Abstract
Protein arginine methyltransferase 5 (PRMT5) targets nuclear and cytoplasmic proteins. Here, we identified a nuclear protein, called cooperator of PRMT5 (COPR5), involved in the nuclear functions of PRMT5. COPR5 tightly binds to PRMT5, both in vitro and in living cells, but not to other members of the PRMT family. PRMT5 bound to COPR5 methylates histone H4 (R3) preferentially when compared with histone H3 (R8), suggesting that COPR5 modulates the substrate specificity of nuclear PRMT5-containing complexes, at least towards histones. Markedly, recombinant COPR5 binds to the amino terminus of histone H4 and is required to recruit PRMT5 to reconstituted nucleosomes in vitro. Consistently, COPR5 depletion in cells strongly reduces PRMT5 recruitment on chromatin at the PRMT5 target gene cyclin E1 (CCNE1) in vivo. Moreover, both COPR5 depletion and overexpression affect CCNE1 promoter expression. We propose that COPR5 is an important chromatin adaptor for PRMT5 to function on a subset of its target genes.
Introduction
Arginine methylation has been identified as one of the important modifications that participates in gene regulation by methylation on histone and non-histone proteins (Bedford & Richard, 2005). More than 200 proteins have been identified as potential targets of protein arginine methyltransferase 1–9 (PRMT1–9; Boisvert et al, 2003). These enzymes monomethylate and dimethylate their substrates by using S-adenosylmethionine (SAM) as a methyl donor. PRMT5, PRMT7 and PRMT9 are classified as type II enzymes, as they symmetrically dimethylate their substrates, whereas type I PRMTs asymmetrically dimethylate their substrates (Bedford & Richard, 2005; Lee et al, 2005; Cook et al, 2006).
These enzymes have been shown to affect several cellular processes (Bedford & Richard, 2005). Recent studies have implicated PRMT5 in various cytoplasmic processes, including modulation of signalling cascades and small nuclear ribonucleoprotein particle biogenesis (Pollack et al, 1999; Friesen et al, 2001). By contrast, the nuclear functions of PRMT5 have only just begun to be explored. Although it is known that PRMT5 regulates gene transcription by methylating histones H3 (R8) and H4 (R3), and by associating with several nuclear complexes (Fabbrizio et al, 2002; Kwak et al, 2003; Pal et al, 2003, 2004; Amente et al, 2006; Ancelin et al, 2006), the mechanisms by which PRMT5 is recruited to chromatin is unclear.
Here, we isolated a new PRMT5 partner, called cooperator of PRMT5 (COPR5), which is present in a subset of nuclear PRMT5-containing complexes and is involved in the regulation of the cyclin E1 (CCNE1) promoter. COPR5 binds to the amino terminus of histone H4 and nucleosomes, suggesting that it is as an important adaptor for PRMT5 to function on a subset of its target genes.
Results And Discussion
COPR5 is a new nuclear partner of PRMT5
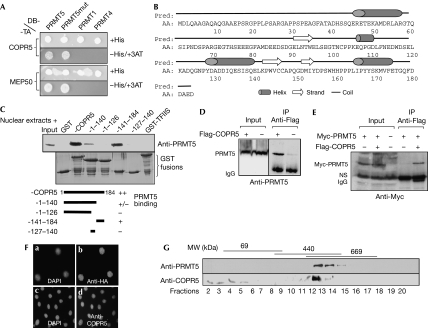
Two interactors were identified from a yeast two-hybrid screen using PRMT5 as bait: (i) the methylosome-associated protein MEP50, previously shown to interact with PRMT5 (Friesen et al, 2001), and (ii) the uncharacterized protein HSA272196, called COPR5 (Fig 1A). Yeast two-hybrid re-testing assays showed that COPR5 also interacted with an enzymatically inactive form of PRMT5 (PRMT5mut; Fig 1A). Human COPR5 spans 184 amino acids, is rich in acidic residues and shows neither a canonical protein domain nor significant similarity with other proteins (Fig 1B). Databases and northern blot analyses showed that COPR5 is expressed in most tissues and cell lines, and that COPR5 orthologues exist in other mammals and chicken (supplementary Fig S1 online). Pull-down assays confirmed a specific association between PRMT5 and COPR5. Glutathione-S-transferase (GST)-COPR5 efficiently pulled down: (i) in vitro-translated PRMT5 but not PRMT1 or PRMT4, two other nuclear PRMTs involved in transcription and chromatin remodelling (supplementary Fig S2 online), and (ii) endogenous PRMT5 from U2OS nuclear extracts (Fig 1C). Similar assays performed using various carboxy-terminal deletions of COPR5 indicated that the last 44 C-terminal residues were required to bind to PRMT5 (Fig 1C). Both ectopically expressed PRMT5 and endogenous PRMT5 were also co-immunoprecipitated from nuclear extracts with a Flag-COPR5-expressed protein (Fig 1D,E). A human COPR5 antibody (EF1) was developed (supplementary Fig S3 online) and used to localize COPR5 by immunofluorescence in U2OS cells. Both endogenous COPR5 and haemagglutinin (HA)-tagged COPR5 (overexpressed at low levels from a retroviral vector) were localized in the nucleus (Fig 1F). Finally, glycerol gradient size fractionation of nuclear extracts was probed with PRMT5 and COPR5 antibodies. Endogenous PRMT5 and COPR5 were distributed in overlapping high-molecular-weight fractions (Fig 1G), suggesting that they might be part of the same nuclear complex.
Figure 1.
COPR5, a nuclear protein that binds to PRMT5. (A) Yeast two-hybrid assay of preys (TA) coding for COPR5 and MEP50, a known PRMT5-associated protein, with baits (DB fusions) coding for PRMT5, an enzymatically inactive form of PRMT5 (PRMT5mut), PRMT1 and PRMT4. Interaction-dependent reporter activation (GAL1:HIS3-dependent prototrophy) was assessed in the presence of 50 mM 3-aminotriazole (3AT). (B) Predicted (Pred) primary and secondary structures (AA) of human COPR5. (C) Cellular PRMT5 is pulled down by GST-COPR5 proteins. Similar amounts of GST, GST-TFIIS, GST-COPR5 or truncated COPR5 fusion proteins (lower panel) were bound to beads and incubated with U2OS nuclear extracts. Cellular proteins retained on beads and a fraction of the nuclear extract (input) were probed by immunoblotting with a PRMT5 antibody (upper panel). (D) Co-immunoprecipitation (IP) of ectopically expressed PRMT5 and COPR5. IPs were performed with a Flag antibody on the nuclear extracts from U2OS cells co-transfected with Myc-tagged PRMT5 together with either Flag-tagged COPR5 or control vector. Three per cent of input extracts (input) and precipitates (IP anti-Flag) were immunoblotted with a Myc antibody. (E) Endogenous PRMT5 co-immunoprecipitates with ectopically expressed COPR5. IPs were performed with a Flag antibody on the nuclear extracts from U2OS cells transfected with either Flag-COPR5 or control vectors. Three per cent of input extracts (input) and precipitates (IP anti-Flag) were immunoblotted with a PRMT5 antibody. (F) Nuclear localization of endogenous and ectopically expressed COPR5. (a,b) HA-COPR5-expressing cells or (c,d) control U2OS cells were labelled with (a,c) DAPI and probed by immunofluorescence with (b) HA or (d) COPR5 antibodies, respectively. (G) Size fractionation of nuclear complexes containing COPR5 or PRMT5. U2OS nuclear extracts and high-molecular-weight (MW) markers were fractionated on a glycerol gradient. Fractions were immunoblotted with PRMT5 and COPR5 antibodies. COPR5, cooperator of PRMT5; DAPI, 4,6-diamidino-2-phenylindole; GST, glutathione-S-transferase; HA, haemagglutinin; MEP50, methylosome-associated protein 50; NS, non specific; PRMT5, protein arginine methyltransferase 5; TFIIS, transcription factor IIS.
Together these results indicate that COPR5 is a new nuclear partner of PRMT5.
COPR5 favours H4R3 PRMT5-dependent methylation
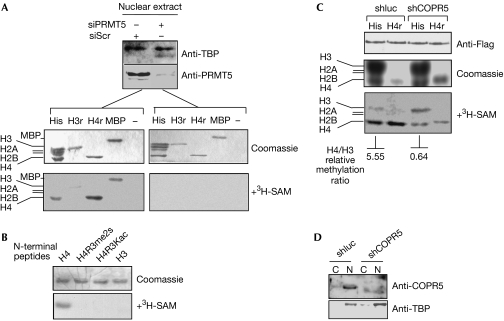
Next, we investigated the effect of COPR5 on the enzymatic activity of PRMT5. First, in vitro methylation assays were performed on nuclear PRMT5 pulled down by GST-COPR5 beads. SAM-dependent methyltransferase activity was detected towards myelin basic protein (MBP) and histone H4 but not, or only very faintly, towards histone H3, although all three proteins have been described as PRMT5 substrates (Fig 2A). GST-COPR5 was not methylated under this condition, although it contains several RG motifs, a sequence targeted by PRMT5 in other proteins. This activity was dependent on PRMT5 (supplementary Fig S4 online), as it was not detected in either GST-COPR5, which lacks the PRMT5-binding site (data not shown) or COPR5 beads incubated with PRMT5-depleted nuclear extracts (Fig 2A). This preference of the COPR5–PRMT5 complex towards histone H4R3 was confirmed, by using, as substrates, N-terminal histone H3 and H4 synthetic peptides that included the PRMT5 methylatable residues H3R8 and H4R3 (Fig 2B; Pal et al, 2004). Consistent with data on total histones, methylation of only the H4 peptide was observed and was blocked by modifications or substitutions at position R3 (Fig 2B).
Figure 2.
Specificity of methylation mediated by the COPR5–PRMT5-containing complex. (A) GST-COPR5 beads were incubated with nuclear extracts from U2OS cells treated with either siRNA directed against PRMT5 (siPRMT5) or scrambled siRNA (siScr). PRMT5 depletion was assessed by immunoblotting (upper panel). Cellular proteins bound to COPR5 beads were used in 3H-SAM-dependent methylation assays in vitro in the absence (−) or presence of substrates: bulk of purified histones (His), recombinant H3r and H4r and MBP. Substrates were visualized by Coomassie staining (middle panels) and their methylation analysed by fluorography (lower panels). (B) COPR5-bound PRMT5 methylates histone H4 in R3. Methylation assays were performed as in (A) towards synthetic peptides corresponding to the amino termini of H4, H4 modified on R3 by dimethylation (H4R3me2s), H4 with an acetylated lysine in the place of R3 (H4R3Kac) or H3. Similar amounts of peptides (upper panel) were methylated and analysed by fluorography (lower panel). (C) Cellular depletion of COPR5 modified the activity and specificity of nuclear PRMT5 towards histones. U2OS cells were infected with retroviral shRNAs directed against COPR5 (shCOPR5) or control target (shluc), and transfected with Flag-PRMT5. Anti-Flag immunoprecipitation of PRMT5 was performed on nuclear extracts prepared from these cells (upper panel) and precipitates were used to methylate either total histones (His) or recombinant H4r in vitro. Substrates were analysed for protein content by Coomassie staining (middle panel) and for methylation by fluorography (lower panel). (D) COPR5 depletion by shCOPR5 was assessed by immunoblotting on nuclear (N) and cytoplasmic extracts (C). As a control of fractionation, samples were also probed for the nuclear protein TBP. COPR5, cooperator of PRMT5; GST, glutathione-S-transferase; MBP, myelin basic protein; PRMT5, protein arginine methyltransferase 5; SAM, S-adenosylmethionine; shRNA, short hairpin RNA; siRNA, short interfering RNA; TBP, TATA-binding protein.
To evaluate the effect of COPR5 on the substrate specificity of PRMT5 in vivo, we compared the nuclear methyltransferase activity of a Flag-PRMT5 construct transfected in cells treated with either control (shluc) or COPR5 short hairpin RNAs (shRNAs). Flag-PRMT5 proteins were immunoprecipitated from nuclear extracts and histone methylation assays were performed on precipitates. As described by Pal et al (2004), the pool of nuclear PRMT5 immunoprecipitated from control cells (shluc) could trigger both H3 and H4 methylation, although more efficiently on H4 than on H3. By contrast, we reproducibly observed that this preference for H4 was abrogated in COPR5-depleted (shCOPR5) extracts and the methyltransferase activity of PRMT5 precipitates towards H4 was strongly reduced, whereas that directed towards H3 was either unaffected or increased (Fig 2C,D).
Together, these results suggest that COPR5 modulates the substrate specificity and/or activity of nuclear PRMT5-containing complexes towards histones.
CCNE1 promoter repression by PRMT5 involves COPR5
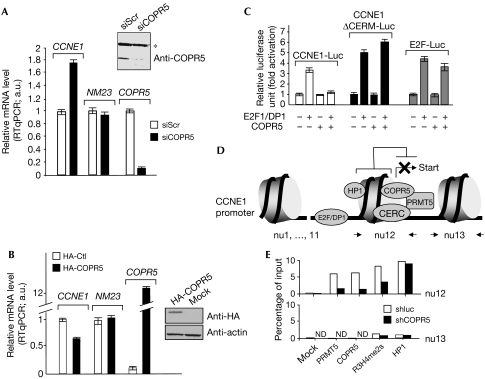
PRMT5 was found in several nuclear complexes that regulate transcription. Notably, its presence was detected on the transcription start site of the CCNE1 gene, where it contributes to repression through CERC, an atypical E2F-containing repressor complex (Fabbrizio et al, 2002), and on the promoter of the tumour suppressor gene NM23, which is regulated by a PRMT5-containing human SWI–SNF chromatin remodelling complex (Pal et al, 2004). To study the role of COPR5 in the nuclear functions of PRMT5, we monitored the effect of COPR5 depletion or overexpression on CCNE1 and NM23 gene expression. COPR5 short interfering RNA (siRNA)-mediated depletion led to a less than twofold increase (1.8 ×) in the levels of CCNE1 messenger RNA (Fig 3A), whereas infection of cells with a retroviral vector encoding COPR5 led to a significant, although incomplete, reduction in the levels of CCNE1 mRNA (Fig 3B). By contrast, no such variation was detected in the levels of NM23 mRNA (Fig 3A,B), which is consistent with the involvement of COPR5 in some but not all nuclear functions of PRMT5. These effects led us to investigate the effects of COPR5 on the promoter activity of CCNE1. Luciferase reporter plasmids driven by the mouse CCNE1 promoter were co-transfected with their activators, transcription factors E2F1/DP1, in the presence or absence of COPR5. shCOPR5-mediated depletion of COPR5 potentiated transactivation of the CCNE1 promoter by E2F/DP1 (supplementary Fig S5 online), whereas its overexpression led to a marked decrease in E2F-stimulated transcription of the CCNE1 reporter (Fig 3C). This was not observed with a CCNE1 promoter construct mutated on the CERC-binding site (CERM (Cyclin E Repressor Module); required to detect the effect of PRMT5 on CCNE1 promoter) or with an E2F site-driven synthetic promoter that does not respond to PRMT5 (Fabbrizio et al, 2002).
Figure 3.
COPR5 affects CCNE1 gene transcription and is required for the recruitment of PRMT5 to this gene. (A) COPR5 depletion enhances CCNE1 messenger RNA levels. Relative mRNA levels of COPR5 and of two PRMT5 target genes, CCNE1 and NM23, measured in U2OS cells treated for 72 h by either scrambled (siScr) or COPR5-specific (siCOPR5) siRNAs are shown. RTqPCR values were normalized to RPLP0 RNA levels and expressed as fold activation of siScr-treated samples. Depletion of COPR5 was confirmed by immunoblotting. The asterisk indicates nonspecific bands. (B) COPR5 overexpression decreases CCNE1 mRNA levels. Relative mRNA levels were determined as in (A) in cells infected with either HA-COPR5 or mock retroviral particles (HA-Ctl). HA-COPR5 expression was analysed by immunoblotting. Protein samples were prepared 72 h after infection and puromycin selection. (C) COPR5 overexpression inhibits E2F-dependent activation of the CCNE1 promoter. U2OS cells were transfected with a combination of vectors for E2F1/DP1 and COPR5, together with CMV-β-galactosidase (βgal) and luciferase reporters driven by wild-type CCNE1 (CCNE1Luc), mutated CCNE1 (ΔCERM-Luc) or synthetic E2F (E2F-Luc) promoters. Results, normalized to βgal activity, are expressed as fold activation of the values obtained with the reporter genes alone. (D) Nucleosomal structure of the CCNE1 promoter regions nu12 and nu13, and proteins reported to associate with this region. (E) COPR5 depletion decreases PRMT5 recruitment at the CCNE1 start site region in vivo. ChIPs were performed using the indicated antibodies on chromatin prepared from shCOPR5- or shluc-treated U2OS cells. DNA–protein immunoprecipitates and 5% of input chromatin were deproteinized and analysed by qPCR for human CCNE1 promoter regions nu12 and nu13. PCR values of ChIPs are presented as a percentage of the PCR values of input. Data correspond to a single experiment representative of three independent experiments. CCNE1, cyclin E1; ChIP, chromatin immunoprecipitation; COPR5, cooperator of PRMT5; Ctl, control; HA, haemagglutinin; ND, not detectable; nu12, nucleosome 12; nu13, nucleosome 13; PRMT5, protein arginine methyltransferase 5; RTqPCR, real-time quantitative PCR; shCOPR5, short hairpin RNA directed against COPR5; siRNA, short interfering RNA.
Finally, chromatin immunoprecipitations (ChIPs) were performed to investigate, in vivo, the presence of COPR5 at the CCNE1 gene and its role in the recruitment of PRMT5. In previous studies, we mapped nucleosome arrays in the CCNE1 promoter region (Morrison et al, 2002; Fig 3D). By using ChIPs, we investigated the nucleosome 12 (nu12) region—which contains CERM and the transcription start site of CCNE1—and, as a control, the adjacent nu13 region. HP1 immunoprecipitates were also included as a control in this analysis, as it was shown to associate with a region overlapping nu12 (Nielsen et al, 2001). This analysis confirmed the presence of PRMT5 and HP1 on nu12 and, for the first time, to the best of our knowledge, the presence of COPR5- and PRMT5-mediated histone H4R3 symmetrical dimethylation (Fig 3E). Markedly, COPR5 depletion strongly reduced PRMT5 recruitment, whereas that of HP1 was unaffected. As a consequence of PRMT5 loss, a decreased H4R3me2s mark at nu12 was concomitantly observed.
These results indicate that the recruitment and function of PRMT5 at the CCNE1 gene requires COPR5.
COPR5 links PRMT5 to histones
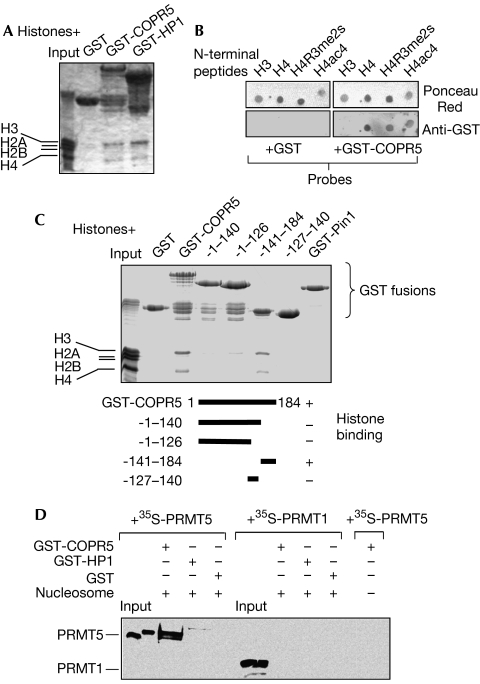
While studying the mechanism by which COPR5 might recruit PRMT5 on chromatin, we observed that COPR5 was a histone-binding protein as potent as the archetypal histone-binding protein HP1 to pull down histone H3 and H4 dimers from a bulk of purified histones (Fig 4A). Overlay experiments on N-terminal H3 and H4 peptides spotted onto membranes confirmed the direct interaction of COPR5 with H4 but not H3 (Fig 4B). Notably, neither an R3 dimethylated nor a K-acetylated peptide affected this histone H4–COPR5 interaction (Fig 4B). However, we cannot rule out the possibility that this association could be modulated by other modifications or interactions with the histone core. Mapping of the histone-binding domain of COPR5 suggests that the last 44 amino acids are essential for this interaction (Fig 4C). As this sequence also corresponds to the PRMT5-binding region, we suggest that COPR5 might function as an adaptor protein that bridges PRMT5 to chromatin. A similar bridging function has been proposed for HP1, which connects nucleosomal histones to the lysine methyltransferase SUV39H1, which can promote repression of genes such as CCNE1 through H3K9 methylation (Nielsen et al, 2001). To gain more insights into this, nucleosomes were reconstituted in vitro around a 5′-biotinylated DNA corresponding to CCNE1 nu12 and then bound to streptavidin-agarose beads to pull down GST-COPR5, GST-HP1 or GST alone. PRMT5, but not PRMT1, was retained on GST-COPR5-coated nucleosomes, whereas neither GST- nor GST-HP1-coated nucleosomes interacted with PRMT5 (Fig 4D).
Figure 4.
COPR5 is a histone-binding protein and recruits PRMT5 to nucleosomes in vitro. (A) Histone dimers H4 and H3 are retained on GST-COPR5 beads. GST, GST-COPR5 or GST-HP1 proteins bound to glutathione beads were incubated with total histones. Bound fractions were analysed by SDS–polyacrylamide gel electrophoresis and Coomassie blue staining. (B) COPR5 binds to the amino terminus of histone H4 in vitro. GST and GST-COPR5 proteins were used as probes for far-western overlays performed on N-terminal H3 and H4 synthetic peptides spotted on membranes. GST fusion proteins retained on membranes were detected using a GST antibody (lower panels). Ponceau Red staining (upper panels) shows peptides bound to membranes. (C) Mapping of COPR5 domain required for histone binding. Full-length and deleted GST, GST-COPR5 and GST-Pin1 proteins bound to beads were incubated with histones and analysed as in (A). (D) COPR5 triggers PRMT5 recruitment on reconstituted nucleosomal particles. Mononucleosomes were reconstituted in vitro using purified histones and a biotinylated DNA fragment corresponding to the start site region of the CCNE1 gene. Biotinylated nucleosomes were then bound to streptavidin-agarose beads and incubated with GST-COPR5, GST-HP1 or GST proteins. After stringent washings, beads were incubated with in vitro-translated 35S-labelled PRMT1 or PRMT5. PRMTs retained on beads were visualized by fluorography. Input: 20% of the PRMTs added to beads. COPR5, cooperator of PRMT5; GST, glutathione-S-transferase; PRMT5, protein arginine methyltransferase 5.
These results indicate that COPR5 binding to the N terminus of histone H4 promotes the association of PRMT5 with chromatin.
In conclusion, we have identified a new PRMT5 nuclear partner, COPR5, required for PRMT5 recruitment at the CCNE1 locus. We propose that COPR5, through its histone-binding domain, functions as an adaptor between chromatin and the nuclear PRMT5 complexes involved in H4R3 symmetrical dimethylation at specific loci, including CCNE1, a crucial regulator of cell proliferation. This opens new directions aiming to investigate the role of COPR5–PRMT5-containing nuclear complexes in chromatin modifications and cell proliferation.
Methods
Two-hybrid screening. Yeast strain MAV103 expressing pPC97-PRMT5 (human PRMT5 (aa 5–637) fused to GAL4DB) was transfected with a WI38 human fibroblast complementary DNA pPC86 preys library. Interactors were selected from 2 × 106 yeast colonies in the presence of 50 mM 3-aminotriazole, essentially as described previously (Sardet et al, 1995). Preys were retested against pPC97-PRMT5mut (pPC97-PRMT5 with mutations G367A and R368A in PRMT5) or full-length human PRMT1 and PRMT4 in pPC97.
Plasmids, sh/siRNAs, antibodies and reagents. Details of all constructs are available on request. Full-length and deleted COPR5-GST fusions were in pGEXPL2. The pSIREN (BD, Erembodegem, Belgium) retroviral vector was used to direct the synthesis of shRNA against human COPR5 (CAGTCTGTTGTTGTTCTTA). For Flag- or HA-tagged overexpression of COPR5, full-length COPR5 was cloned in pBABE-HA or pBABE-Flag retroviral vectors and viral particles were produced using standard protocols. CCNE1-Luc, E2F-Luc and CCNE1ΔCERM-Luc reporter plasmids and Myc- or Flag-tagged pCDNA3-PRMT expression vectors were as described previously (Fabbrizio et al, 2002; El Messaoudi et al, 2006). siRNAs against human COPR5 and PRMT5 are (+) GGCUAUGGAUCGACUAGCCdTdT and (+) CCGCUAUUGCACCUUGGAAdTdT, respectively. Purified bulk histones, recombinant histones H3 and H4, and MBP were purchased from Upstate (UK). The following antibodies were used: Myc (9E10), Flag (M2; Sigma-Aldrich, Saint Quentin, Fallavier, France), HP1α and HP1γ (1519s2 and 42s2; Upstate), H4R3me2S (5823; Abcam, Cambridge, UK), PRMT5 (07405; Upstate; 3766; Abcam), HA (12CA5) and COPR5 (EF1, directed against the C-terminal sequence of COPR5 (PYYSKMVFETGQFDDAED); see supplementary Fig S3 online). N-terminal H3 and H4 peptides were synthesized by Eurogentec (Seraing, Belgium).
Cell culture, transfections, RTqPCR and reporter assays. All cells were grown in DMEM (Sigma-Aldrich)/10% FBS. Transfection, retroviral infection, luciferase reporter assays and real-time quantitative PCR (RTqPCR) in U2OS cells were performed as described previously (El Messaoudi et al, 2006). The following oligonucleotides were used for RTqPCR: COPR5f (TGGAACACAGAGCATTCCTAATGA), COPR5r (TCATCCATGGCAAAGCCTTTC), NM23f (GCGTACCTTCATTGCGATCAA), NM23r (CCTTTCTGCTCAAAACGCTTG), RPLP0f (CGACCTGGAAGTCCAACTAC) and RPLP0r (CCTTTCTGCTCAAAACGCTTG). CCNE1 oligonucleotides have been described previously (El Messaoudi et al, 2006).
Chromatin immunoprecipitation. ChIPs were performed as described previously (El Messaoudi et al, 2006). Formaldehyde-crosslinked chromatin was immunoprecipitated with indicated antibodies and probed by PCR using CCNE1 primers nu12f (TGAGGGGCTCGCAGCCCTCG), nu12r (CCCGGCTTCGAGCGGGACAT), nu13f (GGTGTAGGGGCAGGCGC) and nu13r (CCCGGCAGGCGGCGGCGG).
GST pull-down assay, immunoprecipitations and immunofluorescence. GST pull-down assays: 10 μg of each recombinant GST fusion protein, bound to glutathione Sepharose 4B (Amersham Pharmacia, Buckinghamshire, UK), was incubated in PBS and 0.05% Tween 20 with (i) in vitro-translated [35S]methionine-labelled proteins synthesized (TNT kit; Promega, Charbonnieres, France) from 1 μg of pCDNA3-PRMTs, or (ii) 300 μg of U2OS nuclear extracts or (iii) 10 μg of bulk histones. Bound proteins were separated by SDS–polyacrylamide gel electrophoresis and analysed by autoradiography, immunoblotting or Coomassie staining, as indicated. For GST pull-down in the presence of histones, washing was performed in PBS, 0.5 M NaCl and 0.05% Tween 20. Co-immunoprecipitations of PRMT5 and COPR5 from cells were carried out on 1 mg of U20S nuclear extracts. For COPR5 localization, formalin/methanol-fixed cells were probed with a COPR5 (EF1) or HA (12CA5) antibody, and with suitable Texas-Red-labelled secondary antibodies (Jackson ImmunoResearch Lab, Montlucon, France).
Nucleosome reconstitution, methylation and overlay assays. Nucleosomes were reconstituted in vitro around a DNA fragment of the CCNE1 promoter encompassing nu12 (amplified by PCR with nuCEf biot-GTAAAAGAACACGCCCCCCG and nuCEr TGTCGAGCCGGCTGCTCCTG) and bound to streptavidin beads M280 (Dynal, Oslo, Norway), as detailed in the supplementary information online. Methylation and overlay assays on H3 and H4 proteins, and N-terminal peptides are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Figs S1–S5
Acknowledgments
We thank E. Julien and L. Le Cam for helpful discussions. This work was realized with the institutional support of the CNRS and the support of grants to C.S. from the French Ministry of Sciences, the Agence National pour la Recherche, the Association pour la Recherche contre le Cancer and the La Ligue Contre le Cancer. S.E.M. was supported by a fellowship from La Ligue Contre le Cancer. M.L. was supported by a Ministère de l'Enseignement Supérieur et de la Recherche (MESR) scholarship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amente S, Napolitano G, Licciardo P, Monti M, Pucci P, Lania L, Majello B (2006) Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett 579: 683–689 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation: an emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Richard S (2003) A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics 2: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S (2006) FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun 342: 472–481 [DOI] [PubMed] [Google Scholar]
- El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford M, Sardet C (2006) PRMT4/CARM1 is a cyclin E1 gene regulator. Proc Natl Acad Sci USA 36: 13351–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrizio E et al. (2002) Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep 3: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G (2001) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21: 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB (2003) Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell 11: 1055–1066 [DOI] [PubMed] [Google Scholar]
- Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S (2005) PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem 280: 3656–3664 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Sardet C, Herrera RE (2002) Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol Cell Biol 22: 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S (2003) mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol 23: 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24: 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem 274: 31531–31542 [DOI] [PubMed] [Google Scholar]
- Sardet C, Vidal M, Cobinik D, Geng Y, Onufrik C, Chen A, Weinberg RA (1995) E2F4 and E2F5, two members of the E2F family are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA 92: 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Figs S1–S5