Abstract
The E3 ubiquitin ligase synoviolin (SYVN1) functions as an anti-apoptotic factor that is responsible for the outgrowth of synovial cells during the development of rheumatoid arthritis. The molecular mechanisms underlying SYVN1 regulation of cell death are largely unknown. Here, we report that elevated SYVN1 expression correlates with decreased levels of the protein inositol-requiring enzyme 1 (IRE1)—a pro-apoptotic factor in the endoplasmic reticulum (ER)-stress-induced apoptosis pathway—in synovial fibroblasts from mice with collagen-induced arthritis (CIA). SYVN1 interacts with and catalyses IRE1 ubiquitination and consequently promotes IRE1 degradation. Suppression of SYVN1 expression in synovial fibroblasts from CIA mice restores IRE1 protein expression and reverses the resistance of ER-stress-induced apoptosis of CIA synovial fibroblasts. These results show that SYVN1 causes the overgrowth of synovial cells by degrading IRE1, and therefore antagonizes ER-stress-induced cell death.
Keywords: rheumatoid arthritis, synoviolin, IRE1, ubiquitination, ER-stress-induced cell death
Introduction
Synoviolin (SYVN1), the mammalian orthologue of yeast Hrd1 (3-hydroxy-3-methylglutaryl reductase degradation), is a multispanning membrane protein with its carboxy-terminal RING-H2 finger domain located in the cytoplasm (Hampton et al, 1996). Many studies have shown that SYVN1 is an E3 ubiquitin ligase for endoplasmic reticulum (ER)-associated degradation (ERAD), a process involved in cellular quality control and adaptation of cells to ER stress induced by degrading misfolded proteins (Friedlander et al, 2000; Gardner & Swarbrick, 2000). ERAD has been implicated in many human diseases, including neurodegenerative diseases (such as Parkinson disease, cerebral ischaemia/hypoxia and Alzheimer disease) and diabetes (Yoshida, 2007).
SYVN1 has recently been identified as a rheumatoid regulator, and its expression is highly associated with the development of rheumatoid arthritis. Transgenic SYVN1 mice with overexpressed SYVN1 develop spontaneous arthropathy, whereas mice with reduced SYVN1 (SYVN1+/− mice) are resistant to collagen-induced arthritis (CIA; Amano et al, 2003). Recently, we showed that the pro-inflammatory cytokines interleukin-1β and tumour necrosis factor-α induce SYVN1 expression in mouse synovial fibroblasts through the ERK1/2–ETS1 pathway (Gao et al, 2006). The elevated expression of SYVN1 triggers synovial fibroblast outgrowth, which results from its anti-apoptotic activities (Amano et al, 2003; Yamasaki et al, 2005).
3-Hydroxy-3-methylglutaryl-coenzyme A reductase is the first, to our knowledge, identified substrate of Hrd1 and is tightly regulated by metabolic feedback control (Hampton et al, 1996). Hrd1 is also involved in ERAD of other ER proteins, such as CPY* and Sec61-2 (Bordallo et al, 1998; Taxis et al, 2002). Hrd1 uses the ubiquitin-conjugating enzyme 7 (Ubc7) as a major E2 but also cooperates with Ubc6 and Ubc1 in ERAD (Gardner & Swarbrick, 2000). To recruit misfolded proteins, Hrd1 forms a 1:1 complex with Hrd3, an ERAD factor with a large ER luminal domain that interacts with misfolded substrates (Denic et al, 2006; Gauss et al, 2006). Recently, it was reported that SYVN1 targets p53 for ubiquitination and degradation to regulate the cell cycle and apoptosis (Yamasaki et al, 2007), suggesting that SYVN1 can also target proteins that are not directly involved in ERAD for ubiquitination to regulate cell death and survival.
CIA, similar to rheumatoid arthritis, is characterized by overgrowth of articular synovial cells, the so-called ‘pannus'. At present, the underlying mechanisms of synovial hyperplasia are poorly understood. Here, we show that increased expression of SYVN1 correlates strongly with reduced inositol-requiring enzyme 1 (IRE1) and CHOP expression in the synovial fibroblasts during the development of CIA in mice. Interestingly, SYVN1 is physically associated with and catalyses IRE1 ubiquitination. IRE1 has kinase and RNase activities and transmits an ER stress signal to the cytosol by targeting proteins involved in ERAD. As a consequence, IRE1 regulates ER-stress-induced apoptosis (Mori et al, 1993; Shamu & Walter, 1996; Welihinda & Kaufman, 1996). These data help us to explain the novel mechanism by which SYVN1 regulates cell survival and cell death.
Results And Discussion
Decreased IRE1 and CHOP expression in CIA synovial cells
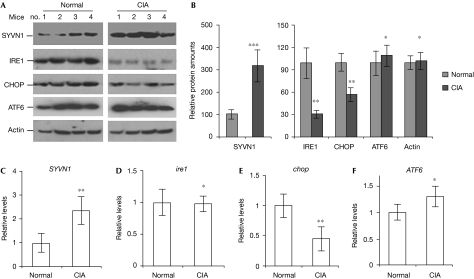
As an E3 ubiquitin ligase, SYVN1 might regulate cell proliferation and apoptosis by downregulating the proteins that are involved in synovial cell apoptosis. Thus, we determined the expression levels of proteins involved in ER-stress-induced cell death in synovial fibroblasts isolated from CIA and naive control mice. Intriguingly, we found that the expression levels of the pro-apoptotic proteins IRE1 and CHOP were significantly decreased (65% and 48% reduction, respectively) in CIA synovial cells. However, the expression of activating transcription factor 6 (ATF6), which is involved in ER stress responses, was not affected by CIA. In addition, the expression of PERK, a PKR (RNA-dependent protein kinase)-like ER kinase that attenuates protein translation in response to ER stress, did not change in CIA synovial fibroblasts. Consistent with previous reports (Yamasaki et al, 2005; Gao et al, 2006), the protein expression levels of SYVN1 increased about threefold in CIA synovial fibroblasts when compared with normal synovial fibroblasts (Fig 1A,B; supplementary Fig 1 online). These results indicate that upregulation of SYVN1 correlates with the decreased expression of the anti-apoptotic factors IRE1 and CHOP in synovial fibroblasts from CIA mice.
Figure 1.
Decreased IRE1 protein expression in synovial fibroblasts from mice with collagen-induced arthritis. (A) The expression of SYVN1, IRE1, CHOP, ATF6 and actin in the lysates of synovial fibroblasts from normal and CIA mice were determined by western blotting. (B) The band densities of each protein in (A) were quantified and shown. (C–F) Total RNA from synovial cells was purified and reverse transcribed into complementary DNA using oligo-dT primers. The cDNA levels of (C) SYVN1, (D) ire1, (E) chop and (F) ATF6 were determined by real-time PCR. Error bars represent data from three independent experiments. Student's t-test was used for statistical analysis; *P>0.05; **P<0.05; ***P<0.001. CIA, collagen-induced arthritis; IRE1, inositol-requiring enzyme 1; SYVN1, synoviolin.
Protein expression could be regulated at either messenger RNA or protein levels. Therefore, we examined the mRNA levels of ire1, chop, ATF6 and SYVN1 in normal and CIA synovial fibroblasts by using real-time PCR with the β-actin gene as a control. The mRNA expression levels of both ire1 and ATF6 were comparable between the two types of synovial fibroblast (Fig 1D,F), whereas chop mRNA levels were reduced in CIA fibroblasts (Fig 1E). By contrast, SYVN1 mRNA levels were increased about twofold in CIA fibroblasts (Fig 1C). These results indicate that the altered expression of both CHOP and SYVN1 is regulated at the mRNA level, whereas IRE1 expression is affected at post-mRNA levels during CIA.
SYVN1 interacts with IRE1
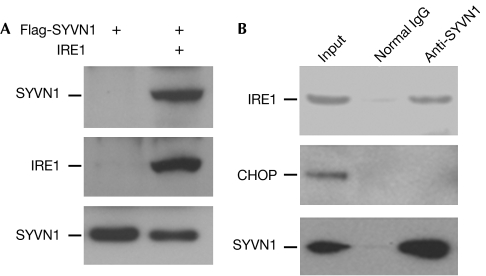
Our findings that the IRE1 protein, but not its mRNA, is decreased in CIA synovial fibroblasts in which SYVN1 expression is increased, prompted us to investigate whether IRE1 could be a target protein for SYVN1-induced ubiquitination. E3 ligases confer specificity to the ubiquitin system by directly interacting with the substrate proteins and helping to transfer ubiquitin to them. Thus, we investigated whether SYVN1 interacts with IRE1. IRE1 was detected in the immunoprecipitates from human embryonic kidney (HEK)293 cells when Flag-SYVN1 was co-transfected, but was not detected in control cells. This indicates that SYVN1 interacts with IRE1 (Fig 2A). This was confirmed by the observation that endogenous IRE1 but not CHOP interacted with SYVN1 in mouse synovial fibroblasts (Fig 2B), providing the possibility that SYVN1 might catalyse IRE1 ubiquitination.
Figure 2.
Synoviolin interacts with IRE1. (A) Flag-tagged SYVN1 and IRE1 expression plasmids were co-transfected into human embryonic kidney 293 cells. IRE1 protein in the lysates of transfected cells was immunoprecipitated with an IRE1 antibody. The interaction of SYVN1 with IRE1 was detected with a Flag antibody (top panel). The same membrane was reprobed with an IRE1 antibody (middle panel). The expression levels of SYVN1 in the whole-cell lysates were determined using a Flag antibody (bottom panel). (B) SYVN1 protein was immunoprecipitated from the lysate of synovial cells using anti-SYVN1 or normal rabbit IgG. The interaction of IRE1 (top panel) or CHOP (middle panel) with SYVN1 was detected with IRE1 and CHOP antibodies, respectively. The same membrane was reprobed with a SYVN1 antibody (bottom panel). IRE1, inositol-requiring enzyme 1; SYVN1, synoviolin.
SYVN1 induces IRE1 ubiquitination
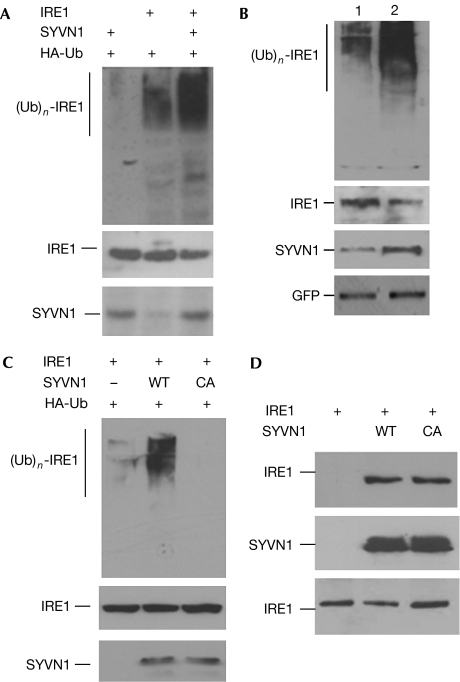
Next, we examined whether IRE1 is ubiquitinated by SYVN1 in HEK293 cells. Transient expression of IRE1 with haemagglutinin (HA)-Ub alone resulted in the formation of a slowly migrating species of IRE1 that was recognized by HA antibody, indicating that IRE1 can be ubiquitinated. Coexpression of IRE1 and SYVN1 markedly enhanced IRE1 ubiquitination (Fig 3A). Conversely, suppression of SYVN1 in mouse synovial fibroblasts by short interfering RNA (siRNA) inhibited endogenous IRE1 ubiquitination (Fig 3B). These data indicate that IRE1 is polyubiquitinated and that SYVN1 can act as an E3 ligase for IRE1.
Figure 3.
Synoviolin induces ubiquitination of IRE1. (A) Different combinations of IRE1, HA-Ub and SYVN1 expression plasmids were co-transfected into human embryonic kidney 293 cells. IRE1 proteins in the lysates of transfected cells were immunoprecipitated with an IRE1 antibody. The conjugation of ubiquitin onto IRE1 was detected with an HA antibody (top panel). The same membrane was reprobed with an IRE1 antibody (middle panel). The protein expression of SYVN1 in the whole-cell lysates was detected with a SYVN1 antibody (bottom panel). (B) Synovial fibroblasts from normal mice were transfected with SYVN1 short interfering RNA expression plasmids (lane 1) or control plasmids (lane 2). Two days after transfection, GFP+ cells were sorted, reseeded and then transfected with HA-Ub expression plasmid. IRE1 ubiquitination in those transfected cells was determined as described in (A) (top panel). The expression levels of IRE1 (second panel), SYVN1 (third panel) and GFP (bottom panel) in the whole-cell lysates were analysed by western blotting. (C) IRE1 and HA-Ub expression plasmids were co-transfected with SYVN1 or its C307A (CA) mutant. IRE1 ubiquitination in the transfected cells was determined as described in (A). (D) IRE1 expression plasmids were co-transfected with SYVN1 or SYVN1/CA mutant. SYVN1 proteins in the lysates of transfected cells were immunoprecipitated with a Flag antibody and the interacting IRE1 was detected with an IRE1 antibody (top panel). The same membrane was reprobed with a SYVN1 antibody (middle panel). The expression levels of IRE1 in the whole-cell lysates were used as controls (bottom panel). GFP, green fluorescent protein; HA, haemagglutinin; IRE1, inositol-requiring enzyme 1; SYVN1, synoviolin; WT, wild type.
The RING finger domain harbours the E3 ligase activity by recruiting ubiquitin-carrying E2s, and mutation of the conserved cysteine to alanine in the RING finger can disrupt its ligase activity (Joazeiro et al, 1999). We then assessed whether mutation of the conserved cysteine 307 to alanine in SYVN1 (SYVN1/C307A) affected its ligase activity towards IRE1 ubiquitination. Coexpression of SYVN1/C307A completely abolished its ability to promote IRE1 ubiquitination (Fig 3C). Analysis of these cell lysates showed comparable levels of protein expression between wild-type SYVN1 and its C307A mutant. The SYVN1/C307A mutant functioned in a dominant-negative manner because its expression inhibited IRE1 ubiquitination mediated by the endogenous SYVN1 (Fig 3C). This C307A mutant of SYVN1 did not affect its interaction with IRE1 (Fig 3D). These results indicate that the RING finger of SYVN1 is required for catalysing IRE1 ubiquitination.
SYVN1 contains many transmembrane regions in its amino terminus, as well as a RING finger and several proline-rich regions in its C terminus. A region between the transmembrane and RING finger, known as 53BD (p53-binding domain), recruits p53 for p53 ubiquitination and degradation (Yamasaki et al, 2007; supplementary Fig 2A online). The C terminus of SYVN1 is required for recruiting IRE1 because the RING finger with the C terminus of SYVN1 was sufficient to mediate its interaction with IRE1 and to promote IRE1 ubiquitination (supplementary Fig 2B,C online). Deletion of the C terminus of SYVN1 completely abolished its ability to promote IRE1 ubiquitination as well as its interaction with IRE1 (supplementary Fig 2D,E online). Thus, SYVN1 recruits IRE1 and p53 through different regions for their ubiquitination in the regulation of cell death.
SYVN1 promotes IRE1 degradation
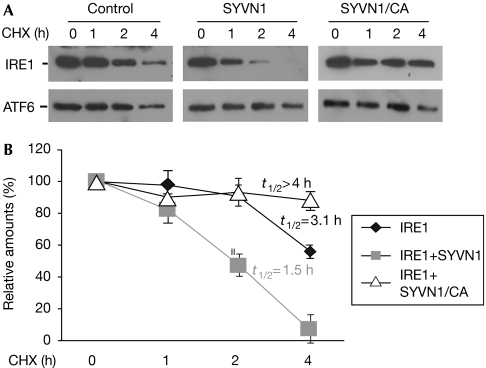
Ubiquitin conjugation to protein substrates can induce their degradation. We examined whether the coexpression of SYVN1 could affect the stability of IRE1. Fig 4 shows that IRE1 was gradually degraded in HEK293 cells with a half-life of about 3.1 h. Coexpression of SYVN1 facilitated IRE1 degradation by shortening IRE1 half-life to about 1.5 h. Together with our findings that SYVN1 catalyses IRE1 ubiquitination, these results indicate that SYVN1 promotes IRE1 degradation through the ubiquitin pathway. This was supported by the observation that expression of the RING finger mutant of SYVN1, which failed to mediate IRE1 ubiquitination, did not promote IRE1 degradation and even stabilized IRE1, possibly because this mutant dominant-negatively blocks endogenous SYVN1-mediated IRE1 degradation. As for the controls, expression of neither SYVN1 nor its RING finger mutant did not affect the stability of ATF6.
Figure 4.
Synoviolin promotes IRE1 degradation. (A) IRE1 and ATF6 expression plasmids were co-transfected with or without SYVN1 or SYVN1/C307A (CA) expression plasmids into human embryonic kidney 293 cells. Transfected cells were treated with cycloheximide (CHX) for different periods, as indicated. The protein stability of IRE1 (top panels) and ATF6 (bottom panels) was determined by western blotting. (B) The band densities of western blotting were quantified using Phosphorimager software. Error bars represent data from three independent experiments (mean±s.d.). The half-lives (t1/2) of IRE1 were calculated. IRE1, inositol-requiring enzyme 1; SYVN1, synoviolin.
SYVN1 knockdown restores IRE1 in CIA synovial cells
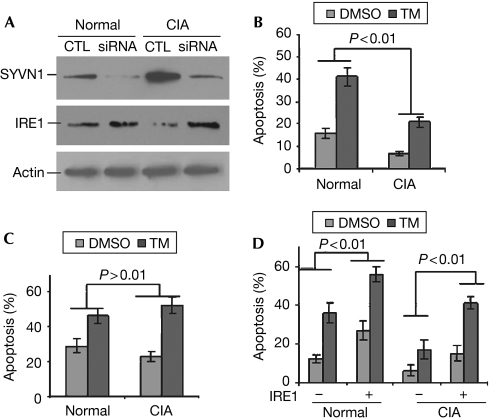
To determine further whether elevated SYVN1 expression in the synovial fibroblasts from CIA mice is responsible for reduced IRE1 expression, we used siRNA to knock down SYVN1 expression. siRNA significantly inhibited SYVN1 expression in the synovial fibroblasts from both normal and CIA mice. Interestingly, knockdown of SYVN1 restored IRE1 expression in those CIA synovial fibroblasts (Fig 5A). This finding suggests that elevated SYVN1 protein expression is responsible for the reduced IRE1 protein expression.
Figure 5.
Suppression of synoviolin expression in synovial cells of mice with collagen-induced arthritis restores IRE1 protein expression. (A) SYVN1 siRNA expression plasmids or control (CTL) plasmids were transfected into synovial fibroblasts from normal and CIA mice. Three days after transfection, GFP-positive cells were sorted and the expression of SYVN1 (top panel) and IRE1 (middle panel) was determined by SYVN1 and IRE1 antibodies, respectively. The protein levels of actin were analysed as loading controls (bottom panel). (B,C) Synovial fibroblasts, transfected with either (B) control plasmids or (C) SYVN1 siRNA expression plasmids, were treated with or without tunicamycin (TM; 10 μg/ml) for 18 h. Apoptosis was determined by annexin V staining by using flow cytometry. (D) Synovial fibroblasts from normal and CIA mice were transfected with or without IRE1 expression plasmids. Three days after transfection, cells were treated with TM or dimethyl sulphoxide (DMSO) as a control. The percentages of apoptotic cells were analysed as described in (B) and (C). Error bars (mean±s.d.) represent data from three independent experiments. Student's t-test was used for statistical analysis. CIA, collagen-induced arthritis; IRE1, inositol-requiring enzyme 1; siRNA, short interfering RNA; SYVN1, synoviolin.
SYVN1 is an anti-apoptotic factor the elevated expression of which is responsible for the outgrowth of synovial tissue during arthritis development (Amano et al, 2003). Here, we confirmed that the synovial fibroblasts from CIA mice were resistant to ER-stress-induced apoptosis when compared with normal mice (Fig 5B). Interestingly, when SYVN1 expression was inhibited, CIA synovial fibroblasts were susceptible to apoptosis induced by ER stress (Fig 5C). Together, these results indicate that elevated SYVN1 expression is involved in the reduced cell death of synovial fibroblasts, possibly by degrading IRE1 protein. We also noticed that suppression of SYVN1 expression in normal synovial fibroblasts only slightly increased their apoptosis induced by ER stress (Fig 5C). Complete loss of SYVN1 in mouse embryonic fibroblasts significantly enhanced ER-stress-induced apoptosis (Yagishita et al, 2005). This discrepancy could be due to the difference in cell type and/or the existence of a small fraction of SYVN1 in these knockdown cells.
Transient expression of IRE1 significantly enhanced apoptosis of synovial fibroblasts from both normal and CIA mice (Fig 5D), indicating that SYVN1 suppresses apoptosis of CIA synovial cells through IRE1 degradation. We also found that IRE1 overexpression had more robust effects on the apoptosis of synovial fibroblasts from normal mice (Fig 5D). This is possibly because the protein levels of the IRE1 E3 ligase, SYVN1, are much higher in CIA synovial cells, and SYVN1 suppresses apoptosis of synovial cells from CIA mice by targeting, either directly or indirectly, several substrates, such as CHOP.
Here, we discovered that the rheumatoid factor SYVN1 targets IRE1 for ubiquitination and degradation in the synovial fibroblasts. This targeted suppression of IRE1 function protects synovial cells from ER-stress-induced apoptosis. In addition, SYVN1 might indirectly inhibit CHOP expression. Both IRE1 and CHOP are pro-apoptotic factors during ER-stress-induced cell death (Mori et al, 1993; Welihinda & Kaufman, 1996). Thus, elevated SYVN1 expression facilitates the hyperproliferation of synovial tissues by antagonizing IRE1 and CHOP functions during the development of arthritis. The hyperproliferation of synovial tissues, which results in the increased production of synovial pro-inflammatory cytokines and the direct invasions and destruction of the bone and cartilage, is essential for rheumatoid arthritis development. Therefore, suppression of SYVN1-mediated IRE1 ubiquitination has great therapeutic potential for treating rheumatoid arthritis.
Methods
Induction of CIA in DBA/1 mice and isolation of synovial fibroblasts. CIA induction in DBA/1 mice and the isolation of synovial fibroblasts were performed as described previously (Gao et al, 2006). A detailed protocol is provided in the supplementary information online.
Co-immunoprecipitation, SDS–polyacrylamide gel electrophoresis and western blotting. A detailed protocol is provided in the supplementary information online.
Real-time PCR analysis. Synovial fibroblasts were collected and lysed in TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was extracted and was reverse transcribed using 200 U of Moloney murine leukaemia virus reverse transcriptase (Invitrogen) and oligo-dT primers. Quantitative real-time PCR was performed on a Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA, USA). The β-actin gene was used as a reference for sample normalization. Mouse SYVN1 primers and probe, as well as all β-actin primers and probe, were purchased as forms of Assay-on-Demand (Applied Biosystems). The standard amplification protocol of the manufacturer was followed.
Suppression of SYVN1 expression by siRNA. The 19-nt sequence of mouse Syn, 668-GGTCCTGCTGTACATGGCC-686, was inserted into the pSuper small RNA-expressing vector. A non-silencing control vector (pSuper-control) was constructed using a 19-nt sequence (GCGCGCTTTGTAGGATTCG) with no significant homology to any mammalian gene sequence (Brummelkamp et al, 2002). These plasmids were confirmed by DNA sequencing. Each plasmid was transfected into mouse synovial fibroblasts and the protein level of SYVN1 was detected by western blotting.
Apoptosis analysis. Synovial fibroblasts were plated in a 24-well dish at 5 × 104 cells per well 1 day before treatment. The cells were either left untreated or treated with 10 μg/ml tunicamycin for 18 h. Treated cells were trypsinized and collected with the supernatants, and cell apoptosis was determined by annexin V (Sigma-Aldrich, St Louis, MO, USA) staining by using flow cytometry.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
We thank Dr C.H. Evans (Center for Molecular Orthopaedics, Harvard Medical School, Boston, MA, USA) for providing protocols for mouse synovial fibroblast isolation. This work was partly supported by National Institutes of Health grant ES015010-01 to D.D.Z. and a research board grant from the University of Missouri, and by an investigator research award from the Arthritis Foundation to D.F.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amano T et al. (2003) Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev 17: 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9: 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2: 379–384 [DOI] [PubMed] [Google Scholar]
- Gao B, Calhoun K, Fang D (2006) The proinflammatory cytokines IL-1β and TNF-α induce the expression of Synoviolin, an E3 ubiquitin ligase, in mouse synovial fibroblasts via the Erk1/2–ETS1 pathway. Arthritis Res Ther 8: R172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM (2000) Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol 151: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E (2006) The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J 25: 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J (1996) Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 7: 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286: 309–312 [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J (1993) A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74: 743–756 [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P (1996) Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J 15: 3028–3039 [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Vogel F, Wolf DH (2002) ER–Golgi traffic is a prerequisite for efficient ER degradation. Mol Biol Cell 13: 1806–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ (1996) The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J Biol Chem 271: 18181–18187 [DOI] [PubMed] [Google Scholar]
- Yagishita N et al. (2005) Essential role of synoviolin in embryogenesis. J Biol Chem 280: 7909–7916 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Yagishita N, Tsuchimochi K, Nishioka K, Nakajima T (2005) Rheumatoid arthritis as a hyper-endoplasmic reticulum-associated degradation disease. Arthritis Res Ther 7: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S et al. (2007) Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin'. EMBO J 26: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H (2007) ER stress and diseases. FEBS J 274: 630–658 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information