construct rF6H-ΔC is identical to rF6H but without the unique C-terminal domain, whereas rF6H-Δ3EC lacks the last four domains. Fragment rFC1 consists of cbEGF41–43 and the furin processed C-terminal domain. Details are described in SI Methods and Table S1. Generation of stable clones in human embryonic kidney cells 293 and purification of the secreted recombinant proteins by Ni2+-chelating chromatography were previously described in detail (13, 22).
Separation and Analysis of Multimerized Fibrillin-1 Fragments.
To separate different multimerization states of recombinant fibrillin fragments, C-terminal nontruncated and deletion constructs of fibrillin-1 (2–3 mg) were subjected to Superose 6 gel filtration chromatography (100-ml column on a high-precision ÄktaPurifier 10; GE Healthcare) equilibrated in 50 mM Tris·HCl (pH 7.4)/5 mM CaCl2 containing either 400 mM NaCl (for binding assays) or 150 mM NaCl (for cell culture assays), or in 500 mM NH4 acetate (pH 7.0) (for electron microscopy) at a flow rate of 0.5 ml/min. No differences in the protein elution profiles were found with these buffer systems. One-milliliter fractions were collected starting with the void volume (≈40 ml). Quantification of relative amounts of monomers, intermediates, and multimers was performed by integration of the individual peak areas from a baseline of zero using the trapezoidal rule (Origin 7.0; Microcal). Protein concentrations were determined with the absorbance at 280 nm using calculated extinction coefficients (see SI Methods and Table S2). Aliquots of the fractions were separated by SDS gel electrophoresis (4–20% acrylamide) and silver-stained. For reduction experiments, the fractions were incubated with 0.005–10 mM DTT for 1 h at 37°C, alkylated with iodoacetamide (Sigma) in a 2.5- to 5-fold excess over DTT for 1 h at 22°C in the dark. The samples were then digested with 0.5 μg of TPCK-treated trypsin (Sigma) for 4 h at 37°C.
Protein Interaction Assays.
Solid-phase binding experiments were performed as described elsewhere with some modifications (22). Briefly, 10 μg/ml (100 μl per well) of the fibrillin-1 (rF16) N-terminal construct was coated in 50 mM Tris·HCl (pH 7.4)/150 mM NaCl (TBS) onto 96-well plates, blocked with 5% nonfat milk in TBS, and incubated with serial dilutions (1:2) of either a control (rF6H, not subjected to gel filtration) or gel filtration fractions of individual recombinant fragments diluted in TBS including 5 mM CaCl2 and 5% nonfat milk (binding buffer). To detect bound proteins, the polyclonal antiserum α-rF6H was used 1:1,000 diluted (30). Because no efficient antibody detection method was available for rFC1, this fragment (1.12 mg/ml) as well as the control fragments rF18 (1.25 mg/ml) (13) and the nonfractionated rF6H (1.56 mg/ml) were biotinylated with Sulfo-NHS-Biotin (Pierce) according to the instructions. After biotinylation, the proteins were dialyzed against TBS/2 mM CaCl2, and final protein concentrations were determined with the BCA protein assay kit (Pierce). The proteins were labeled with three to five biotin molecules per protein molecule. Detection of bound biotinylated proteins was performed by using NeutrAvidin horseradish peroxidase conjugate (Pierce) 1:500 diluted in binding buffer.
Dynamic Light Scattering and Electron Microscopy.
The hydrodynamic radius of size-fractionated protein assemblies was determined in 50 mM Tris·HCl (pH 7.4), 400 mM NaCl, and 5 mM CaCl2 by dynamic light scattering analyses (DynaPro-MS/X; Wyatt Technology). Protein assemblies separated by gel filtration chromatography were rotary shadowed and visualized by electron microscopy (see SI Methods). Measurements were performed with ImageJ version 1.37 (http://rsb.info.nih.gov/ij/).
Immunolabeling of the Extracellular Matrix of Fibroblasts.
Human foreskin fibroblasts (age of donor = 2.5 years; passage number = 4–9) were grown in DMEM including 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) with 10% (vol/vol) FCS at 37°C in a 5% CO2 atmosphere. Cells were trypsinized and seeded at a density of 7.5 × 104 cells per well in DMEM containing 1% (vol/vol) FCS in the presence of sterile-filtrated (0.22-μm filter) multimeric (0.07–7 nM) or monomeric (7 nM) rF6H in eight-well chamber slides. After 3 days, cells were fixed and immunostained as described previously using the antiserum against rF6H (1:1,000 diluted) or control proteins as indicated. Cell densities were monitored by DAPI staining, and all images were recorded from areas with similar cell densities on an Axiocam microscope (Zeiss).
Supplementary Material
Acknowledgments.
We thank Christine Fagotto and Guy Charette for excellent technical contributions and Drs. Lynn Sakai and Takako Sasaki for providing fibrillin-1 and fibulin-2 antibodies. This work was supported by Canadian Institutes of Health Research Grant MOP-68836, the Canadian Marfan Association (D.P.R.), the German Academic Exchange Service (D.H.), and the Shriners of North America (E.R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706335105/DCSupplemental.
References
- 1.Hubmacher D, Tiedemann K, Reinhardt DP. Fibrillins: From biogenesis of microfibrils to signaling functions. Curr Top Dev Biol. 2006;75:93–123. doi: 10.1016/S0070-2153(06)75004-9. [DOI] [PubMed] [Google Scholar]
- 2.Robinson P, et al. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43:769–787. doi: 10.1136/jmg.2005.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai LY, Keene DR, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol. 1986;103:2499–2509. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kielty CM. Elastic fibres in health and disease. Expert Rev Mol Med. 2006;8:1–23. doi: 10.1017/S146239940600007X. [DOI] [PubMed] [Google Scholar]
- 5.Henderson M, Polewski R, Fanning JC, Gibson MA. Microfibril-associated glycoprotein-1 (MAGP-1) is specifically located on the beads of the beaded-filament structure for fibrillin-containing microfibrils as visualized by the rotary shadowing technique. J Histochem Cytochem. 1996;44:1389–1397. doi: 10.1177/44.12.8985131. [DOI] [PubMed] [Google Scholar]
- 6.Ramirez F, Sakai LY, Dietz HC, Rifkin DB. Fibrillin microfibrils: Multipurpose extracellular networks in organismal physiology. Physiol Genomics. 2004;19:151–154. doi: 10.1152/physiolgenomics.00092.2004. [DOI] [PubMed] [Google Scholar]
- 7.Keene DR, Maddox BK, Kuo HJ, Sakai LY, Glanville RW. Extraction of extendable beaded structures and their identification as fibrillin-containing extracellular matrix microfibrils. J Histochem Cytochem. 1991;39:441–449. doi: 10.1177/39.4.2005373. [DOI] [PubMed] [Google Scholar]
- 8.Kielty CM, Cummings C, Whittaker SP, Shuttleworth CA, Grant ME. Isolation and ultrastructural analysis of microfibrillar structures from foetal bovine elastic tissues. J Cell Sci. 1991;99:797–807. doi: 10.1242/jcs.99.4.797. [DOI] [PubMed] [Google Scholar]
- 9.Tiedemann K, Bätge B, Reinhardt DP. In: Marfan Syndrome: A Primer for Clinicians and Scientists. Robinson PN, Godfrey M, editors. Georgetown, TX: Landes Bioscience; 2004. pp. 130–142. [Google Scholar]
- 10.Sakai LY, Keene DR, Glanville RW, Bächinger HP. Purification and partial characterization of fibrillin, a cysteine-rich structural component of connective tissue microfibrils. J Biol Chem. 1991;266:14763–14770. [PubMed] [Google Scholar]
- 11.Downing AK, et al. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: Implications for the Marfan syndrome and other genetic disorders. Cell. 1996;85:597–605. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- 12.Liu WG, et al. Mutant fibrillin-1 monomers lacking EGF-like domains disrupt microfibril assembly and cause severe Marfan syndrome. Hum Mol Genet. 1996;5:1581–1587. doi: 10.1093/hmg/5.10.1581. [DOI] [PubMed] [Google Scholar]
- 13.Reinhardt DP, et al. Fibrillin 1: Organization in microfibrils and structural properties. J Mol Biol. 1996;258:104–116. doi: 10.1006/jmbi.1996.0237. [DOI] [PubMed] [Google Scholar]
- 14.Qian RQ, Glanville RW. Alignment of fibrillin molecules in elastic microfibrils is defined by transglutaminase-derived cross-links. Biochemistry. 1997;36:15841–15847. doi: 10.1021/bi971036f. [DOI] [PubMed] [Google Scholar]
- 15.Baldock C, et al. The supramolecular organization of fibrillin-rich microfibrils. J Cell Biol. 2001;152:1045–1056. doi: 10.1083/jcb.152.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SS, et al. Structure of the integrin binding fragment from fibrillin-1 gives new insights into microfibril organization. Structure (Cambridge, UK) 2004;12:717–729. doi: 10.1016/j.str.2004.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldock C, et al. Nanostructure of fibrillin-1 reveals compact conformation of EGF arrays and mechanism for extensibility. Proc Natl Acad Sci USA. 2006;103:11922–11927. doi: 10.1073/pnas.0601609103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo CL, et al. Effects of fibrillin-1 degradation on microfibril ultrastructure. J Biol Chem. 2007;282:4007–4020. doi: 10.1074/jbc.M606370200. [DOI] [PubMed] [Google Scholar]
- 19.Reinhardt DP, Gambee JE, Ono RN, Bächinger HP, Sakai LY. Initial steps in assembly of microfibrils. Formation of disulfide-cross-linked multimers containing fibrillin-1. J Biol Chem. 2000;275:2205–2210. doi: 10.1074/jbc.275.3.2205. [DOI] [PubMed] [Google Scholar]
- 20.Ashworth JL, Kelly V, Wilson R, Shuttleworth CA, Kielty CM. Fibrillin assembly: Dimer formation mediated by amino-terminal sequences. J Cell Sci. 1999;112:3549–3558. doi: 10.1242/jcs.112.20.3549. [DOI] [PubMed] [Google Scholar]
- 21.Trask TM, Ritty TM, Broekelmann T, Tisdale C, Mecham RP. N-terminal domains of fibrillin 1 and fibrillin 2 direct the formation of homodimers: A possible first step in microfibril assembly. Biochem J. 1999;340:693–701. [PMC free article] [PubMed] [Google Scholar]
- 22.Lin G, et al. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J Biol Chem. 2002;277:50795–50804. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- 23.Marson A, et al. Homotypic fibrillin-1 interactions in microfibril assembly. J Biol Chem. 2005;280:5013–5021. doi: 10.1074/jbc.M409029200. [DOI] [PubMed] [Google Scholar]
- 24.Fleischmajer R, et al. Elastin-associated microfibrils (10 nm) in a three-dimensional fibroblast culture. J Invest Dermatol. 1991;97:638–643. doi: 10.1111/1523-1747.ep12483132. [DOI] [PubMed] [Google Scholar]
- 25.Wright DW, Mayne R. Vitreous humor of chicken contains two fibrillar systems: An analysis of their structure. J Ultrastruct Mol Struct Res. 1988;100:224–234. doi: 10.1016/0889-1605(88)90039-0. [DOI] [PubMed] [Google Scholar]
- 26.Wallace RN, Streeten BW, Hanna RB. Rotary shadowing of elastic system microfibrils in the ocular zonule, vitreous, and ligament nuchae. Curr Eye Res. 1991;10:99–109. doi: 10.3109/02713689109007614. [DOI] [PubMed] [Google Scholar]
- 27.Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J Cell Physiol. 2002;193:154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- 28.Maddox BK, Sakai LY, Keene DR, Glanville RW. Connective tissue microfibrils. Isolation and characterization of three large pepsin-resistant domains of fibrillin. J Biol Chem. 1989;264:21381–21385. [PubMed] [Google Scholar]
- 29.Jensen SA, Reinhardt DP, Gibson MA, Weiss AS. MAGP-1, protein interaction studies with tropoelastin and fibrillin-1. J Biol Chem. 2001;276:39661–39666. doi: 10.1074/jbc.M104533200. [DOI] [PubMed] [Google Scholar]
- 30.Tiedemann K, Bätge B, Müller PK, Reinhardt DP. Interactions of fibrillin-1 with heparin/heparan sulfate: Implications for microfibrillar assembly. J Biol Chem. 2001;276:36035–36042. doi: 10.1074/jbc.M104985200. [DOI] [PubMed] [Google Scholar]
- 31.Brinckmann J, et al. Absence of autoantibodies against correctly folded recombinant fibrillin-1 protein in systemic scleroderma patients. Arthritis Res Ther. 2005;7:R1221–R1226. doi: 10.1186/ar1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Hallous E, et al. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adapter function to tropoelastin. J Biol Chem. 2007;282:8935–8946. doi: 10.1074/jbc.M608204200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.