Abstract
A crucial part of a successful systems biology experiment is an assay that provides reliable, quantitative measurements for each of the components in the system being studied. For proteomics to be a key part of such studies, it must deliver accurate quantification of all the components in the system for each tested perturbation without any gaps in the data. This will require a new approach to proteomics that is based on emerging targeted quantitative mass spectrometry techniques. The PeptideAtlas Project comprises a growing, publicly accessible database of peptides identified in many tandem mass spectrometry proteomics studies and software tools that allow the building of PeptideAtlas, as well as its use by the research community. Here, we describe the PeptideAtlas Project, its contents and components, and show how together they provide a unique platform to select and validate mass spectrometry targets, thereby allowing the next revolution in proteomics.
Keywords: systems biology, targeted proteomics, selected reaction monitoring
Introduction
One of the crucial aspects of a successful systems biology study is to perturb a system in a controlled manner to obtain quantitative measurements for each component at each perturbation. Such complete data sets are then used to establish or improve mathematical models that simulate the system and make predictions about its behaviour. So far, gene expression arrays have been the most frequently used data collection technology in systems biology. Modern arrays and related protocols are able to provide accurate, reproducible transcript abundances for each of the genes in the system being studied.
For proteomics to have a key role in systems biology experimentation, it must be able to deliver accurate, absolute or relative quantification for all relevant proteins for each perturbation performed. However, given the current technical limitations, this has not been feasible using the standard method of shotgun proteomics practiced in the field. Although shotgun proteomics has revolutionized the high-throughput study of proteins, and has allowed the identification and quantification of thousands of proteins per experiment, it suffers from several drawbacks that hinder its successful application in systems biology experiments. First, the dynamic range of protein abundance observation in shotgun proteomics experiments is still limited to just a few orders of magnitude; therefore, it is often difficult to observe low-abundance proteins of interest among high-abundance proteins. Second, even at observable abundances, proteins present in a sample are often not observed in a shotgun experiment owing to various technical limitations. An undesirable consequence of this is the inability to determine a reliable detection threshold and thereby provide reasonably accurate upper limits for proteins not observed in a sample. This often leaves multiple-sample perturbation experiments, such as time-course or dose-response experiments, with missing measurements for several proteins, which severely hinders the desired abundance analysis. Third, as the brute-force nature of the method essentially prevents the researcher from pre-determining which proteins to observe, much time and effort can be wasted on acquiring and analysing data that will probably not answer the biological question being investigated. Therefore, although shotgun proteomics has been highly successful in determining the protein composition of biological samples and for suggesting hypotheses about their function, it is not an optimal platform for systems biology or any other scenario that requires quantitative and reproducible data sets.
Emerging targeted proteomics workflows provide a compelling solution to the problem (Kuster et al, 2005). Targeted proteomics represents a different approach to obtaining proteome-wide qualitative and quantitative information. Rather than simply programming the instrument to collect data on whatever ions are detectable—which is analogous to expressed sequence tag (EST) sequencing in the field of genomics—the targeted proteomics approach starts with a list of precise elements that will be probed, as is the case for microarray experiments in transcriptomics. The mass spectrometer is set to monitor unique signals from targets specified before the experiment, eliminating the inherent redundancy in data collection and analysis in discovery-oriented experiments. This not only results in an increase in sensitivity, but also ensures that the same targets can be measured across many runs, providing valuable opportunities for qualitative and quantitative sample comparisons essential for answering interesting biological questions. Therefore, targeted proteomics workflows, described below, hold great promise in the transition of proteomics from a discovery-oriented technique to a robust and quantitative method suitable for hypothesis-driven studies in systems biology.
Targeted proteomics workflows
A targeted proteomics workflow has essentially two requirements. The first is a method by which specific peptides or proteins can be reliably quantified across several experiments. Faster mass spectrometers with more advanced instrument control software are now becoming available (Stahl-Zeng et al, 2007) allowing studies in which the relative abundances of hundreds of peptides can be measured by selected reaction monitoring (SRM; also known as multiple reaction monitoring (MRM)) techniques with remarkable sensitivity and throughput.
Although SRM is not a new technique for mass spectrometry, it has recently emerged as a valuable technique for proteomics. In SRM, the instrument—typically a triple quadrupole—is instructed to repeatedly sweep through a list of precursor, product ion m/z pairs, called transitions, and to record the intensity of fragments that pass through both isolation windows. Assuming that each transition, or set of few transitions, uniquely identifies a peptide, this allows the instrument to monitor a specific set of target peptides of interest instead of blindly sequencing the most intense peaks. SRM yields an ion chromatogram for each transition, and the area under the curve of the chromatogram provides a quantitative measurement for each desired peptide and protein. The instrumental aspect of targeted proteomics has been reviewed previously (Domon & Aebersold, 2006; Kuster et al, 2005).
The other requirement of a targeted proteomics workflow is a method to compile the list of target proteins and peptides, and the necessary attributes of these targets to facilitate the measurements. In the context of SRM techniques, this suggests a mechanism to generate high-quality lists of targets and corresponding SRM transitions to feed into the instruments. This procedure can be divided into several steps.
The first step is target protein selection. Ideally, one would like to target a whole proteome in an experiment to be able to answer systems-wide biological questions. It is, however, not a feasible goal in this early stage of proteomics, just as it was not possible to assay all genes in the early days of microarray transcriptomics. How the target protein list is defined depends on the aim of the study.
The second step is target peptide selection. From the list of targeted proteins, the exact target peptides must be determined. This is not a trivial problem because it is well known that not all peptides derived from a protein can be easily observed in mass spectrometry platforms. In addition, some peptides are common to multiple proteins or protein isoforms and so cannot be used as conclusive evidence for the presence or quantification of a single protein. Therefore, ideal target peptides must combine the attributes of mass spectrometry observability and unique protein mapping. Determining these so-called proteotypic peptides (Mallick et al, 2007) is one of the main challenges in targeted proteomics.
The third step is SRM transition selection for the target peptides. Primarily this involves the pre-determination of the most intense and most reproducible fragment ions that can uniquely identify the target. In addition, for increased throughput, the approximate chromatographic retention time of the peptide can be used to limit the time span in which the instrument is set to monitor the transitions, freeing it up to detect other peptides at other times. Such scheduled SRM workflows have been shown to increase markedly the number of transitions that can be monitored without compromising sensitivity (Stahl-Zeng et al, 2007).
The final step of the ideal targeted proteomics assay is the ability to obtain absolute quantification of the target peptide and hence the protein of interest. The most commonly practiced approach to achieving this goal involves injecting reference peptides, or synthetic reference proteins, of known concentration together with the sample to be analysed, so that the absolute quantification of the target peptide can be inferred from the relative signal intensities of the target and reference peptides. These reference peptides are usually isotopically heavy forms of the targeted peptides and can be synthesized in various ways (Gerber et al, 2003; Pratt et al, 2006), although spiking in synthetic reference proteins before digestion can yield more reliable results. An additional consideration is how much of the reference peptide should be injected for optimal quantification accuracy.
These challenges, which are mostly information-based in nature, must be met for targeted proteomics to be a general and effective strategy for systems biology approaches. We believe that the necessary components to address the above challenges are already in place in PeptideAtlas. PeptideAtlas is a compendium of observations of peptides and associated annotations, based on a large number of contributed data sets that have been reprocessed through a single processing pipeline that includes search–result–validation using the latest tools. As such, it provides the necessary functionalities to facilitate every step of the above process of target selection.
In the subsequent sections, we first describe the creation and maintenance of the PeptideAtlas resource, and then discuss how its various components support the aforementioned targeted proteomics workflow by leveraging the large amount of data collected and assembled.
Building of the PeptideAtlas
With the rapidly increasing number of installed tandem mass spectrometers able to generate large amounts of tandem mass spectrometry (MS/MS)-based proteomics data, we perceive there to be significant value in collecting and combining many of these data sets. Expected benefits from such an endeavour include higher coverage of a proteome, sufficient data density for meaningful statistics and the possibility to contribute extensive observational data back to genome annotation projects. The PeptideAtlas Project thus began as a compendium of peptides observed in a group of human and Drosophila shotgun MS/MS data sets, along with annotations describing in which samples the peptides and proteins were observed, in which modified forms and how frequently the peptides were observed, and how these peptides mapped onto the genome (Desiere et al, 2004).
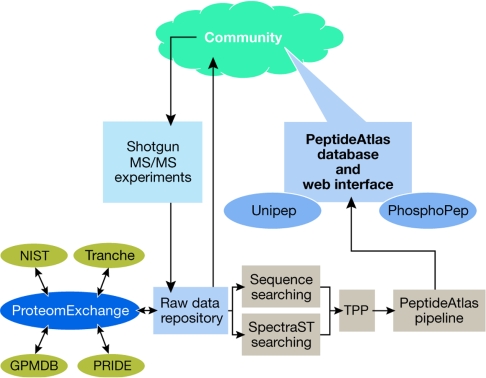
The build process of the PeptideAtlas has evolved since it was initially described by Desiere et al (2006). As shown in Fig 1, raw mass spectrometer output files for MS/MS experiments are collected from the community and processed through a consistent analysis pipeline that performs sequence database searching and automated validation of the results using the Trans Proteomic Pipeline (TPP; Keller et al, 2005). This begins with conversion to a common mzXML file format, then sequence searching with either SEQUEST (Eng et al, 1994) or X!Tandem (Craig & Beavis, 2004), followed by validation of the top hits with PeptideProphet (Keller et al, 2002), a programme that models the correct and incorrect spectrum-peptide match populations, and assigns a probability of being correct to each match.
Figure 1.
An overview of the build process of PeptideAtlas. Shotgun tandem mass spectrometry (MS/MS) experimental data are contributed by the community to the PeptideAtlas raw data repository, which is linked to other repositories by the ProteomExchange consortium. The raw data are processed through an evolving but consistent analysis and validation pipeline (Trans Proteomic Pipeline (TPP)) and loaded into the PeptideAtlas database, and made available to the community. Tranche, Global Proteome Machine Database (GPMDB), National Institute of Standards and Technology (NIST) and Protein Identifications Database (PRIDE) are currently the main participants in the ProteomExchange consortium.
All PeptideProphet results are then combined using ProteinProphet (Nesvizhskii et al, 2003), a programme that uses the spectrum-peptide match models from PeptideProphet to derive protein-level probabilities, as well as to adjust the peptide-level probabilities based on the information available from the ensemble of experiments. Given a set of high-scoring spectra, the spectral library-building tool SpectraST is used to create a consensus spectrum library comprising all observed peptide ions. As part of the library building process, the spectrum-match quality filters reject some high scoring but incorrect identifications. Then all raw data are subjected to a second round of searching, this time by the spectral library-searching component of SpectraST. This allows the identification of many more spectra from the available data, with a higher sensitivity and lower error (Lam et al, 2007). Output of SpectraST is validated in the same manner as described above with PeptideProphet and ProteinProphet.
All peptides are then mapped to a single reference Ensembl (Hubbard et al, 2007) build (if available for the species) and mapped to the genome. All this information is loaded into the PeptideAtlas database for browsing or downloading.
The result of each build process is also made publicly available at the PeptideAtlas web site (see Table 1 for URLs) in several formats. The front-end web site software is distributed as part of the Systems Biology Experiment Analysis System (SBEAMS) framework (Marzolf et al, 2006). A summary of the current state of the various PeptideAtlas builds is provided in Table 2. In the following sections we describe in greater detail some of the components of the PeptideAtlas Project that are important for targeted proteomics.
Table 1.
URLs or references for tools and databases associated with proteomics
Table 2.
Summary of public PeptideAtlas builds
| Build | Number of experiments | Number of MS runs | Searched spectra | IDs P > 0.9 | Distinct peptides | Distinct proteins |
|---|---|---|---|---|---|---|
| Human—all | 219 | 54 k | 49 M | 5.6 M | 97 k | 12,141 |
| Human—plasma | 76 | 48 k | 16 M | 1.8 M | 18 k | 2,486 |
| Drosophila | 43 | 1,769 | 7.5 M | 498 k | 72 k | 9,124 |
| Drosophila PhosphoPep | 4 | 448 | 0.9 M | 170 k | 10 k | 4,583 |
| Yeast | 53 | 2,957 | 6.5 M | 1.1 M | 36 k | 4,336 |
| Mouse | 59 | 3,097 | 10 M | 1.4 M | 51 k | 7,686 |
| Halobacterium | 88 | 497 | 0.5 M | 76 k | 12 k | 1,518 |
| Streptococcus pyogenes | 5 | 64 | 215 k | 52 k | 7 k | 1,068 |
MS, mass spectrometry.
A crucial component of PeptideAtlas is a data repository in which raw data and search results are made available to the community. The PeptideAtlas data repository has had an important role in the advancement of research using high-throughput technologies, acting as data provider to several projects, including the spectrum library building at the National Institute of Standards and Technology (NIST), the PepSeeker database, as well as large-scale genome annotation efforts (Tanner et al, 2007). In addition to PeptideAtlas, several repositories for proteomics data have emerged during the past few years, including the Proteomics Identifications Database (PRIDE; Martens et al, 2005), Open Proteomics Database (OPD; Prince et al, 2004), Tranche (Falkner & Andrews, 2007) and Global Proteome Machine Database (GPMDB; Craig et al, 2004). These repositories have various strengths and fill different niches, but it is obvious that the greatest benefit can be gained if all the repositories share data and metadata to allow users to access information from the same experiments using the repository that best meets their requirements. PeptideAtlas is actively participating in the formation of the ProteomExchange consortium that attempts to facilitate this interoperability between the repositories.
However, most of the aforementioned repositories are largely passive—that is, results are stored and can be queried or downloaded, but the remaining untapped potential within the primary data is not extracted with continually advancing analysis tools. Typically, only a small fraction of acquired MS/MS spectra are confidently identified in the first attempt. Although many of the unidentified spectra are of inadequate quality to ever be identified, a considerable fraction can be identified with more effort and newer techniques (Nesvizhskii et al, 2006). PeptideAtlas aims to be an active repository in which only raw data are accepted and these raw data are periodically reprocessed with more advanced techniques for identification and statistical validation as they become available. The results of this advancing analysis of the raw data are then made available to the community in forms that allow additional research, specifically with tools that support the new targeted proteomics workflows.
Using PeptideAtlas to perform targeted workflows
The challenges of targeted proteomics workflows, as discussed above, involve the selection of targets and the determination of their relevant attributes to facilitate detection in the mass spectrometer. Without a resource such as PeptideAtlas, one possible solution is to run a series of shotgun experiments to determine the optimal peptide targets for a specific protein set, as well as the optimal transitions for each target peptide. However, ready access to hundreds of previously run experiments in PeptideAtlas should transform this problem from the costly acquisition and analysis of preparatory data to a relatively simple informatics problem, for the most popular species at least.
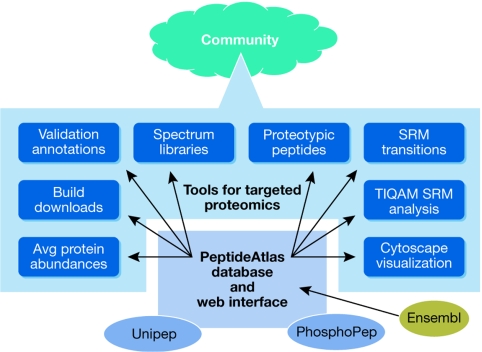
The various ways of using PeptideAtlas in support of targeted proteomics studies are summarized in Fig 2 and described in detail in the following sections.
Figure 2.
An overview of the features provided by PeptideAtlas that allow targeted proteomics workflows. PeptideAtlas and its specialized builds, Unipep (containing N-glycosylation sites) and PhosphoPep (containing phosphorylation sites), allow the community to: select proteotypic peptides for targeting; select SRM transitions for targeting; annotate and view annotations for these peptides and transitions; visualize peptides and proteins with Cytoscape; interface with Targeted Identification for Quantitative Analysis by MRM (TIQAM) for experimental design; obtain approximate protein abundances for spiking in synthetic peptides; obtain spectrum libraries for search and verification; and download PeptideAtlas builds for new projects. MRM, multiple reaction monitoring; SRM, selected reaction monitoring.
Selecting proteotypic peptides for targeted proteins
As discussed above, proteotypic peptides are ideal target peptides. In PeptideAtlas we calculate an empirical observability score (EOS), which acts as an approximate likelihood that, if protein X were detected using shotgun techniques within a given sample, it would be detected through peptide A. Peptides with a high EOS that map uniquely within the proteome are the most suitable—that is, the most proteotypic—peptides to target for any given protein.
For proteins not yet observed in the PeptideAtlas, we provide information for possible follow-up peptide targets from calculations based purely on their sequence. PeptideSieve (Mallick et al, 2007) and DetectabilityPredictor (Tang et al, 2006) calculate an observability score based purely on the physiochemical properties conducive to detection for all the tryptic peptides of a protein. The two algorithms are not in complete agreement, but correlate acceptably. The average scores are presented in a subsection in the PeptideAtlas web interface, allowing one to select the highest scoring peptides as the most suitable peptides for targeting.
Selecting transitions for SRM using PeptideAtlas
For each target peptide to be assayed, the instrument needs two sets of information. First, the expected chromatographic retention time of the target peptide so that the mass spectrometer can be tuned to look for it at a specific time. PeptideAtlas provides a measure of hydrophobicity, namely the Sequence-Specific Retention factor computed using the SSRCalc 3.0 algorithm (Krokhin et al, 2004).for each peptide in the database. These values can be scaled to a given specific instrumental setup and gradient programme, with a typical accuracy of a few minutes. Second, a list of transitions that uniquely and sensitively identify the target peptide ion must be specified. The ideal transitions for the parent peptide ion are fragment ions that are consistently present at high intensity and a high signal-to-noise ratio. Although the fragment peak intensities of a targeted peptide can be predicted to some degree from its sequence, it is too simplistic for effective transition selection. Therefore, a more reliable and effective approach for transition selection is to rely on experimentally observed spectra.
PeptideAtlas provides several features that turn transition selection into an informatics task. In a consensus spectral library building process, performed by the software SpectraST, MS/MS spectra confidently identified from all the data sets contained in PeptideAtlas are first extracted and grouped by their identifications. Next, whenever there are multiple spectra, known as replicates, identified for the same peptide ion they are combined to generate a consensus spectrum, which has more representative peak intensities owing to averaging across observations. The consensus spectrum for each observed peptide ion is loaded into PeptideAtlas and can be visualized by the user. In addition, rules for transition selections from consensus spectra can be specified to generate transition lists automatically. These are also loaded into PeptideAtlas as recommended transitions for each peptide and made accessible through user-defined queries. In addition to the web interface that is already available, a desktop Java application called Targeted Identification for Quantitative Analysis by MRM (TIQAM) is available (Lange et al, 2008) to facilitate the selection of peptides and transitions with a more responsive user interface.
Peptide and transition annotations in the PeptideAtlas
Most of the resources described above are generated automatically, allowing greater throughput. Most PeptideAtlas builds have millions of identified spectra from tens of millions of raw spectra searched. However, as users in the community use these tools to design targeted proteomic experiments, first-hand experience will be gained on the suitability of individual peptides and transitions. Indeed, several recent papers have published lists of validated transitions (Anderson & Hunter, 2006; Lange et al, 2008). To facilitate the reusability of validated transitions, we have implemented a peptide annotation system that allows users to annotate individual peptides with this additional information, such as comments on the suitability of individual peptides, which peptides have synthesized versions available for purchase, which transitions have been validated and which to avoid.
Approximate protein abundances from spectral counting
One technique for obtaining absolute protein abundance measurements involves the use of spiked-in reference peptides, typically a heavy version of the targets (Gerber et al, 2003; Lu et al, 2004; Stahl-Zeng et al, 2007). For optimal results, it is helpful to spike-in the synthetic peptides at a concentration similar to that expected in the sample. PeptideAtlas provides an approximate estimate of the absolute abundance of each protein computed by spectral counting of the represented data sets (N. Zhang, E.W.D., H.L., P. Picotti, L. Mendoza, H. Mirzaei, J. Watts & R.A., unpublished data). These globally calibrated protein abundances can be used to assist in determining suitable spike-in concentrations of synthetic peptides.
Conclusion
The PeptideAtlas Project encompasses more than just a database of observed peptides. It also brings together several related informatics technologies to create an active proteomics repository designed to allow the full potential of targeted proteomic techniques.
PeptideAtlas supports targeted proteomics workflows, such as SRM, by allowing the researcher to identify suitable proteotypic peptides to target and to estimate approximate retention time for the target peptides. Through the building of consensus spectral libraries, which coalesce multiple observations of the same peptide ions to obtain reliable and representative fragmentation patterns, PeptideAtlas also allows the selection of high-quality transition lists for SRM experiments.
The next revolution in proteomics is its transformation from an exploratory field to a robust quantitative discipline. Proteomic experiments that are able to deliver complete, quantitative measurements for thousands of proteins, suitable for correlation with the quantitative transcriptomic measurements already routinely performed, will transform systems biology. The PeptideAtlas Project brings together the necessary informatics capabilities to allow targeted proteomics workflows.

Eric W. Deutsch

Henry Lam

Ruedi Aebersold
Acknowledgments
We thank D. Campbell, L. Mendoza, D. Shteynberg, J. Tasman and N. Zhang for their invaluable efforts in developing the PeptideAtlas resource and the Trans Proteomics Pipeline software on which it depends. Furthermore, we thank all of the investigators who have contributed data to the PeptideAtlas Project. This work has been funded in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract number N01-HV-28179, from PM50 GMO76547/Center for Systems Biology and with funds from grant number 31000-10767 from the Swiss National Science Foundation (to R.A.).
References
- Anderson L, Hunter CL (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 5: 573–588 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC (2004) Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3: 1234–1242 [DOI] [PubMed] [Google Scholar]
- Desiere F et al. (2004) Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol 6: R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R (2006) The PeptideAtlas project. Nucleic Acids Res 34: D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon B, Aebersold R (2006) Mass spectrometry and protein analysis. Science 312: 212–217 [DOI] [PubMed] [Google Scholar]
- Eng J, McCormack AL, Yates JR (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Falkner JA, Andrews PC (2007) Tranche: secure decentralized data storage for the proteomics community. J Bio Tech 18: 3 [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 100: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ et al. (2007) Ensembl 2007. Nucleic Acids Res 35: D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Keller A, Eng J, Zhang N, Li XJ, Aebersold R (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol 1: 2005. 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin OV, Craig R, Spicer V, Ens W, Standing KG, Beavis RC, Wilkins JA (2004) An improved model for prediction of retention times of tryptic peptides in ion pair reversed-phase HPLC: its application to protein peptide mapping by off-line HPLC-MALDI MS. Mol Cell Proteomics 3: 908–919 [DOI] [PubMed] [Google Scholar]
- Kuster B, Schirle M, Mallick P, Aebersold R (2005) Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6: 577–583 [DOI] [PubMed] [Google Scholar]
- Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7: 655–667 [DOI] [PubMed] [Google Scholar]
- Lange V et al. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Bottari P, Turecek F, Aebersold R, Gelb MH (2004) Absolute quantification of specific proteins in complex mixtures using visible isotope-coded affinity tags. Anal Chem 76: 4104–4111 [DOI] [PubMed] [Google Scholar]
- Mallick P et al. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125–131 [DOI] [PubMed] [Google Scholar]
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R (2005) PRIDE: the proteomics identifications database. Proteomics 5: 3537–3545 [DOI] [PubMed] [Google Scholar]
- Marzolf B, Deutsch EW, Moss P, Campbell D, Johnson MH, Galitski T (2006) SBEAMS-microarray: database software supporting genomic expression analyses for systems biology. BMC Bioinformatics 7: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Roos FF, Grossmann J, Vogelzang M, Eddes JS, Gruissem W, Baginsky S, Aebersold R (2006) Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol Cell Proteomics 5: 652–670 [DOI] [PubMed] [Google Scholar]
- Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ (2006) Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc 1: 1029–1043 [DOI] [PubMed] [Google Scholar]
- Prince JT, Carlson MW, Wang R, Lu P, Marcotte EM (2004) The need for a public proteomics repository. Nat Biotechnol 22: 471–472 [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics 6: 1809–1817 [DOI] [PubMed] [Google Scholar]
- Tang H, Arnold RJ, Alves P, Xun Z, Clemmer DE, Novotny MV, Reilly JP, Radivojac P (2006) A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics 22: e481–e488 [DOI] [PubMed] [Google Scholar]
- Tanner S, Shen Z, Ng J, Florea L, Guigo R, Briggs SP, Bafna V (2007) Improving gene annotation using peptide mass spectrometry. Genome Res 17: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]