Abstract
Gadd45 proteins are recognized as tumor and autoimmune suppressors whose expression can be induced by genotoxic stresses. These proteins are involved in cell cycle control, growth arrest, and apoptosis through interactions with a wide variety of binding partners. We report here the crystal structure of Gadd45γ, which reveals a fold comprising an αβα sandwich with a central five-stranded mixed β-sheet with α-helices packed on either side. Based on crystallographic symmetry we identified the dimer interface of Gadd45γ dimers by generating point mutants that compromised dimerization while leaving the tertiary structure of the monomer intact. The dimer interface comprises a four-helix bundle involving residues that are the most highly conserved among Gadd45 isoforms. Cell-based assays using these point mutants demonstrate that dimerization is essential for growth inhibition. This structural information provides a new context for evaluation of the plethora of protein–protein interactions that govern the many functions of the Gadd45 family of proteins.
Keywords: crystallography, growth inhibition, tumor suppressor
The Gadd45 proteins are small, acidic proteins located primarily in the nucleus (1). Three isoforms, α, β, and γ, have been identified, and amino acid sequence identities among the three isoforms range from 50% to 60% (2). The Gadd45 proteins are recognized as important tumor and autoimmune suppressors (3). Expression of these proteins is induced in response to a number of DNA-damaging agents and genotoxic stresses including hyperosmotic stress and UV irradiation. They have been implicated in diverse processes including growth arrest, cell cycle control, DNA repair, terminal differentiation, and apoptosis. Some of these roles are common among the three isoforms, whereas others are isoform- and cell-type-specific (4). Although these tumor suppressors are rarely mutated in cancers, Gadd45β and Gadd45γ are epigenetically repressed in some cancers by methylation of CpG islands in promoter regions of their genes (5–7). Repression of Gadd45α and Gadd45γ may be a common survival mechanism allowing cancer cells to escape from apoptosis (5, 7–9).
Gadd45 proteins are best known for their role in growth arrest after genotoxic stress. Their role in the G2/M cell cycle checkpoint is attributed to their capacity to bind and inhibit Cdc2, displacing cyclin B1 (10–14). The importance of their up-regulation in response to stress is supported by the fact that reduction of transcript levels of Gadd45α or Gadd45γ by siRNA (8) or by other posttranscriptional means (15) abrogates growth arrest. DNA repair roles for Gadd45 proteins are suggested by their ability to directly interact with histone core proteins (16), with the repair endonuclease XPG to effect DNA demethylation (17), and with proliferating cell nuclear antigen (PCNA) (18–21). The Gadd45 proteins also are involved in p38 MAPK and other signal transduction pathways (22–27).
We report here the crystal structure of Gadd45γ, the first structure reported for a member of the Gadd45 family of proteins. Because a monomer–oligomer transition of Gadd45 proteins was proposed to regulate their function (28), identification of the dimerization interface is critical for understanding Gadd45 interactions and functions. To date, the dimerization interface and the interactions of Gadd45 proteins with many of their binding partners have been characterized by genetic and biochemical approaches, many of which involved deletion constructs. Based on the structural analysis presented here, it is evident that, for many of these deletion constructs, the integrity of the tertiary structure of the protein would be severely compromised, thereby limiting interpretation. We define here the dimerization interface by demonstrating that point mutations that we designed based on the observed crystal packing affect dimerization without compromising the structural integrity of the monomer. In contrast to previous work that suggested oligomerization to be inhibitory to some Gadd45 functions (28), we show that dimerization is essential for growth inhibition of HepG2 cells. This structure provides a new context enabling dissection of the role of dimerization in the interactions of Gadd45 with partner proteins.
Results
Crystallization and Structure Solution.
A truncated Gadd45γ construct containing residues 16–159 was crystallized, and the structure was solved by Se-SAD methods. The model was refined by using diffraction data to 1.7-Å resolution to an Rwork of 22.4% and Rfree of 27.2%. The final model includes chains A and B and consists of residues 16A–105A (18B–106B), 116A–126A (116B–125B), and 130A–159A (131B–159B) plus 233 water molecules. The data collection, refinement, and validation statistics are reported in supporting information (SI) Table S1.
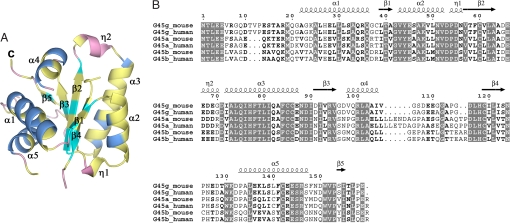
The structure of the monomer consists of a three-layer αβα sandwich [CATH classification 3.30.1330 (29, 30)] with a five-stranded mixed β-sheet at the core (Fig. 1). Strand β5, defined by hydrogen bonds typical of parallel β-strands involving the carbonyl oxygen atom of Pro-153 and the backbone nitrogen atom of Ile-155, comprises only three residues. Helices α2 and α3 pack on one side of the β-sheet with the helix axes running roughly parallel to the β-strands, whereas helices α1, α4, and α5 pack on the opposite side of the β-sheet with helix axes running transverse to the β-strands. Short 310 helices are located within the α2–β2 and β2–α3 connecting loops. Residues that are highly conserved among Gadd45 isoforms are concentrated on one side of the monomer in helices α2 and α3 and the residues in the β-sheet with which these helices interact (Fig. 1).
Fig. 1.
Monomeric structure and sequence conservation of Gadd45 isoforms. (A) Ribbon representation of the Gadd45γ monomer. Secondary structural elements are marked, and residues that are identical among all Gadd45 isoforms are colored yellow. This figure was created by using PyMOL (53). (B) Amino acid sequence alignment of Gadd45 family members. Identical residues are boxed in gray. Secondary structural elements observed in Gadd45γ crystals are marked above the sequences. This figure was created by using ESPript (54, 55).
Identification of Dimerization Interface.
Design of point mutants.
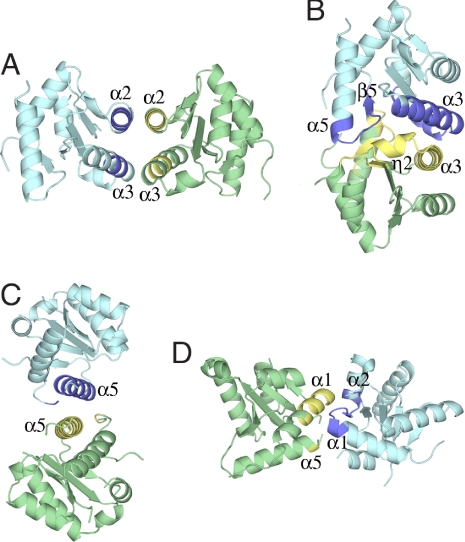
The asymmetric unit of the crystals obtained contains two monomers of Gadd45γ. Inspection of the crystal packing revealed four possible interactions of monomers that could generate the expected dimer. Notably, none of the four possible dimer interfaces involves both of the two putative oligomerization regions previously identified by deletion analysis (28). Although the crystal packing revealed four possible dimer interactions (Fig. 2) we focused on the two interactions with the largest interfacial areas as the most probable dimerization modes. The interfacial contact areas of interactions 1 (Fig. 2A) and 2 (Fig. 2B) are ≈465 Å2 and ≈750 Å2, respectively. Although the larger interfacial contact area implies that interaction 2 is the more probable interaction, both interactions 1 and 2 involve highly conserved residues and contain one of the putative oligomerization regions (28), so both putative dimer models were considered further.
Fig. 2.
Ribbon representation of four possible dimer interfaces. In each case interacting regions marked in dark blue for one monomer interact with regions colored yellow in the monomer colored green. Secondary structural elements are marked as in Fig. 1A.
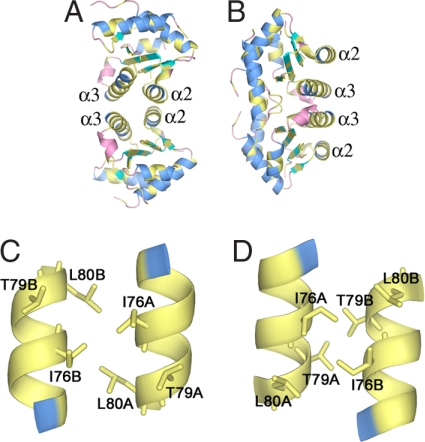
Both interactions 1 and 2 involve helix α3 (Fig. 3). Interaction 1 also involves helix α2, forming an interface comprising a four-helix bundle. The two monomers of the dimer in interaction 1 are related by a twofold symmetry axis that is perpendicular to the helix axes, generating antiparallel α2–α2 and α3–α3 contacts between monomers. In contrast, interaction 2 involves, in addition to helix α3, the residues following helix α5 including those of strand β5. Fig. 3 C and D compares the α3–α3 interactions for both interactions 1 and 2. To distinguish which of these possibilities is correct, we looked for point mutations that would be expected to prevent one mode of oligomerization without influencing the other. Virtual alanine scanning analysis (31, 32) indicates that Leu-80 is one of the most important residues only in interaction 1. In contrast, the juxtaposition of Thr79A and Thr79B in interaction 2 indicates that replacement of Thr-79 with a larger residue should block only interaction 2. Accordingly, we chose to mutate Leu-80 and Thr-79 to glutamate. This should result in one or the other of these two modes of dimerization being impaired sterically by the longer side chain and/or electrostatically by the introduction of neighboring negative charges. The helical conformation of α3 is expected to be preserved upon mutation because of the high helix potential of glutamate.
Fig. 3.
Ribbon representations of interactions 1 and 2. The color coding is the same as in Fig. 1A. (A) Dimer generated by interaction 1. (B) Dimer generated by interaction 2. (C) Close-up view of the interactions of helices α3 in dimer 1. (D) Close-up view of the interactions of helices α3 in dimer 2.
Because the α2–α2 interaction is involved only in interaction 1, mutations in α2 that might disrupt dimerization were also considered. Analysis of the interaction shows, however, that most of the interactions of these helices involve backbone and Cβ interactions, perhaps rendering interaction 1 robust to modifications of side chains in helix α2. Nevertheless, because one of the few side chain interactions present in this region is a hydrogen bond between Lys-48 and Glu-45, we mutated Lys-48 to glutamate to examine the effects of reversing the charge at this position. Because both interactions 1 and 2 are twofold symmetric, each of the three point mutations discussed above introduces two modifications to the potential dimer interface.
Biophysical characterization of point mutants.
Wild-type protein and the three point mutants were expressed and purified in parallel. The presence of the mutations was confirmed both by sequencing the clones and by electrospray mass spectrometry of the purified protein (Table S2). The CD spectra of wild-type and mutant Gadd45γ were indistinguishable (Fig. S1), and the melting points of the four forms as monitored by molar ellipticity at 222 nm were also essentially identical, ranging from 44°C to 46°C (Fig. S1b), confirming that the structural integrity of the mutants was not compromised.
The oligomerization state of each mutant form was assessed by gel filtration and by dynamic light scattering (DLS) (Table 1). As expected, wild-type protein was clearly dimeric under all conditions tested, with an apparent molecular weight of 33,000–35,000. The L80E mutation, which we predicted to disrupt interaction 3, completely abolished the dimerization of Gadd45γ, as indicated by the apparent molecular weight of 15,000–20,000. In fact, the monomeric state of the L80E mutant was found to be independent of the protein concentration in the range used in these experiments (≈0.5–8 mg/ml). These L80E results strongly support interaction 1 as the mode of dimerization for Gadd45γ in solution. However, the K48E mutation, which we also suggested as having the potential to disrupt interaction 1, did not affect the dimerization state of the protein. This result is explained by our observation that most of the interactions between the two α2 helices in interaction 3 involve backbone and Cβ interactions, indicates that relatively little binding energy is provided by the hydrogen bond between the Lys-48 and Glu-45 side chains, and remains consistent with our conclusion based on the L80E mutant that interaction 1 is the dimerization mode. In view of these results, that the molecular weight of the T79E mutant would appear to be concentration-dependent, was unexpected. As determined by DLS, at a concentration of 0.5 mg/ml the estimated molecular weight was only slightly larger than that which would be expected for a monomer, whereas at a concentration of 7 mg/ml the estimated molecular weight suggested dimerization. Furthermore, the molecular weight of T79E estimated by size exclusion was reproducibly smaller than that of the wild-type and K48E proteins but was not consistent with the T79E mutant being monomeric. These results suggest the presence of a dynamic equilibrium between monomeric and dimeric states for this mutant. The concentration-dependent molecular weight of the T79E mutation may indicate that this mutation causes a small perturbation in helix α3, which, because of the importance of helix α3 in interaction 1, affects dimerization. Together, these results strongly support interaction 1 as the mode of dimerization of Gadd45γ in solution. This dimerization interface is formed by a four-helix bundle with predominantly hydrophobic interactions. Residues Ile-76–Leu-80 of helix α3 and residues Tyr-44–Lys-48 of helix α2 contribute the vast majority of the interactions. Importantly, this dimerization interface is formed by residues that are highly conserved among Gadd45 isoforms (Fig. 1B).
Table 1.
Molecular weight estimates of wild-type and mutant Gadd45γ
| Construct | Calculated molecular weight | Estimated molecular weight |
||
|---|---|---|---|---|
| Size-exclusion chromatography | DLS high concentration | DLS low concentration | ||
| WT | 15,788.6 | 32,960 | 35,000 | 34,000 |
| L80E | 15,804.6 | 20,130 | 19,000 | 15,200 |
| T79E | 15,816.6 | 26,959 | 33,250 | 20,200 |
| K48E | 15,789.6 | 34,503 | 34,300 | 29,300 |
Functional Consequences of Dimerization.
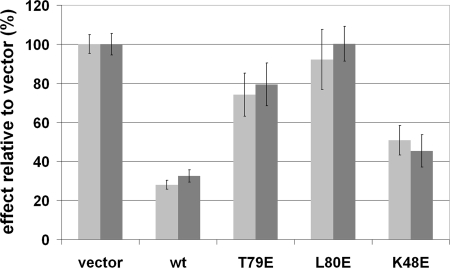
The functional consequences of the mutations were assessed by monitoring effects on cell growth in HepG2 cells, a cell line that is known to have epigenetically down-regulated Gadd45γ (7). We evaluated the effect of expressing wild-type or mutant forms of Gadd45γ on cell growth by quantifying viable cells using a metabolic dye and by manually counting cell colonies after transfection with appropriate expression vectors (see Materials and Methods). Fig. 4 shows that these two cell growth measurements yielded similar results. As expected, expression of wild-type Gadd45γ reduced cell growth by ≈70%. The K48E mutant also reduced cell growth significantly (by 50–60%). The T79E mutant weakly inhibited cell growth (≈20%), whereas the L80E mutant had no detectable effect on cell growth. These effects of wild-type and mutant forms of Gadd45γ on cell growth parallel their abilities to dimerize, indicating that dimerization is essential for growth inhibition. Similar results were obtained when evaluating the activity of these mutants in HeLa cells (data not shown).
Fig. 4.
Growth inhibition of HepG2 cells by wild-type and mutant Gadd45γ. Growth inhibition was assayed by using alamarBlue staining and by manual counts of colonies after crystal violet staining. Results shown are an average of three to four experiments with associated standard errors of the mean.
Discussion
Homologous Structures.
The Gadd45 proteins have been classified based on sequence similarity with the ribosomal proteins of the L7Ae/L30e/S12e family (Pfam entry PF01248) (33, 34). The ModBase database of homology models (35) has five models of Gadd45γ derived from three different ribosomal and spliceosomal protein templates that share ≈17–20% amino acid sequence identity to the Gadd45 proteins. The rmsds in backbone atom positions between the crystal structure reported here and these three templates range from 1.43 to 1.6 Å for 300–352 atoms. A homology model of Gadd45β based on one of these templates was recently reported (36). The main differences between the homology-modeled secondary structural elements of Gadd45β and those observed in the crystal structure of Gadd45γ involve the two 310 helices observed in the crystal structure (residues 54–57 and 66–70) that were not predicted by homology modeling. No regular secondary structural elements were modeled in these loops for Gadd45β. Another difference is the presence of the short-strand β5 in the crystal structure, which is not predicted in the homology models.
Homodimerization and Heterodimerization.
Based primarily on results from deletion mutants, residues in the regions from 33 to 61 and from 133 to 165 were identified as the oligomerization interface of Gadd45α (28). However, none of the four possible dimer interfaces observed in our Gadd45γ crystals involves this combination of residues at the interface. The L80E point mutation made in our study did not compromise the integrity of the monomer as measured by CD, but dimerization was blocked, clearly identifying the four-helix bundle involving helices α2 and α3 as the dimerization interface. Although it is possible that Gadd45α and Gadd45γ dimerize differently, the sequence similarity among Gadd45 family members suggests that this is unlikely. Hetero-oligomerization of Gadd45 isoforms has been demonstrated by coimmunoprecipitation (28). This is consistent with the fact that the four-helix bundle involves the most highly conserved regions in the Gadd45 family sequences (Fig. 1B). Dimerization through these highly conserved residues leaves the less conserved regions and loops exposed to effect isoform-specific interactions and functions.
Oligomerization has been proposed to decrease the ability of Gadd45 to protect nucleosomes from nuclease digestion (28). In contrast, our results clearly indicate that both growth inhibition and dimerization depend on residue Leu-80, implying that this Gadd45 function requires dimerization. The reduced growth-inhibitory activity of the T79E mutant may also be due to the fact that its dimerization is compromised, although not eliminated. Alternatively, Thr-79 may be required for interaction of Gadd45γ with other proteins involved in cell cycle control such that the compromised growth-inhibitory activity of the T79E mutant may be derived from compromised Gadd45γ interaction(s) rather than effects on dimerization. In this regard, it is interesting to note that the equivalent residue is implicated in interactions of Gadd45β with MKK7 (36).
Interactions with Other Proteins.
The many cellular functions that have been attributed to Gadd45 proteins are thought to result from their ability to interact with a wide variety of other proteins. Deletion mutants have been used previously in studies designed to identify regions of Gadd45 family proteins that are necessary for interactions with other proteins (14, 19, 28, 37–40). Although some consistencies emerge from these different investigations, some contradictions exist regarding the regions of Gadd45 that are proposed to be required for PCNA binding (19, 37, 38). The interactions of Gadd45 members with Cdc2 (14), p21 (40), p38 (41), and histones (42) all have been mapped to the central region of Gadd45. This region includes the dimerization interface that we identified. Because we have demonstrated here that residues that play a critical role in dimerization are also essential for growth inhibition, it is possible that the studies mapping binding regions of Gadd45 isoforms for Cdc2, p21, and p38 have simply identified interactions for which dimerization is necessary.
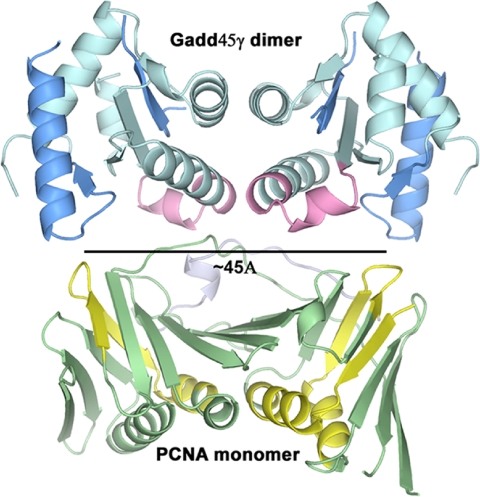
The interaction with PCNA is one of the most documented interactions of Gadd45 proteins. Several studies have mapped the interacting regions of both proteins using deletion mutants and peptide binding (18, 19, 37, 38) and have suggested that the stoichiometry of the interaction is 2 Gadd45:1 PCNA. Mapping of the proposed interacting regions onto the three-dimensional structure of PCNA and our structure of Gadd45γ is suggestive. In Fig. 5, the regions of Gadd45β that reportedly interact with PCNA (38) are mapped onto a Gadd45γ dimer, and the regions of PCNA that reportedly interact with Gadd45 (19) are highlighted. A distance of ≈45 Å separates both the PCNA-binding regions of Gadd45 and the Gadd45-binding regions of PCNA. The PCNA monomer displays pseudotwofold symmetry that relates two β-α-β-β motifs (43). Juxtaposed as in Fig. 5, the twofold rotation axis that relates the two monomers of the Gadd45γ dimer is parallel to the pseudotwofold axis of the PCNA monomer. This arrangement demonstrates how one dimer of Gadd45 may bind to one monomer of PCNA and is entirely consistent with the reported 2:1 stoichiometry (19). None of the other potential dimeric interactions observed in our Gadd45γ crystals could produce a reasonable match to the proposed docking regions, again supporting interaction 1 as the mode of dimerization of Gadd45γ in solution. An arrangement similar to that depicted in Fig. 5 also is consistent with observations indicating that Gadd45 binding to PCNA is competitive with that of p21WAF1/CIF (18) and with the mapping of the Gadd45α–p21 interaction to the central portion of Gadd45 (40).
Fig. 5.
Potential interaction of Gadd45γ with PCNA. Portions of PCNA proposed to be involved in Gadd45 interactions are marked in yellow. Proposed PCNA-binding regions of Gadd45 isoforms are mapped in dark blue onto a Gadd45γ dimer. The potential binding surfaces on PCNA and on the Gadd45γ dimer are spaced ≈45 Å apart. The regions of Gadd45 reported to bind p21 are shown in pink, and p21WAF1/CIF1 is shown as a light blue ribbon. PCNA-p21WAF1/CIF1 coordinates are PDB entry 1AXC (2, 43).
Summary.
This work reveals that the dimerization interface of Gadd45γ involves helices α2 and α3, the most highly conserved regions of the molecule. We demonstrate that the L80E point mutation is sufficient to block dimerization and growth inhibition without compromising the three-dimensional fold of the monomer. Mutants of this type will serve as useful tools to define which of the many interactions with other proteins, and which of the cellular roles of Gadd45 isoforms, depend on dimerization. This structure provides a new context for dissecting the structure–activity relationships of Gadd45 proteins.
Materials and Methods
Cloning.
Mouse Gadd45γ cDNA was obtained from American Type Culture Collection (MGC-5695, IMAGE Clone ID 3493618). A construct containing amino acid residues 16–159 was subcloned into a pGEX vector derivative modified to contain a TEV cleavage site between the GST and the coding sequence for Gadd45γ. BL21(DE3) and B834 strains of Escherichia coli were transformed with the resulting plasmid. Point mutations were made by using the QuikChange kit from Stratagene according to the manufacturer's instructions. Mutations were confirmed by sequencing the resulting clones and by mass spectrometry of the purified protein.
Expression and Purification.
The transformed BL21(DE3) cells were cultured by using Terrific Broth, and B834 cells were grown in LeMaster medium supplemented with 100 μg/ml ampicillin. B834 cultures were also supplemented with 250 μM l-selenomethionine. At an A600 of 0.6–0.8 the temperature was reduced to 15°C and recombinant protein expression was induced by addition of IPTG to a final concentration of 1 mM. Cells were harvested 16–20 h after induction by centrifugation, and the cell pellets were lysed by sonication in PBS. The lysate was cleared by centrifugation. Recombinant protein was purified by affinity, ion-exchange, and size-exclusion chromatographies. Details can be found in SI Text.
Crystallization.
Sparse matrix screening and optimization were done by using hanging drop vapor diffusion. The final conditions consisted of 1.9 M NaH2PO4 and 100 mM Hepes (pH 7.5). The crystals are orthorhombic and belong to space group C2221 with unit cell dimensions a = 43.0 Å, b = 122.5 Å, and c = 105.4 Å. There are two molecules in the asymmetric unit.
Data Collection and Structure Solution.
Crystals were transferred in two steps into 15% xylitol in 1.9 M NaH2PO4, and the crystals were flash-frozen in a nitrogen gas stream at −180°C. Diffraction data from native crystals were collected on a Q315 detector (ADSC) at beamline X25 at National Synchrotron Light Source, Brookhaven National Laboratory. Data were processed by using either HKL2000 (44) or D*trek (45). Phases were calculated by using single-wavelength anomalous dispersion (SAD) methods from data collected at the peak of the Se absorption at beamline X8C on a Quantum 4 detector (ADSC). Heavy atom sites were located by using BnP (46), and solvent flattening was done with RESOLVE (47). Most of the model was built by using ARP/wARP (48), and refinement was done with Refmac5 (49, 50). Final adjustments to the model were made by using O (51). The model has been refined to 1.7-Å resolution with Rwork 22.4% and Rfree 27.2%. No interpretable density is observed for residues 106–115 or 127–130 of chain A or for residues 107–115 or 126–130 of chain B. The quality of the model was evaluated by using PROCHECK (52).
Size-Exclusion Chromatography.
Analytical size-exclusion chromatography analysis of wild-type and mutant Gadd45γ was done on a Superdex 75 10/300 column (GE Healthcare). The column was calibrated by using the low-molecular-weight kit from GE Healthcare. For one calibration aprotinin (Roche Diagnostics) was also included.
DLS.
DLS analysis was done on a DynaPro Plate reader (Wyatt Technology). The protein concentrations ranged from 0.5 to 10 mg/ml, and analysis was done by using Dynamics 6.7 software provided by the manufacturer.
Circular Dichroism.
Circular dichroism was measured on a Jasco J-815-150S spectrometer at 23°C. The buffer was exchanged by size-exclusion chromatography to 20 mM Na,K PO4 (pH 7.6), 200 mM NaF, and 1 mM DTT. Aliquots of the peak fractions were diluted ≈100-fold to 0.01 mg/ml in 10 mM Na,K PO4 (pH 7.6), 100 mM NaF, and 0.5 mM DTT immediately before CD measurements. Two scans over the wavelength range of 350–190 nm were averaged for each sample, and the contribution of buffer was subtracted. The melting point of each protein was measured by monitoring the ellipticity at 222 nm. The temperature was increased at a rate of 2°C per minute.
Electrospray Mass Spectrometry.
Molecular masses of wild-type and mutant Gadd45γ were measured on an Agilent Technologies 1100 series LC/MS. Five-microliter samples at 0.1 mg/ml were injected directly into the spectrometer at a flow rate of 0.3 ml/min in 20% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid. The scan covered a m/z range of 600–1,900.
Growth Inhibition Assay.
Full-length mouse Gadd45γ wild type and point mutants were subcloned into the BglII/EcoRI sites of the bicistronic mammalian expression vector pIRES2-DSred-Express (BD Biosciences). HepG2 cells were transfected by using Lipofectamine (Gibco BRL). Transfection efficiency was monitored by DSRed expression, and stably transfected HepG2 cells were selected by using 1 mg/ml G418. Stably transfected HepG2 cells were grown in six-well plates in 2.5 ml of DMEM-10 with changes of medium twice per week. After 3 weeks viable cells were quantitated after addition of 250 μl of the metabolic dye alamarBlue (Biosource) in a CytoFluor multiwell plate reader (Applied Biosystems). Cells were washed twice with ice-cold PBS, fixed for 10 min in ice-cold methanol, and stained for 15 min with 0.5% crystal violet.
Supplementary Material
Acknowledgments.
We thank Christine Munger for performing the mass spectrometry analysis and Dr. Edmund Ziomek (Biotechnology Research Institute, National Research Council Canada) for assistance with the CD spectroscopy. We also gratefully acknowledge advice from Yunge Li in crystallization, advice from Dr. Ante Tocilj regarding crystallographic software, and the critical evaluation of the manuscript by Dr. Mirek Cygler. We also thank Leon Flaks and Michael Becker (National Synchrotron Light Source, Brookhaven National Laboratory) for assistance at beamlines X8C and X25, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3CG6).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800086105/DCSupplemental.
References
- 1.Zhan Q, et al. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang W, et al. CR6: A third member in the MyD118 and Gadd45 gene family which functions in negative growth control. Oncogene. 1999;18:4899–4907. doi: 10.1038/sj.onc.1202885. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, et al. Gadd45 beta and Gadd45 gamma are critical for regulating autoimmunity. J Exp Med. 2005;202:1341–1347. doi: 10.1084/jem.20051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mak SK, Kultz D. Gadd45 proteins induce G2/M arrest and modulate apoptosis in kidney cells exposed to hyperosmotic stress. J Biol Chem. 2004;279:39075–39084. doi: 10.1074/jbc.M406643200. [DOI] [PubMed] [Google Scholar]
- 5.Bahar A, Bicknell JE, Simpson DJ, Clayton RN, Farrell WE. Loss of expression of the growth inhibitory gene GADD45gamma, in human pituitary adenomas, is associated with CpG island methylation. Oncogene. 2004;23:936–944. doi: 10.1038/sj.onc.1207193. [DOI] [PubMed] [Google Scholar]
- 6.Qiu W, et al. Hypermethylation of growth arrest DNA damage-inducible gene 45 beta promoter in human hepatocellular carcinoma. Am J Pathol. 2004;165:1689–1699. doi: 10.1016/s0002-9440(10)63425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ying J, et al. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res. 2005;11:6442–6449. doi: 10.1158/1078-0432.CCR-05-0267. [DOI] [PubMed] [Google Scholar]
- 8.Zerbini LF, et al. NF-kappa B-mediated repression of growth arrest- and DNA-damage-inducible proteins 45alpha and gamma is essential for cancer cell survival. Proc Natl Acad Sci USA. 2004;101:13618–13623. doi: 10.1073/pnas.0402069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerbini LF, Libermann TA. GADD45 deregulation in cancer: Frequently methylated tumor suppressors and potential therapeutic targets. Clin Cancer Res. 2005;11:6409–6413. doi: 10.1158/1078-0432.CCR-05-1475. [DOI] [PubMed] [Google Scholar]
- 10.Jin S, et al. The GADD45 inhibition of Cdc2 kinase correlates with GADD45-mediated growth suppression. J Biol Chem. 2000;275:16602–16608. doi: 10.1074/jbc.M000284200. [DOI] [PubMed] [Google Scholar]
- 11.Jin S, et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene. 2002;21:8696–8704. doi: 10.1038/sj.onc.1206034. [DOI] [PubMed] [Google Scholar]
- 12.Vairapandi M, Balliet AG, Hoffman B, Liebermann DA. GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J Cell Physiol. 2002;192:327–338. doi: 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- 13.Wang XW, et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan Q, et al. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- 15.Sakaue M, Adachi H, Jetten AM. Post-transcriptional regulation of MyD118 and GADD45 in human lung carcinoma cells during 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid-induced apoptosis. Mol Pharmacol. 1999;55:668–676. [PubMed] [Google Scholar]
- 16.Carrier F, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 18.Chen IT, Smith ML, O'Connor PM, Fornace AJ., Jr Direct interaction of Gadd45 with PCNA and evidence for competitive interaction of Gadd45 and p21Waf1/Cip1 with PCNA. Oncogene. 1995;11:1931–1937. [PubMed] [Google Scholar]
- 19.Hall PA, et al. Characterisation of the interaction between PCNA and Gadd45. Oncogene. 1995;10:2427–2433. [PubMed] [Google Scholar]
- 20.Paunesku T, et al. Proliferating cell nuclear antigen (PCNA): Ringmaster of the genome. Int J Radiat Biol. 2001;77:1007–1021. doi: 10.1080/09553000110069335. [DOI] [PubMed] [Google Scholar]
- 21.Smith ML, et al. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 22.Mita H, Tsutsui J, Takekawa M, Witten EA, Saito H. Regulation of MTK1/MEKK4 kinase activity by its N-terminal autoinhibitory domain and GADD45 binding. Mol Cell Biol. 2002;22:4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyake Z, Takekawa M, Ge Q, Saito H. Activation of MTK1/MEKK4 by GADD45 through induced N-C dissociation and dimerization-mediated trans autophosphorylation of the MTK1 kinase domain. Mol Cell Biol. 2007;27:2765–2776. doi: 10.1128/MCB.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 25.Ungefroren H, Groth S, Ruhnke M, Kalthoff H, Fandrich F. Transforming growth factor-beta (TGF-beta) type I receptor/ALK5-dependent activation of the GADD45beta gene mediates the induction of biglycan expression by TGF-beta. J Biol Chem. 2005;280:2644–2652. doi: 10.1074/jbc.M411925200. [DOI] [PubMed] [Google Scholar]
- 26.Thyss R, et al. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 2005;24:128–137. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvador JM, Mittelstadt PR, Belova GI, Fornace AJ, Jr, Ashwell JD. The autoimmune suppressor Gadd45alpha inhibits the T cell alternative p38 activation pathway. Nat Immunol. 2005;6:396–402. doi: 10.1038/ni1176. [DOI] [PubMed] [Google Scholar]
- 28.Kovalsky O, Lung FD, Roller PP, Fornace AJ., Jr Oligomerization of human Gadd45a protein. J Biol Chem. 2001;276:39330–39339. doi: 10.1074/jbc.M105115200. [DOI] [PubMed] [Google Scholar]
- 29.Orengo CA, et al. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 30.Pearl FM, et al. The CATH database: An extended protein family resource for structural and functional genomics. Nucleic Acids Res. 2003;31:452–455. doi: 10.1093/nar/gkg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004:l2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 33.Finn RD, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieper U, et al. MODBASE: A database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papa S, et al. Insights into the structural basis of the GADD45beta-mediated inactivation of the JNK kinase, MKK7/JNKK2. J Biol Chem. 2007;282:19029–19041. doi: 10.1074/jbc.M703112200. [DOI] [PubMed] [Google Scholar]
- 37.Azam N, Vairapandi M, Zhang W, Hoffman B, Liebermann DA. Interaction of CR6 (GADD45gamma) with proliferating cell nuclear antigen impedes negative growth control. J Biol Chem. 2001;276:2766–2774. doi: 10.1074/jbc.M005626200. [DOI] [PubMed] [Google Scholar]
- 38.Vairapandi M, Azam N, Balliet AG, Hoffman B, Liebermann DA. Characterization of MyD118, Gadd45, and proliferating cell nuclear antigen (PCNA) interacting domains. PCNA impedes MyD118 AND Gadd45-mediated negative growth control. J Biol Chem. 2000;275:16810–16819. doi: 10.1074/jbc.275.22.16810. [DOI] [PubMed] [Google Scholar]
- 39.Yang Q, et al. Identification of a functional domain in a GADD45-mediated G2/M checkpoint. J Biol Chem. 2000;275:36892–36898. doi: 10.1074/jbc.M005319200. [DOI] [PubMed] [Google Scholar]
- 40.Zhao H, et al. The central region of Gadd45 is required for its interaction with p21/WAF1. Exp Cell Res. 2000;258:92–100. doi: 10.1006/excr.2000.4906. [DOI] [PubMed] [Google Scholar]
- 41.Bulavin DV, Kovalsky O, Hollander MC, Fornace AJ., Jr Loss of oncogenic H-ras-induced cell cycle arrest and p38 mitogen-activated protein kinase activation by disruption of Gadd45a. Mol Cell Biol. 2003;23:3859–3871. doi: 10.1128/MCB.23.11.3859-3871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hildesheim J, Fornace AJ., Jr Gadd45a: An elusive yet attractive candidate gene in pancreatic cancer. Clin Cancer Res. 2002;8:2475–2479. [PubMed] [Google Scholar]
- 43.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. In: Macromolecular Crystallography, Part A. Carter CW Jr, Sweet RM, editors. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 45.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 46.Weeks CM, et al. Towards automated protein structure determination: BnP, the SnB-PHASES interface. Z Kristallogr. 2002;217:686–693. [Google Scholar]
- 47.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 48.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 51.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 53.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 54.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 55.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.