Abstract
Polycyclic terpenoid lipids such as hopanes and steranes have been widely used to understand ancient biology, Earth history, and the oxygenation of the ocean–atmosphere system. Some of these lipids are believed to be produced only by aerobic organisms, whereas others actually require molecular oxygen for their biosynthesis. A persistent question remains: Did some polycyclic lipids initially evolve in response to certain environmental or metabolic stresses, including the presence of oxygen? Here, we identify tetracyclic isoprenoids in spores of the bacterium Bacillus subtilis. We call them sporulenes. They are produced by cyclization of regular polyprenes, a reaction that is more favorable chemically than the formation of terpenoids such as hopanoids and steroids from squalene. The simplicity of the reaction suggests that the B. subtilis cyclase may be analogous to evolutionarily ancient cyclases. We show that these molecules increase the resistance of spores to a reactive oxygen species, demonstrating a specific physiological role for a nonpigment bacterial lipid biomarker. Geostable derivatives of these compounds in sediments could thus be used as direct indicators of oxidative stress and aerobic environments.
Keywords: biomarker, spores, squalene-hopene cyclase, lipid, peroxide resistance
Polycyclic terpenoids are a class of organic molecules that includes cholesterol and numerous other related lipids. Their molecular fossils, which are ubiquitous in the sedimentary rock record, are used as records of ancient bacterial and eukaryotic life. For example, the timing of the onset of oxygenic photosynthesis has been reported to be as early as 2.7 billion years ago (1) based on the detection of polycyclic terpenoids presumably derived from Cyanobacteria (2). However, recent studies have questioned this assumption and have urged caution about making taxonomic associations of specific lipid biomarkers with specific microbial groups (3, 4).
Here, we take a different approach to this debate by showing definitively that some polycyclic terpenoids reflect a specific physiological function. The association of molecular fossils with physiological functions reduces the dependence of molecular fossils on specific organisms and taxonomic groups. Significantly, we demonstrate in vivo the protective role of a class of polycyclic terpenoids against a reactive oxygen species. Geostable derivatives of these compounds, called sporulenes, could become more definitive geologic markers for aerobic bacteria and environments. Future studies of the specific biological roles of other polycyclic terpenoids may greatly inform the interpretations of the past, including the oxygenation of the Earth.
Biosynthesis of Polycyclic Terpenoids by Bacillus subtilis
The best-studied polycyclic terpenoids, tetracyclic sterols and pentacyclic hopanoids, are produced enzymatically from acyclic 30-carbon (C30) precursors by a family of squalene and oxidosqualene cyclases (5, 6). A gene (sqhC) encoding a putative squalene cyclase enzyme (SqhC) is present in a two-gene operon in B. subtilis (Fig. 1A). Maximum-likelihood analysis of this SqhC relative to other bacterial true squalene-hopene cyclases (SHCs) shows that SqhCs of all bacilli form a coherent outgroup between sterol oxidosqualene cyclases and SHCs [supporting information (SI) Fig. S1]. At the amino acid level, SqhC is most similar (>90%) to proteins in six other Bacillus species, none of which is known to produce hopanoids. However, B. subtilis SqhC contains functional motifs responsible for some critical steps of cyclase activity: a domain rich in aspartic acid initiates the protonation of a polyprene precursor, and aromatic residues are present that would stabilize the first two cyclohexyl rings (5) (SI Text). In contrast to most known squalene-hopene cyclases, however, SqhC does not contain residue F601 that would enable formation of the third cyclohexyl ring from a squalene precursor (7, 8). Thus, B. subtilis would be expected to produce cyclic polyprenoids, but not hopanoids.
Fig. 1.
Expression of sqhC and the intracellular localization of SqhC. (A) Organization of the sqhC operon. Hairpin symbols represent transcriptional terminators. (B) Accumulation of colorimetric marker-enzyme β-galactosidase expressed from PsqhC-lacZ in a wild-type strain (amyE::PsqhC-lacZ, TB12; open diamond) and in a strain lacking the sporulation-dependent RNA polymerase subunit σE but harboring PsqhC-lacZ (spoIIGA::tet, amyE::PsqhC-lacZ, TB19; filled square). Samples were collected at the indicated times after the beginning of sporulation (hour 0). Error bars correspond to 1σ error on the mean value from triplicate samples. (C) Fluorescence micrograph of representative cells producing green fluorescent protein (GFP) fused in-frame to the C terminus of SqhC (amyE::PHyspank-shqC-gfp, TB29) 4 h after the beginning of the sporulation. The expression of the fusion was driven by 1 mM IPTG 2.5 h after the induction of sporulation. (D) The same field of view as in C in transmitted light.
Indeed, previous studies failed to detect hopanoids in vegetative B. subtilis (9), prompting us to examine the expression of sqhC in more detail and search for the lipid products of SqhC. A recent study detected a sporulation-specific increase in the transcription levels of sqhC (10). We measured the activity of the promoter of sqhC by a colorimetric assay (PsqhC-lacZ fusion) (Fig. 1B). This assay confirmed that the expression of sqhC depended on the sporulation-specific RNA polymerase subunit σE that controls the expression of a number of genes during sporulation (Fig. 1B). The intracellular localization of SqhC was also sporulation-specific: in fluorescence micrographs, SqhC fused to green fluorescent protein (GFP) surrounded the developing spore (Figs. 1 C–D and Fig. S2), suggesting that the lipid products of SqhC might be found in forespores and/or in spores.
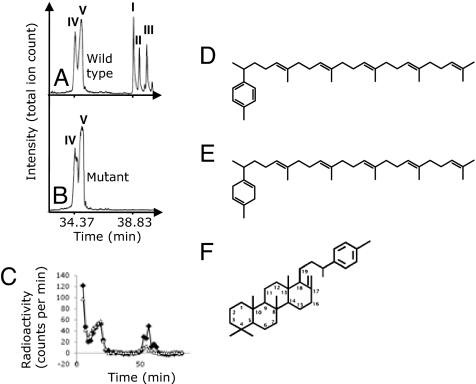
We searched for the natural products of SqhC by comparing the lipids extracted from wild-type spores (strain PY79) to those extracted from spores of a mutant strain that lacked sqhC (strain TB10). Wild-type spores contained at least 0.1 mg of three polycycloprenyls of molecular formula C35H54 per 1 g of dry spores (Fig. 2A), a level typical for other organisms that produce polycyclic terpenoids (9). These compounds were not detectable in the spores of mutant strain TB10 (Fig. 2B).
Fig. 2.
SqhC produces polycyclic terpenoids. (A) Total ion chromatogram of the lipid extract of wild-type spores (PY79). Three conspicuous peaks (I, II, and III) were never present in the extracts of the strain lacking SqhC (ΔsqhC ΔsodF::tet, TB10). (B) Acyclic tetraprenyl curcumenes (IV and V) were abundant in the spores of both strains. (C) The radioactivity of eighty lipid fractions extracted from the cell lysates of the strain overexpressing SqhC (ΔsqhC ΔsodF::tet amyE::PHyperspank-sqhC, TB28; filled diamond) and the strain mutant for sqhC (TB10; open triangle) after incubation with 3H-FPP. Fractions derived from TB28 that contained I–III were three times as radioactive as the corresponding fractions derived from TB10 (*). These fractions from TB10 still contained a nonnegligible amount of radioactivity that is probably due to the presence of unidentified polyprenoid lipids other than I–III. (D) Structure of acyclic C-35 polyprenoid tetraprenyl-ar-curcumene (IV) detected in the spores of B. subtilis both in the presence and the absence of the putative cyclase. (E) Structure of acyclic C-35 polyprenoid tetraprenyl-β-curcumene (IV) detected in the spores of B. subtilis in the presence and the absence of the putative cyclase. (F) Proposed structure of isomer (II) of tetracyclic C-35 polyprenoids. Isomers I–III are detected only in B. subtilis spores that contained the putative squalene hopene cyclase.
To confirm that these compounds are synthesized de novo, we incubated 3H-farnesyl pyrophosphate, a possible precursor to longer acyclic isoprenyls (11) that can be precursors to polycycloprenyls, with cell lysates of a B. subtilis strain overexpressing SqhC (TB28) and with mutant strain TB10 (Fig. 2C). B. subtilis contains a heptaprenyl pyrophosphate synthetase that can use FPP as a precursor (11). The lipid fractions that contained the polycycloprenyls were significantly more radioactive than the corresponding fractions from the mutant strain, in which these compounds were undetectable (Fig. 2C). We infer that SqhC is responsible for the formation of these polycycloprenyls.
Chemical and mass-spectral properties of these molecules are most consistent with the formation from head-to-tail linked (regular) polyprenoids, and not from squalene (which contains a tail-to-tail linkage) (12, 13) (Fig. S3 and SI Text). In addition, incubation of the potential substrate, squalene, with cell lysates of Escherichia coli overexpressing SqhC (TB34) yielded no detectable products, suggesting that the enzyme is not active toward this tail-tail polyprene (see SI Text). Our interpretation of a regular polyprene precursor is further supported by concurrent detection of the recently identified regular polyprenoids tetraprenyl-ar-curcumene (IV) and tetraprenyl-β-curcumene (V) (12) in both strains (TB10 and PY79; Fig. 2 and Fig. S3). The three most abundant products of SqhC (I–III) all have identical molecular ions (m/z 474). The mass spectral signatures of I–III and their fully hydrogenated forms are consistent with the presence of five total rings and four total double bonds (Fig. 2F and Fig. S3). All three compounds contain four cyclohexyl rings in the backbone, whereas the fifth one is a methylphenyl group terminating the side chain (Fig. 2F). The isomers I–III appear to be distinguished by the position of the double bond in the fourth ring; the proposed structure of isomer II is shown (Fig. 2F). From here onward we will call these compounds sporulenes (Fig. S4).
Protective Role of Sporulenes
Clues to the physiology of these unusual molecules came from sodF, the second gene in the two-gene operon that contains sqhC (Fig. 1A). sodF encodes a putative superoxide dismutase, an enzyme that disproportionates toxic superoxide into oxygen and hydrogen peroxide (H2O2) (14). Because pentacyclic hopanoids can regulate membrane permeability and fluidity in vitro (15), we hypothesized that the lipid products of SqhC could limit the permeability of forespores or spores to reactive oxygen species.
The absence of SqhC previously was reported not to affect sporulation under standard conditions (10). To test this and detect very small sporulation defects, we grew and sporulated mixed cultures of the colorimetrically abeled wild-type strain (TB38, lacZ) and the mutant strain lacking sqhC (TB10) for eight consecutive cycles. After each cycle, we determined the ratio of heat-resistant spores of the two strains and used purified spores to inoculate the next cycle. The initial ratio (10% TB38, 90% TB10) did not change (Fig. 3), confirming that lack of SqhC does not affect its sporulation under standard experimental conditions and background levels of reactive oxygen species.
Fig. 3.
Spores containing sqhC are more resistant to hydrogen peroxide. Wild-type colorimetrically marked cells (amyE::PSpac-lacZ) (TB38) and cells lacking sqhC (ΔsqhC ΔsodF::tet) (TB10) were mixed in 10:90 initial ratio in liquid sporulation medium. Purified spores were treated with heat and hydrogen peroxide. A small aliquot of spores was used to determine the ratio of wild-type and mutant cells in the population and the rest was used to inoculate the next cycle of the experiment for eight consecutive cycles. The treatment included the incubation of spores in the presence of 1% H2O2 (open squares) and the absence of H2O2 (filled triangles). The same experiment was repeated with a mixture of TB38 cells and cells mutant for sqhC and sodF that contained a functional copy of sqhC (ΔsqhC ΔsodF::tet, thrC::PsqhC-sqhC) (TB71, filled squares). The observed trend was not influenced by the presence of the gene encoding for the colorimetric marker lacZ (Fig. S5).
Alternatively, sporulenes could be involved in the protection of spores against prolonged or elevated exposure to reactive oxygen species. Again, we tested this hypothesis in a mixed culture containing 90% mutant (TB10) and 10% wild type (TB38). We treated the purified spores with H2O2, a neutral reactive oxygen species that damages the heat resistance of B. subtilis spores, presumably by affecting the inner spore membrane (16). To then select for spores more resistant to H2O2, we exposed the mixtures to elevated heat and repeated the experiment in eight consecutive cycles. The ratio of the sqhC-lacking mutant (TB10) to wild type (TB38) decreased by 11 ± 1% per cycle (Fig. 3). Because sodF removes superoxide and produces H2O2, we expected that sodF was not responsible for the resistance of spores to the H2O2 treatment. To test this, we repeated the experiment by using a strain that lacked the sqhC-sodF operon but was complemented with a copy of sqhC (TB71). The reintroduction of sqhC significantly restored the resistance of spores to H2O2: the ratio of the complemented strain (TB71) to wild type (TB38) decreased only by 4 ± 1% in each successive cycle (Fig. 3), confirming a specific H2O2-mitigating role for sqhC.
Finally, spores of a strain whose sqhC had been mutated to abolish the hypothesized initiation site of cyclization (7, 17) had an equally reduced resistance to H2O2 as the sqhC-lacking mutant TB10 (Fig. S5). These results demonstrate that the formation of sporulene lipids contributes to the resistance of spores to H2O2. The mechanism of their action requires further exploration.
Biological and Environmental Implications
The presence of a gene encoding a putative terpenoid cyclase in B. subtilis was recognized (10, 18), but the function and products of the enzyme remained unknown. Here, we have demonstrated that this enzyme is a cyclase that produces C-35 terpenes with tetracyclic skeletons, from regular acyclic polyprenes. The cyclization of regular polyprenes requires no stabilization of secondary (2°) cations and therefore is a more favorable reaction than the cyclization of squalene and oxidosqualene. Such reactions are thought to have evolved earlier than the reactions that form hopanoids and sterols (19, 20). Our results show that regular polyprenes are natural substrates for the B. subtilis SqhC, and that this enzyme does not appear to be able to cyclize squalene in vitro (see SI Text). In contrast, studies of the squalene-hopene cyclase of Alicyclobacillus acidocaldarius and the tetrahymanol synthase of Tetrahymena pyriformis show that these enzymes are able to cyclize both squalene and regular polyprenes in vitro (12, 13).
Until now, natural tetracyclic compounds derived from regular polyprenes, known as scalaranes, had been reported only from some marine sponges (21, 22). Similar tricyclic structures (cheilanthanes) are known in the geologic record in association with deposits of Tasmanites algae (23). The apparent association of these molecular fossils only with eukaryotic sources long has been puzzling and inconsistent with the hypothesized early evolution of the cyclization of regular polyprenes (19). Our discovery of the bacterial formation of tetracyclic backbones from regular acyclic polyprenes was not unexpected, however, given that several other skeletons (bicyclic, tricyclic, and octacyclic) derived from the cyclization of regular polyprenes had been reported in bacteria (24–26). The detection of bacterial sporulenes shows uniquely that tetracyclic regular polyprenoids are not restricted to eukaryotes. More cyclase sequences and biochemical studies are needed to further examine the evolutionary position of regular polyprenyl cyclases in relation to bacterial squalene and oxidosqualene cyclases.
Some hopanoids have been used as taxonomic markers for specific groups of Bacteria, e.g., cyanobacteria (2); but these empirical associations do not always hold (3) or are not sufficient to ascertain the metabolic attributes of the organisms that left biomarkers in the rock record, i.e., their oxygen-evolving capacity (4). In contrast, sporulenes are potentially geostable lipids that also may reflect a specific geologically relevant physiology that may extend beyond Bacillus spp. as a taxonomic group. If sporulenes are found in other bacteria, they would be likely to have a protective role against oxidative stress, although our data do not preclude other functions for these molecules as well.
Our study has experimentally demonstrated in vivo a specific physiological role for a nonpigment bacterial lipid biomarker. A diversity of similar roles has been proposed for pentacyclic hopanoids, based on other analytical approaches. For example, hopanoids can modify the fluidity of artificial membranes in vitro (27), and this modification may explain the observed correlation between temperature and the percentage of hopanoids in the thermoacidophile A. acidocaldarius (28). The nonessential protective role of sporulenes also differs from the essential role of hopanoids in some mesophilic bacteria, where the inhibition of hopanoid biosynthesis is lethal but the mechanisms of lethality remain unknown (29). Intriguingly, our finding that sporulenes increase the fitness of B. subtilis spores by protecting them against a reactive oxygen species resembles the hypothesized inhibition of oxygen diffusion by bacteriohopanetetrol phenylacetate monoesters in the nitrogen-fixing Gram-positive root symbiont, Frankia (30). Our study thus complements these previous suggestions that polycyclic isoprenoids may be more than mere membrane reinforcers (19, 30, 31) and inspires future studies that will test the relationship between the diversity and the function of other bacterial polycyclic terpenoids.
Our results also encourage the search for polycyclic compounds derived from regular polyprenes both in other modern microbes and the rock record. Although tricyclic cheilanthanes are abundant in sediments of all ages, their modern sources remain unknown. However, although some modern (eukaryotic) sources of tetracyclic scalaranes are known, these compounds have not yet been reported in ancient sediments. This is surprising, given the abundance of Bacillus spores in aerosols (32, 33), the long record of marine sponges (34), and an even longer record of microbial life (35). These compounds may have been overlooked to date because spores may not contribute a large fraction of the total carbon in sediments, but we expect that an informed search will identify diagenetic products of ancient sporulenes and tetraprenyl curcumenes. Last, because bacterial sporulenes provide enhanced resistance to conditions of oxidative stress, detection of geostable derivatives of these compounds in sediments can be used as an experimentally verified indicator of oxic environments, oxidative stress response, and, possibly, the rising levels of atmospheric oxygen in the past.
Materials and Methods
Bacterial Strains and Growth Conditions.
B. subtilis strains and plasmids used in this study are listed in Table S1. E. coli DH5α (36) was used for cloning experiments. All strains are derivatives of B. subtilis PY79 (37). The sporulation was induced either by exhaustion in DSM medium or resuspension in Sterlini–Mandelstam medium (38). B. subtilis competent cells were prepared as described in ref. 39. The construction of specific strains is described in SI Text and Table S2.
Spore Preparation.
To obtain mixed cultures, B. subtilis strains were grown from single colonies in two separate cultures in 2 ml of LB, mixed in the desired proportion (1:9), washed with DSM twice, and inoculated into 20 ml of DSM to an OD600 of 0.05. This mixed culture was then sporulated by exhaustion for 2 days. The spores were harvested by centrifugation, washed, and incubated for 1 h in 2 ml of Tris-EDTA buffer with 10 mg/ml lysozyme at 37°C before the addition of 0.4 ml of 10% SDS, incubated for an additional 20 min at 37°C, washed with 0.01% Tween (Sigma–Aldrich), washed with dI-H2O, and resuspended in dI-H2O. Clean spore preparations were stored at 4°C in 15-ml conical tubes covered by aluminum foil and placed on a rocking platform.
Hydrogen Peroxide Treatment.
The spore preparations of mixed cultures were resuspended in dI-H2O to an OD600 of 0.6–0.8 and incubated at 80°C for 10 min to remove all germinated spores. The spore suspensions were then cooled and resuspended in 967 μl of 50 mM KH2PO4 buffer. Thirty-three microliters of 30% hydrogen peroxide (Sigma–Aldrich) was added to the suspension and incubated at 37°C for the amount of time necessary to kill at least 80% and not >99.9% of the spores (generally ≈30 min, but determined more precisely by taking samples at 25, 30, and 35 min for each spore preparation). Ten-microliter samples were taken at the beginning of the incubation and at the later three time points, added to 990 μl of 100 units/ml catalase (Sigma–Aldrich) in 50 mM phosphate buffer, incubated for 10 min to remove H2O2, incubated at 80°C for 10 min to kill the damaged spores, diluted, and plated on LB agar plates with 100 μl/ml of X-galactoside (Sigma–Aldrich). The ratio of strains in mixed cultures was determined by blue-white counts after at least 24 h of growth at 37°C. The catalase/phosphate solution was removed by centrifuging, and the cells were resuspended in 20 ml of DSM and sporulated.
Microscopy.
The production of SqhC-GFP was induced by the addition of isopropyl thiogalactopyranoside (IPTG; Sigma–Aldrich) to a 1 mM final concentration at 2.5 h after the beginning of the sporulation. Cells were harvested during vegetative growth, and at 4 and 24 h after the beginning of sporulation, centrifuged, resuspended in 50 μl of PBS with 1 μg/ml of the membrane dye FM4–64 (Invitrogen). Three microliters of the stained cells was immobilized by a coverslip treated with poly(l-lysine). The equipment and analysis used in imaging has been described (40). We repeated the imaging experiment three times.
β-Galactosidase Activity Assay.
Strains harboring PsqhC-lacZ fusion were sporulated by resuspension, and 1-ml samples were taken during sporulation, centrifuged, and frozen at −80°C. The specific activity of β-galactosidase in these samples was determined with o-nitrophenyl-β-galactopyranoside (ONPG; Sigma–Aldrich) as a substrate (38). We repeated the experiment twice.
Farnesyl Pyrophosphate Incorporation Assay.
A single colony of the strain mutant for sqhC but with a copy of sqhC under the control of the IPTG-inducible promoter (ΔsqhC sodF::tet amyE::PHyperspank-sqhC, TB28) was grown to midlog phase, then resuspended in 100 ml of the growth medium and grown overnight at 30°C. This culture was used to inoculate 1 liter of growth medium and grown at 37°C to OD600 0.5–0.8. The cells were collected by centrifugation at 6,000 × g for 10 min at room temperature and resuspended in the same volume of the resuspension medium to induce sporulation. At 2.5 h after the resuspension, 1 ml of 1 M IPTG was added to the culture. After an additional 2 h of incubation (4.5 h after the beginning of the sporulation), the cells were collected by centrifugation. The cell pellet was resuspended in 20 ml of 54 mM Na2HPO4, 10 mM KH2PO4, 2 mM EDTA, 10 mM β-mercaptoethanol, and 0.2 mg/ml lysozyme, incubated for 45 min at 37°C, and sonicated 10 times for 30 s on ice. MgCl2 and cold FPP (Sigma–Aldrich) were then added to the lysate to a final concentration of 20 mM and 10 μM, respectively. 3H-FPP [23 nCi (1 Ci = 37 GBq)] (Sigma–Aldrich) was added to the reaction mix and the crude cell lysate was incubated with shaking at 37°C for 12 h. The incubation was stopped by adding 10% concentrated HCl and by placing the lysate at −80°C. Crude cell lysates of the strain with the deleted sqhC operon (ΔsqhC sodF::tet) were prepared and incubated with FPP and 3H-FPP in the same manner. Before the lipid extraction, the acidified reaction mixtures were incubated at 70°C for 2 h. The lipids were extracted by a two-phase chloroform/methanol extraction, dissolved in dichloromethane (DCM) and stored at −20°C (38). Lipids were separated by reversed-phase Zorbax Eclipse XDB-C18 column (5 μm, 4.6 × 250 mm) at 30°C and 1 ml/min flow on an Agilent Technologies 1100 Series high-performance liquid chromatograph. We used a 120-min binary gradient method (SI Text and Table S3), collecting the fractions from 5 to 85 min. The radioactivity of 3H-FPP in each fraction was measured. Subsamples of 1-min fractions from 47 to 68 min were analyzed by GC-MS as described below. We incubated the cell lysates of TB28 and TB10 with 3H- PP twice and once, respectively, and analyzed the lipids from the cold cell lysates of TB28 and TB10 by GC/MS in four and three independent experiments, respectively. Although the dilution of the radioactive substrate by a large amount of cold FPP most likely reduced the incorporation of radioactive FPP into isoprenoids, we were able to use the results of this assay to identify the products of SqhC. An optimized assay could be used in the future biosynthetic studies of the SqhC activity.
Lipid Extraction from Spores.
A single colony of the wild type was grown in 100 ml of growth medium at 37°C to midlog phase, then resuspended in 1 liter of DSM medium and grown at 37°C for 72 h. Spores were collected by centrifugation at 6,000 × g for 10 min at room temperature and the pellet was incubated in 100 ml of 12% HCl at 80°C for 36 h. The acidified spore resuspension was then bead-beaten (BeadBeater and 0.1-mm diameter glass beads; Biospec Products) 10 times in 1-min intervals with 1-min pauses. Examination by light microscopy confirmed that spores were thoroughly lysed. The lysate was extracted by vigorous shaking with DCM and the extract was separated on a SiO2-gel gravity column (32–64 μm) prepared in DCM by using hexane (GC grade; EMD Chemicals, an affiliate of Merck). The hexane fraction was dried down and androstanol (Sigma–Aldrich) was added as a quantification standard for analysis by GC-MS. Preliminary 1H-NMR screening was performed on purified aliquots of individual lipids extracted from 4-liter cultures of strain TB28. The production of SqhC was induced by 1 mM IPTG at 3 h into the sporulation and the lipids were extracted from the spores and separated on a silica column in the manner described above. To obtain purified aliquots, compounds I–V were separated by preparative capillary gas chromatography by using a Gerstel Preparative Fraction Collector (PFC; Gerstel GmbH & Co) by using methods similar to those described (41). We also extracted and examined the lipids from 4 liters of the sporulated culture of the strain mutant for sqhC (TB10) as a negative control.
GC-MS Analyses.
Lipids were analyzed on an Agilent Technologies 6890N gas chromatograph with Agilent DB5-MS (5% phenyl) column (30 m × 0.25 mm, 0.25 μm) by using He as the gas phase (1.2 ml/min). Eluted compounds were detected by using an Agilent 5973 inert mass spectrometer detector operated at 70 eV (GC program; Table S4). Lipids were analyzed as trimethylsilyl ethers, as acetate derivatives (prepared in 1:1 pyridine/acetic anhydride), and as underivatized samples. All preparations yielded the same results, indicating the substances contained no alcohol or acid functional groups. GC-MS analyses were repeated at least two times per sample. The detection limit for all cyclization products was 5 ng.
Supplementary Material
Acknowledgments.
We thank J. J. Brocks, S. C. Brassell, and R. E. Summons for help with mass spectral interpretations; B. VanMooy and H. Fredricks for use of their HPLC; N. Drenzek for his help with PCGC; R. A. Butcher and R. Kontnik for preliminary NMR; and B. Weiss and two anonymous reviewers for comments. T.B. was supported by a Harvard Microbial Sciences Initiative Postdoctoral Fellowship. This work was supported by the National Science Foundation-Earth Sciences and a Packard Fellowship in Science in Engineering (to A.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800199105/DCSupplemental.
References
- 1.Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- 2.Summons RE, Jahnke LL, Hope JM, Logan GA. 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature. 1999;400:554–557. doi: 10.1038/23005. [DOI] [PubMed] [Google Scholar]
- 3.Rashby SE, Sessions AL, Summons RE, Newman DK. Biosynthesis of 2-methylbacteriohopanepolyols by an anoxygenic phototroph. Proc Natl Acad Sci USA. 2007;104:15099–15104. doi: 10.1073/pnas.0704912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp RE, Kirschvink JL, Hilburn IA, Nash CZ. The paleoproterozoic snowball Earth: A climate disaster triggered by the evolution of oxygenic photosynthesis. Proc Natl Acad Sci USA. 2005;102:11131–11136. doi: 10.1073/pnas.0504878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendt KU, Poralla K, Schulz GE. Structure and function of a squalene cyclase. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 6.Thoma R, et al. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 7.Merkofer T, Pale-Grosdemange C, Wendt KU, Rohmer M, Poralla K. Altered product pattern of a squalene-hopene cyclase by mutagenesis of active site residues. Tetrahedron Lett. 1999;40:2121–2124. [Google Scholar]
- 8.Pale-Grosdemange C, Merkofer T, Rohmer M, Poralla K. Production of bicyclic and tricyclic triterpenes by mutated squalene-hopene cyclase. Tetrahedron Lett. 1999;40:6009–6012. [Google Scholar]
- 9.Rohmer M, Bouvier-Navé P, Ourisson G. Distribution of hopanoid triterpenes in prokaryotes. J Gen Microbiol. 1984;130:1137–1150. [Google Scholar]
- 10.Eichenberger P, et al. The óE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol. 2003;327:945–972. doi: 10.1016/s0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi I, Ogura K, Seto S. Heptaprenyl pyrophosphate synthetase from Bacillus subtilis. J Biol Chem. 1980;255:4539–4543. [PubMed] [Google Scholar]
- 12.Hoshino T, Kumai Y, Kudo I, Nakano S, Ohashi S. Enzymatic cyclization reactions of geraniol, farnesol and geranylgeraniol, and those of truncated squalene analogs having C20 and C25 by recombinant squalene cyclase. Org Biomol Chem. 2004;2:2650–2657. doi: 10.1039/B407001A. [DOI] [PubMed] [Google Scholar]
- 13.Renoux JM, Rohmer M. Enzymatic cyclization of all-trans pentaprenyl and hexaprenyl methyl ethers by a cell-free system from the protozoan Tetrahymena pyriformis—The biosynthesis of scalarane and polycyclohexaprenyl derivatives. Eur J Biochem. 1986;155:125–132. doi: 10.1111/j.1432-1033.1986.tb09467.x. [DOI] [PubMed] [Google Scholar]
- 14.McCord JM, Fridovic I. Superoxide dismutase and enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 15.Kannenberg E, Poralla K, Blume A. A hopanoid from the thermo-acidophilic Bacillus acidocaldarius condenses membranes. Naturwissenschaften. 1980;67:458–459. [Google Scholar]
- 16.Cortezzo DE, Koziol-Dube K, Setlow B, Setlow P. Treatment with oxidizing agents damages the inner membrane of spores of Bacillus subtilis and sensitizes spores to subsequent stress. J Appl Microbiol. 2004;97:838–852. doi: 10.1111/j.1365-2672.2004.02370.x. [DOI] [PubMed] [Google Scholar]
- 17.Hoshino T, Abe T, Kouda M. Unnatural natural triterpenes produced by altering isoleucine into alanine at position 261 in hopene synthase and the importance of having the appropriate bulk size at this position for directing the stereochemical destiny during the polycyclization cascade. Chem Commun. 2000:441–442. [Google Scholar]
- 18.Wendt KU, Schulz GE, Corey EJ, Liu DR. Enzyme mechanisms for polycyclic triterpene formation. Angew Chem Int Ed. 2000;39:2812–2833. [PubMed] [Google Scholar]
- 19.Ourisson G, Rohmer M, Poralla K. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu Rev Microbiol. 1987;41:301–333. doi: 10.1146/annurev.mi.41.100187.001505. [DOI] [PubMed] [Google Scholar]
- 20.Fischer WW, Pearson A. Hypotheses for the origin and early evolution of triterpenoid cyclases. Geobiology. 2007;5:19–34. doi: 10.1111/j.1472-4669.2007.00096.x. [DOI] [PubMed] [Google Scholar]
- 21.Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR. Marine natural products. Nat Prod Rep. 2006;23:26–78. doi: 10.1039/b502792f. [DOI] [PubMed] [Google Scholar]
- 22.Buchanan MS, Edser A, King G, Whitmore J, Quinn RJ. Cheilanthane sesterterpenes, protein kinase inhibitors, from a marine sponge of the genus Ircinia. J Nat Prod. 2001;64:300–303. doi: 10.1021/np0004597. [DOI] [PubMed] [Google Scholar]
- 23.Greenwood PF, Arouri KR, George SC. Tricyclic terpenoid composition of Tasmanites kerogen as determined by pyrolysis GC-MS. Geochim Cosmochim Acta. 2000;64:1249–1263. [Google Scholar]
- 24.Hamano Y, et al. Cloning of a gene cluster encoding enzymes responsible for the mevalonate pathway from a terpenoid-antibiotic-producing Streptomyces strain. Biosci Biotechnol Biochem. 2001;65:1627–1635. doi: 10.1271/bbb.65.1627. [DOI] [PubMed] [Google Scholar]
- 25.Chuck JA, Barrow KD. The isolation of isoagathenediol—A new tricyclic diterpene from the lipids of Rhodospirillum rubrum. Microbiology. 1995;141:2659–2663. [Google Scholar]
- 26.Nakanishi S, et al. KS-505a, a novel inhibitor of bovine brain Ca2+ and calmodulin-dependent cyclic-nucleotide phosphodiesterase from Streptomyces argenteolus. J Antibiot. 1992;45:341–347. doi: 10.7164/antibiotics.45.341. [DOI] [PubMed] [Google Scholar]
- 27.Kannenberg E, Blume A, McElhaney RN, Poralla K. Monolayer and calorimetric studies of phosphatidyl-cholines containing branched-chain fatty acids and of their interaqction with cholesterol and with bacterial hopanoid in model membranes. Biochim Biophys Acta. 1983;733:111–116. [Google Scholar]
- 28.Poralla K, Hartner T, Kannenberg E. Effect of temperature and pH on the hopanoid content of Bacillus acidocaldarius. FEMS Microbiol Lett. 1984;23:253–256. [Google Scholar]
- 29.Flesch G, Rohmer M. Growth inhibition of hopanoid synthesizing bacteria by squalene cyclase inhibitors. Arch Microbiol. 1987;147:100–104. [Google Scholar]
- 30.Berry AM, et al. Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc Natl Acad Sci USA. 1993;90:6091–6094. doi: 10.1073/pnas.90.13.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kannenberg EL, Poralla K. Hopanoid biosynthesis and function in bacteria. Naturwissenschaften. 1999;86:168–176. [Google Scholar]
- 32.Snyder AP, et al. Field detection of bacillus spore aerosols with stand-alone pyrolysis-gas chromatography-ion mobility spectrometry. Field Anal Chem Technol. 1999;3:315–326. [Google Scholar]
- 33.Amato P, et al. Microbial population in cloud water at the Puy de Dome: Implications for the chemistry of clouds. Atmos Environ. 2005;39:4143–4153. [Google Scholar]
- 34.Yang Q, Ma JY, Sun XY, Cong PY. Phylochronology of early metazoans: Combined evidence from molecular and fossil data. Geol J. 2007;42:281–295. [Google Scholar]
- 35.Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 37.Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson WL, Setlow P. Sporulation, germination, and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. New York: Wiley; 1990. p. 581. [Google Scholar]
- 39.Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968;95:1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- 41.Eglinton TI, Aluwihare LI, Bauer JE, Druffel ERM, McNichol AP. Gas chromatographic isolation of individual compounds from complex matrices for radiocarbon dating. Anal Chem. 1996;68:904–912. doi: 10.1021/ac9508513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.