Abstract
Chromatin insulators are thought to restrict the action of enhancers and silencers. The best-known insulators in Drosophila require proteins such as Suppressor of Hairy wing (Su(Hw)) and Modifier of mdg4 (Mod(mdg4)) to be functional. The insulator-related proteins apparently colocalize as nuclear speckles in immunostained cells. It has been asserted that these speckles are ‘insulator bodies' of many Su(Hw)–insulator DNA sites held together by associated proteins, including Mod(mdg4). As we show here using flies, larvae and S2 cells, a mutant Mod(mdg4) protein devoid of the Q-rich domain supports the function of Su(Hw)-dependent insulators and efficiently binds to correct insulator sites on the chromosome, but does not form or enter the Su(Hw)-marked nuclear speckles; conversely, the latter accumulate another (C-truncated) Mod(mdg4) mutant that cannot interact with Su(Hw) or with the genuine insulators. Hence, it is not the functional genomic insulators but rather aggregated proteins that make the so-called ‘insulator bodies'.
Keywords: Drosophila, Mod(mdg4), nuclear speckles, Su(Hw) insulator
Introduction
Insulators are genomic regulatory elements that are defined by two properties: these nucleoprotein complexes can block enhancer action on a promoter when interposed between them, and can protect the transgenes that they flank from chromosomal position effects (for reviews, see Kuhn & Geyer, 2003; Brasset & Vaury, 2005; West & Fraser, 2005; Gaszner & Felsenfeld, 2006). The most studied insulator in Drosophila is the one found in the gypsy retrotransposon (mdg4). It contains 12 degenerate repeats of the binding motif for the zinc-finger protein Suppressor of Hairy wing (Su(Hw)), which is essential for its function (Holdridge & Dorsett 1991; Geyer & Corces 1992). Among the numerous potential Su(Hw)-binding sites dispersed throughout the wild-type genome, rarely three or more motifs occur within reasonable proximity (Parnell et al, 2006; Ramos et al, 2006). However, the 1A2 insulator downstream of the yellow gene, with only two Su(Hw)-binding sites, shows both insulator functions in standard transgene assays (Golovnin et al, 2003; Parnell et al, 2003).
Two more proteins, Modifier of mdg4 (Mod(mdg4)) and Centrosomal protein 190kD (CP190), are required for the gypsy insulator function (Gerasimova et al, 1995; Georgiev & Kozycina, 1996; Pai et al, 2004). Mod(mdg4) is a BTB/POZ protein capable of oligomerization; the Mod(mdg4)-67.2 isoform interacts with Su(Hw) by its unique carboxy-terminal domain (Buchner et al, 2000; Gause et al, 2001; Ghosh et al, 2001).
A decade ago, it was reported by Gerasimova & Corces (1998) that Su(Hw) and Mod(mdg4) colocalized in discrete foci observed by microscopy in the Drosophila interphase cell nucleus. Exclusively on the basis of the disappearance of such immunofluorescent foci and concurrent weakening of a gypsy insulator after a Mod(mdg4)-affecting mutation, these nuclear speckles were named ‘insulator bodies'. Furthermore, it was stated by the same team (Gerasimova et al, 2000; Ghosh et al, 2001; Pai et al, 2004; Capelson & Corces, 2004, 2005; Lei & Corces, 2006) that these bodies represent nuclear matrix-fixed congregations of many genomically remote Su(Hw)–insulator DNA complexes, somehow brought together and held by interactions through Mod(mdg4) and CP190, thereby establishing ‘separate chromatin loop domains' and thus controlling the higher order organization and function of the genome.
In fact, the presumed clustering of distinct insulator DNA sequences within an ‘insulator body' has not been verified over the years. Here, we show, by expressing altered forms of the indispensable Su(Hw)–insulator component Mod(mdg4) in the same objects, that such nuclear speckles are irrelevant to genuine insulators or their function.
Results And Discussion
Structure and properties of the Mod(mdg4) mutants
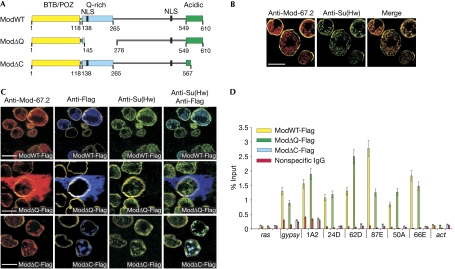
We designed deletions in the Mod(mdg4)-67.2 protein on the basis of published data (Buchner et al, 2000; Gerasimova et al, 2000; Gause et al, 2001; Ghosh et al, 2001; Golovnin et al, 2007). The wild-type protein (ModWT; Fig 1A) has an amino-terminal BTB/POZ domain, an adjacent glutamine(Q)-rich domain and a C-terminal acidic domain that binds to Su(Hw) (Buchner et al, 2000); sequence analysis predicts two nuclear localization signals (NLS). Two Mod derivatives were used in the present study: in ModΔQ, deletion of residues 145–276 removed the Q-rich domain and one NLS; in ModΔC, deletion of 43 C-terminal residues removed most of the Su(Hw)-binding domain (Fig 1A).
Figure 1.
Mod(mdg4) proteins and their distribution in S2 cells. (A) Schematics of Mod(mdg4)-67.2 and its deletion derivatives: the wild-type protein (ModWT), ModΔQ, which lacks the Q-rich domain, and ModΔC, which lacks most of the Su(Hw)-binding domain. (B) Immunostained control cells; scale bar, 5 μm. (C) Transfected cells expressing Flag-tagged Mod variants specified in (A) are the upper three cells in the top row, the central one in the middle row and the right two in the bottom row; scale bar, 5 μm. All images in (B) and (C) include staining for lamin to demarcate the nuclei. (D) Crosslinking chromatin immunoprecipitation of specified chromatin regions with the Mod variants (percentage of input DNA, M±m, n=3); actin and ras coding regions are controls devoid of Su(Hw)-binding sites. Mod(mdg4), Modifier of mdg4; NLS, nuclear localization signals; Su(Hw), Suppressor of Hairy wing.
ModΔQ has been shown to be able to self-associate and interact with ModWT and Su(Hw), as evidenced by the yeast two-hybrid assay and co-immunoprecipitation from transfected S2 cells (supplementary Table S1 and Fig S1 online). As expected, ModΔC could also oligomerize by itself as well as with ModWT, but had completely lost the ability to interact with Su(Hw) in the two-hybrid assay. Nonetheless, Su(Hw) was partly co-precipitated with ModΔC from the S2 nuclear extract (supplementary Fig S1 online)—that is, both proteins were present in some type of agglomerate, although they were incapable of direct binding.
Localization of Mod(mdg4) variants in S2 cells
The nuclei of S2 cells derived from Drosophila embryos showed speckles that stained for Mod(mdg4) and Su(Hw) (Fig 1B). These speckles were similar in size, number and disposition to those reported in flies and named ‘insulator bodies' (Gerasimova & Corces, 1998; Gerasimova et al, 2000). To assess the distribution of the Mod(mdg4) variants, these cells were transfected with plasmids encoding ModWT, ModΔC and ModΔQ tagged at the C termini with triple Flag epitopes. Thus, the plasmid expression of the Flag-tagged wild-type or mutant protein was superimposed on the basal genomic expression.
The results of immunostaining are summarized in Fig 1C. By way of an internal control, each panel shows transfected cells (distinguished by Flag staining and specified in the legend) and nontransfected cells.
Overexpression of ModWT-Flag did not appreciably change the ‘normal' staining patterns: anti-Flag shows exclusively nuclear ‘punctated' (Gerasimova et al, 2000) deposition, incident with that of Su(Hw). Overall, the Mod+Su(Hw) speckles might be more abundant than in the control.
By contrast, intense expression of ModΔQ-Flag gave rise to massive diffuse staining of the protein and Flag in the cytoplasm, but not in the nucleus. The few nuclear speckles stained with anti-Su(Hw) and anti-Mod represent the ‘basal deposits' (or rather those that existed before transfection) and none of these was stained with anti-Flag. Such a pattern could be expected for ModΔQ, which retains the ability to interact with proteins but is handicapped in nuclear targeting: its accumulation/oligomerization in the cytoplasm can further reduce the amount delivered to the nucleus and also trap a considerable amount of the ‘basal' Mod(mdg4) and Su(Hw). The overexpression of ModΔQ clearly shows the marked difference between the Mod variants in their intracellular distribution. The minor drawback is that no cytoplasmic deposition of Su(Hw) can be discerned by simultaneous immunostaining among such overwhelming amounts of Mod, so the cytoplasm appears ‘empty' in the ModΔQ-Flag/anti-Su(Hw) panel. However, co-deposition of Su(Hw) and ModΔQ in the cytoplasm was clearly seen in transgenic flies discussed below, in which the amounts of the two proteins are naturally much closer.
Expression of ModΔC-Flag again resulted in exclusive nuclear deposition of the mutant protein together with Su(Hw); these speckles were somewhat fewer but larger than in the control or with ModWT. Notably, all Flag-positive speckles were also Su(Hw) positive, whereas some (most likely, the pre-existing ones) contained the Mod protein and Su(Hw), but not ModΔC-Flag. The colocalization of ModΔC and Su(Hw)—which cannot interact directly—is in line with their partial co-precipitation (see the end of preceding section) and is not at all surprising; the speckles perhaps also include other proteins that can link them, for example, CP190 is known to interact with Su(Hw) and Mod(mdg4) in vitro (Pai et al, 2004; Golovnin et al, 2007).
It must be noted that among these four cases, the nuclear speckles might vary or not in number and/or dispersion; such variability is not consistent with the crucial structural and functional role proposed for ‘insulator bodies'.
Importantly, the lack of ModΔQ-Flag staining in the nucleus does not mean that there is no protein; it only means that the intranuclear ModΔQ did not aggregate or stick to the existing speckles. Standard subcellular fractionation and Western blotting (supplementary Fig S2 online) clearly showed ModΔQ-Flag and Su(Hw) in the nuclei (although rough estimates ‘per nucleus' were 3–4 times lower than for the ModWT case, as expected). Still more pertinent were the crosslinking chromatin immunoprecipitation (X-ChIP) data (Fig 1D), showing that, overall, the same amounts of ModWT and ModΔQ were bound to the chromatin regions known to contain Su(Hw)-dependent insulators and similar motifs (Golovnin et al, 2003; Parnell et al, 2003, 2006; Ramos et al, 2006). By contrast, the ModΔC level throughout was indistinguishable from the background.
To summarize, ModΔQ, which retains all the properties essential to insulator function, is delivered into Drosophila cell nuclei, perhaps owing to the single NLS, and associates with the chromatin Su(Hw)–insulator sites no less efficiently than the wild-type protein, but it does not form any nuclear speckles or join the existing ones. Conversely, ModΔC, which cannot interact with Su(Hw), completely fails to bind to the correct insulator sites in chromatin but instead consistently colocalizes (perhaps aided by other proteins) with Su(Hw) in the nuclear speckles.
In vivo functional testing of Mod(mdg4) mutants
Next, we compared the functional effects of ModΔQ and ModΔC in flies. The source of ModΔQ was a transgenic line providing UAS-driven ModΔQ expression in a null mod(mdg4)u1 background. Its counterpart expressing ModWT was used as a reference in addition to the wild type. The source of ModΔC was the previously described (Ghosh et al, 2001) mod(mdg4)T6 mutation, which generates the same protein lacking the 43 C-terminal residues.
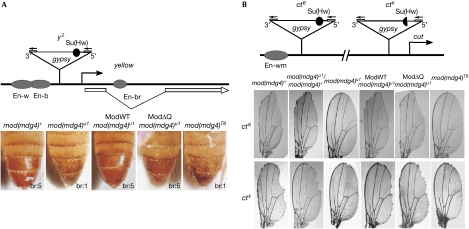
Phenotypic analysis of the competence of Mod variants in insulator function was performed in male flies carrying gypsy-induced alleles in the yellow and cut loci, as in the studies furthering the idea of ‘insulator bodies' (Pai et al, 2004; Capelson & Corces, 2005; Lei & Corces, 2006). Yellow expression determines the cuticular pigmentation and is controlled by several tissue-specific enhancers. In the y2 mutation (Fig 2A), a gypsy element is interposed between the yellow promoter and the wing and body enhancers (Geyer et al, 1986), therefore its insulator blocks these enhancers but not the bristle enhancer in the yellow intron (Geyer et al, 1986; Geyer & Corces, 1992). Phenotypically, this gives a pale abdomen with dark bristles (leftmost panel). The mod(mdg4)u1 mutation alters the y2 phenotype, repressing yellow expression in bristles (Gerasimova et al, 1995; Georgiev & Kozycina, 1996) and partly weakening the gypsy insulator, which results in variegated yellow expression in the body cuticle (Gerasimova & Corces, 1998), as shown by the second-left dappled abdomen with pale bristles. Expression of ModWT as well as ModΔQ completely overrides the mod(mdg4)u1 effect on both traits, indicating that ModΔQ substitutes for the wild-type protein in this insulator-related function. Conversely, the mod(mdg4)T6 mutation expressing ModΔC yields exactly the same phenotype as the null mod(mdg4)u1 mutation, indicating that ModΔC is nonfunctional.
Figure 2.
Testing of Mod(mdg4) variants for insulator function in y2 ct6 male flies. Schematics show the structure of the (A) y2 and (B) ct6 or ctk alleles; beginnings and direction of the yellow and cut genes are shown by arrows; ovals denote wing (w), body (b), bristle (br) and wing margin (wm) enhancers (En); triangles show insertions of gypsy with flanking long terminal repeats and the Su(Hw) insulator as a black (semi)circle. Photographs collate (A) the body cuticle (bottom-right, bristle pigmentation: 5 for wild type, 1 for none) and (B) the wing phenotypes; ModWT and ModΔQ refer to transgenic expression of the variant specified in Fig 1A; mod(mdg4)T6 itself produces ModΔC; mod(mdg4)u1 is the null mutation. Mod(mdg4), Modifier of mdg4; Su(Hw), Suppressor of Hairy wing; y2, yellow mutation associated with gypsy insertion.
In the ct6 and ctk alleles (Fig 2B), gypsy is between the wing margin enhancer and the cut promoter, which are 85 kb apart (Hoover et al, 1992; Gause et al, 2001). In ct6, the insulator completely blocked this enhancer, producing a cut wing phenotype (leftmost in the upper row). The null mod(mdg4)u1 (middle left) and the mod(mdg4)T6 (rightmost) mutations clearly suppressed the ct6 mutant phenotype, indicating that Mod(mdg4)-67.2 is essential for blocking the wing margin enhancer and that ModΔC does not compensate for its loss. By contrast, ModΔQ completely restored the gypsy insulator function in the mod(mdg4)u1 background, similar to ModWT (Fig 2B). The gypsy insulator was weaker in ctk than in ct6, perhaps because it has only 7 instead of 12 Su(Hw) sites (Hoover et al, 1992), and is also more sensitive to the level of Mod(mdg4)-67.2 (Gause et al, 2001): it produced an intermediate cut wing phenotype (leftmost in the bottom row) that was almost completely suppressed with a single dose of mod(mdg4)u1 (second left). ModΔQ restored the activity of this insulator in the null background similar to ModWT, confirming that sufficient amounts of the functional ModΔQ protein bind to insulator sites. Again, ModΔC is ineffective (the mod(mdg4)T6 and null mod(mdg4)u1 wing phenotypes were identical).
Exactly the same pattern of responses was obtained in another system (AS-C; supplementary information and Fig S3 online), which allowed testing the functionality of Mod variants with both gypsy and endogenous Su(Hw)-dependent (1A2) insulators.
These data prove that, in agreement with the properties established in vitro, ModΔQ is functionally equivalent to the wild-type Mod(mdg4)-67.2 protein at the authentic insulators in Drosophila, whereas ModΔC is totally incompetent.
Localization of the Mod(mdg4) mutants in larvae
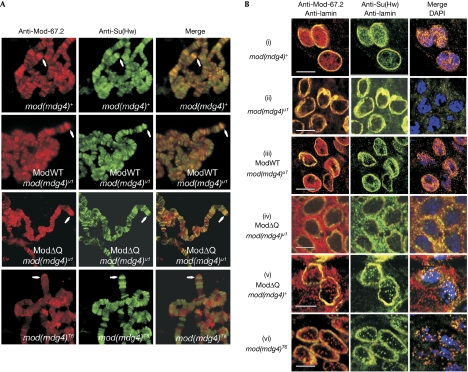
The binding of mutant Mod(mdg4) to insulator sites was analysed by immunostaining on polytene chromosomes, in which ModWT was shown to colocalize with Su(Hw) (Gerasimova & Corces, 1998). Two well-defined locations of Su(Hw) insulators are the gypsy inserts in the y2 and scD1 loci at the tip of the X chromosome (Gerasimova et al, 1995). The two corresponding intense bands for Su(Hw) in the y2scD1 strain are indicated in Fig 3A. As expected, ModΔQ bound to polytene chromosomes exactly as the wild-type protein did, in particular, at the gypsy bands in y2 and scD1. By contrast, ModΔC (mod(mdg4)T6) decorated a considerably smaller number of places that did not coincide with the Su(Hw) insulators. These results further show that ModΔQ, but not ModΔC, interacts with authentic Su(Hw) insulators.
Figure 3.
Localization of Mod(mdg4) variants in Drosophila larvae. (A) Polytene chromosomes of y2scD1 larvae; arrows indicate gypsy inserts at the X-chromosome tip. (B) Imaginal disc cells; scale bar, 5 μm. The mod(mdg4) genotype designations are as in Fig 2. DAPI; 4,6-diamidino-2-phenylindole; Mod(mdg4), Modifier of mdg4; Su(Hw), Suppressor of Hairy wing.
Finally, we examined the immunostaining patterns in imaginal disc cells (Fig 3Bi–vi). In accordance with the published observations (Gerasimova et al, 2000) and our results in S2 cells (Fig 1), the wild-type nuclei contained multiple Mod+Su(Hw)-positive speckles (i). Again, only a cloudy Su(Hw) backdrop was barely visible in the mod(mdg4)u1 cells (ii). As expected, transgenic expression of ModWT in this null background (iii) restituted the wild-type staining pattern.
However, ModΔQ in the null background—already shown to restore all tested Su(Hw) insulator functions and to bind to all correct insulator sites—did not form or enter any nuclear speckles (iv), although we saw pronounced co-deposition of Mod and Su(Hw) in the cytoplasm. Interestingly, the mod(mdg4)+ background for ModΔQ (v) largely restored the double-positive nuclear speckles, attenuated the cytoplasmic Mod staining and virtually abolished cytoplasmic Su(Hw).
Conversely, in the mod(mdg4)T6 cells (vi), the ModΔC variant—which cannot functionally support the Su(Hw) insulators or associate with the corresponding chromatin sites—was seen to colocalize with Su(Hw) in nuclear speckles (which were fewer than in wild type but comparable with ModΔQ/mod(mdg4)+).
Conclusion
We reproduced the basic features and behaviour of putative ‘insulator bodies' (see Introduction) using the same or equivalent objects and experimental approaches. However, on analysis of the structural and functional data obtained using different forms of the essential insulator protein Mod(mdg4) in various genetic environments, we must conclude that the very presence of such bodies in the nucleus (or their absence, let alone their number, size or disposition) is irrelevant to the organization and function of authentic Su(Hw)-dependent genomic insulators, and thus cannot be regarded as evidence for insulator clustering.
It was not the aim of this study to scrutinize the composition, properties or actual purpose of these nuclear inclusions. The various bodies that can be visualized by microscopy in the nucleus (apart from the nucleolus) often appear to be depots for spare components. By analogy to the well-known promyelocytic leukaemia nuclear bodies comprising many unrelated proteins (Bernardi & Pandolfi, 2007), the so-called ‘insulator bodies' in Drosophila are perhaps aggregates of surplus proteins not immediately engaged in any function, and most certainly include proteins other than Mod(mdg4), Su(Hw) and CP190. We have preliminary data that the Drosophila analogue of the vertebrate CTC-binding factor (dCTCF), another zinc-finger protein required for activity of another type of insulator (Mohan et al, 2007), is also present in the same nuclear speckles. The findings recounted here—especially when viewed together with the well-known facts that elimination of Su(Hw) protein affects only female fertility and that the null mod(mdg4)u1 mutation does not apparently affect any trait in fly development—defy the idea of such ‘insulator bodies' as organizers of genome structure and function, notwithstanding the general plausibility and expedience of its higher order organization and management.
Methods
Transformation. The S2 cells cultured as described previously (Georgieva et al, 2001) were transformed using the Effectene Transfection Reagent as recommended by Qiagen (Hilden, Germany). The constructs, Drosophila strains, transgenic manipulations and phenotypic analyses are described in the supplementary information online.
Chromatin immunoprecipitation. The S2 cell suspension was treated with 1% formaldehyde at 20°C for 10 min. The nuclei were washed and lysed, and chromatin was sheared to an average length of 400 bp by sonication. X-ChIP was carried out as recommended by Upstate Biotechnology (Lake Placid, NY, USA), with 4 μg of antibodies against Flag (Sigma, St Louis, MO, USA). The negative control was 4 μg of nonspecific IgG from preimmune serum. The PCR primers are listed in supplementary Table S2 online.
Immunostaining. The S2 cells were grown on coverslips, stained with antibodies against Mod(mdg4)-67.2, Flag, Su(Hw) and lamin as described by Kyrshakova et al (2007), and examined using a Leica TCS SP2 confocal microscope. Squashed salivary gland specimens were prepared and immunostained as described by Platero et al (1996) and co-stained with 4,6-diamidino-2-phenylindole (DAPI). Diploid larval cells were treated according to Gerasimova et al (2000).
Antibodies. The specific antibodies and working dilutions were as follows: chicken anti-Mod(mdg4)-67.2 (1:500), a gift from P. Geyer; mouse anti-Flag (1:300) from Sigma; rabbit or mouse anti-lamin (1:500) and mouse anti-tubulin (1:2,000), gifts from P. Fisher; and rabbit antibodies against the Su(Hw) N-terminal domain (1:200), raised in our laboratory. The secondary antibodies were Cy3-conjugated anti-chicken (Amersham, Little Chalfont, UK), Alexa Fluor 488 anti-rabbit and Cy-5 anti-mouse (Invitrogen, Carlsbad, CA, USA) goat IgG, all used at a dilution of 1:500.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Materials
Acknowledgments
We are grateful to P. Geyer and P. Fisher for antibodies, and to D. Dorsett for Mod(mdg4)-67.2 cDNA. This study was supported by the Russian Foundation for Basic Research (07-04-01076) and the Molecular and Cell Biology Program, RAS; a stipend from the Centre for Medical Studies, Oslo University and the Presidential grant for young scientists (MK-3613.2007.4; to A.G.); and the International Research Scholar Award from the Howard Hughes Medical Institute (to P.G.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Brasset E, Vaury C (2005) Insulators are fundamental components of the eukaryotic genomes. Heredity 94: 571–576 [DOI] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R (2000) Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155: 141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2004) Boundary elements and nuclear organisation. Biol Cell 96: 617–629 [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20: 105–116 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gause M, Morcillo P, Dorsett D (2001) Insulation of enhancer–promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol 21: 4807–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P, Kozycina M (1996) Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S, Nabirochkina E, Dilworth FJ, Eickhoff H, Becker P, Tora L, Georgiev P, Soldatov A (2001) The novel transcription factor e(y)2 interacts with TAFII40 and potentiates transcription activation on chromatin templates. Mol Cell Biol 21: 5223–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG (2000) A chromatin insulator determines the nuclear localisation of DNA. Mol Cell 6: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Geyer PK, Corces VG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 865–873 [DOI] [PubMed] [Google Scholar]
- Geyer PK, Spana C, Corces VG (1986) On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J 5: 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG (2001) Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J 20: 2518–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovnin A, Birukova I, Romanova O, Silicheva M, Parshikov A, Savitskaya E, Pirrotta V, Georgiev P (2003) An endogenous Su(Hw) insulator separates the yellow gene from the Achaete–scute gene complex in Drosophila. Development 130: 3249–3258 [DOI] [PubMed] [Google Scholar]
- Golovnin A, Mazur A, Kopantseva M, Kurshakova M, Gulak PV, Gilmore B, Whitfield WGF, Geyer P, Pirrotta V, Georgiev P (2007) Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila. Mol Cell Biol 27: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdridge C, Dorsett D (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol 11: 1894–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover KK, Gerasimova TI, Chien AJ, Corces VG (1992) Dominant effects of suppressor of Hairy-wing mutations on gypsy-induced alleles of forked and cut in Drosophila melanogaster. Genetics 132: 691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK (2003) Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol 15: 259–265 [DOI] [PubMed] [Google Scholar]
- Kyrshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner PS, Tora L, Georgieva SG (2007) SAGA and novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 26: 4956–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG (2006) RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941 [DOI] [PubMed] [Google Scholar]
- Mohan M et al. (2007) The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26: 4203–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C-Y, Lei EP, Ghosh D, Corces VG (2004) The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK (2003) An endogenous Suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc Natl Acad Sci USA 100: 13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK (2006) Identification of genomic sites that bind the Drosophila suppressor of hairy-wing insulator protein. Mol Cell Biol 26: 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero JS, Sharp EJ, Adler PN, Eissenberg JC (1996) In vivo assay for protein–protein interactions using Drosophila chromosomes. Chromosoma 104: 393–404 [DOI] [PubMed] [Google Scholar]
- Ramos E, Ghosh D, Baxter E, Corces VG (2006) Genomic organisation of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172: 2337–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Fraser P (2005) Remote control of gene transcription. Hum Mol Genet 14: 101–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials