Abstract
Cytokine-inducible protein SSI-1 [signal transducers and activators of transcription (STAT)-induced STAT inhibitor 1, also referred to as SOCS-1 (suppressor of cytokine signaling 1) or JAB (Janus kinase-binding protein)] negatively regulates cytokine receptor signaling by inhibition of JAK kinases. The SSI family of proteins includes eight members that are structurally characterized by an SH2 domain and a C-terminal conserved region that we have called the SC-motif. In this study, we investigated the roles of these domains in the function of SSI-1. Results of reporter assays demonstrated that the pre-SH2 domain (24 aa in front of the SH2 domain) and the SH2 domain of SSI-1 were required for the suppression by SSI-1 of interleukin 6 signaling. Coexpression studies of COS7 cells revealed that these domains also were required for inhibition of three JAKs (JAK1, JAK2, and TYK2). Furthermore, deletion of the SH2 domain, but not the pre-SH2 domain, resulted in loss of association of SSI-1 with TYK2. Thus, SSI-1 associates with JAK family kinase via its SH2 domain, and the pre-SH2 domain is required for the function of SSI-1. Deletion of the SC-motif markedly reduced expression of SSI-1 protein in M1 cells, and this reduction was reversed by treatment with proteasome inhibitors, suggesting that this motif is required to protect the SSI-1 molecule from proteolytic degradation. Based on these findings, we concluded that three distinct domains of SSI-1 (the pre-SH2 domain, the SH2 domain, and the SC-motif) cooperate in the suppression of interleukin 6 signaling.
Cytokines act on a wide variety of cells and mediate multiple functions, but their effects are limited in both strength and duration. It is well established that Janus kinase (JAK) family kinases play central roles in initiating cytokine signal transduction and the subsequent activation of signal transducers and activators of transcription (STAT) family transcription factors (1–6). However, the mechanisms by which cytokine-signal transduction is attenuated have not been clearly identified.
Recently, a new family of proteins has been identified (7–10). The proteins are induced by stimulation with various cytokines and then negatively control cytokine signaling. For this reason they are thought to be classical negative feedback molecules. Eight members of this family have been identified (11–14): CIS1, SSI-1/SOCS-1/JAB, SSI-2/SOCS-2/CIS2, SSI-3/SOCS-3/CIS3, SOCS-4, SOCS-5/CIS6, SOCS-6/CIS4 and SOCS-7/CIS5, where CIS is a cytokine-inducible SH2-containing protein, SSI is STAT-induced STAT inhibitor, JAB is a JAK-binding protein, and SOCS is a suppressor of cytokine signaling. All of these proteins share two homologous domains, an SH2 domain and a C-terminal conserved domain, which we have called the SC-motif (also referred to as SOCS box or CH domain). DNA database searches revealed that the SC-motif/SOCS box/CH domain also is conserved in four new families of proteins: WD-40-repeat-containing proteins (WSA-1 and WSA-2), SPRY domain-containing proteins (SSB-1 to SSB-3), ankyrin repeat-containing proteins (ASB-1 to ASB-3), and a class of small GTPases (13). However, the functions of this motif have yet to be determined.
SSI-1/SOCS-1/JAB, one member of the SSI family, is induced via activation of STAT3 after interleukin (IL) 6 stimulation in murine myeloid leukemia M1 cells. Constitutive expression of SSI-1 interferes with IL-6-mediated differentiation and apoptosis in M1 cells, as well as the tyrosine-phosphorylation of IL-6 signal transducer gp130 and STAT3. Although SSI-1/SOCS-1/JAB does not affect fibroblast growth factor, insulin, or Flt-3 ligand-induced tyrosine phosphorylation of cellular proteins, it inhibits not only IL-6 signaling but also interferon γ, IL-2, IL-3, and growth hormone signaling, which are triggered through activation of JAKs (8–10, 15). It has been shown that SSI-1 interacts with all four JAKs (JAK1, JAK2, JAK3, and TYK2) and associates with the tyrosine-kinase domain (JH1 domain) of JAK2. Thus, SSI-1 appears to be a general inhibitor of JAKs. However, it has not been fully determined which structure is required for the function of SSI-1.
In this study, we used structure-function analysis of wild-type (WT) or mutant SSI-1 to examine the basic mechanism of suppression by SSI of cytokine signaling. We report here that three distinct domains of SSI-1 are required for the suppression of IL-6 signaling: (i) the pre-SH2 domain for the function of SSI-1, (ii) the SH2 domain for association of SSI-1 with JAK family kinase, and (iii) the SC-motif for stabilization of the SSI-1 protein. Our findings suggest the possibility that similar mechanisms are involved in functions of the SSI family proteins.
MATERIALS AND METHODS
Plasmid Construction and DNA Transfection.
A mouse SSI-1 cDNA was subcloned into the mammalian expression vector pEF-BOS and expressed under the control of an elongation factor gene promoter (10). An Arg-105 point mutation of SSI-1 yielding Gln was introduced by using a quick change site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions and designated as R/Q. ΔLH was generated by excising the ApaI–ApaI fragment from SSI-1 cDNA, resulting in the generation of an extra 31 aa on the C terminal of SSI-1 by frame shift. ΔSC was generated by introducing a stop codon linker (Nippon Gene, Toyama, Japan) into the NotI site of SSI-1. The other deletion mutants, Δ23, Δ49, Δ73, ΔPS, and ΔFH, were generated by introducing two SalI sites into two indicated points of the sequence and excising the SalI–SalI fragment. All constructs were cloned into the pEF-BOS vector and confirmed by DNA sequencing. The mutants were transfected into M1 cells by electroporation in combination with pSV2 neo, and neomycin-resistant clones were selected in a growth medium containing Geneticin (GIBCO) at 550 μg/ml. N-acetyl-l-leucinyl-l-leucinyl-norleucinal (LLnL) and N-acetyl-l-leucinyl-l-leucinyl-methioninal (LLM) were purchased from Sigma and were dissolved in dimethyl sulfoxide.
Anti-SSI-1 mAb.
A hybridoma clone producing the mAb 1262B, which reacts with the oligopeptide THFRTFRSHSD, was established by fusing mouse myeloma cells with spleen cells from BALB/c mice immunized with the synthetic oligopeptide CAPAPGDTHFRTFRSHSDYRRITR (amino acids 43–60 in mouse SSI-1). The mAb was purified by protein A affinity chromatography from the ascitic fluid of BALB/c mice. The mAb FL238, which reacts with the SH2 domain of SSI-1, has been described (10).
Luciferase Assay.
The IRF-1 promoter-luciferase reporter gene has been described previously (16). Plasmids of this reporter gene together with either the pEF-BOS vector alone or pEF-BOS-SSI-1 were transfected into COS7 cells by using SuperFect (Qiagen). In addition, plasmids of pRL-SV40 encoding the Renilla luciferase gene, which was expressed under the control of the simian virus 40 early promoter, were included in each transfection as an internal control (Dual-Luciferase Reporter Assay System, Promega). After 18 hr, the cells were stimulated with IL-6 (100 ng/ml) plus soluble IL-6R (50 ng/ml) for 6 hr. Cell extracts were prepared, and luciferase activity then was measured.
Immunoprecipitation and Immunoblot Analysis.
COS7 cells (1 × 106) were transfected with the plasmids encoding mouse SSI-1 (2 μg), mouse JAK1 (10 μg), mouse JAK2 (10 μg), or human TYK2 (10 μg) by using the DEAE-dextran method. After 36 hr, cells were solubilized with RIPA buffer (1% Nonidet P-40/0.1% sodium desoxycholate/0.1% SDS/10 mM Tris, pH 7.4/150 mM NaCl/2 mM Na3VO4/5 μg/ml aprotinin/1 mM phenylmethylsulfonyl fluoride). Immunoprecipitates obtained with anti-JAK1, JAK2, or TYK2 antibody (Santa Cruz Biotechnology) were resolved by SDS/PAGE under reducing conditions, transferred to a nitrocellulose membrane, and immunoblotted with antiphosphotyrosine mAb (4G10, Upstate Biotechnology) or anti-SSI-1 mAb. Blots were visualized with the ECL detection system (Amersham).
Northern Blot Analysis.
Total RNA was extracted from the transfected M1 cells by using RNA zol B (Tel-Test, Friendswood, TX). Twenty micrograms per lane was subjected to agarose gel electrophoresis and transferred to a membrane (Hybond-N+, Amersham). The membrane was hybridized with radiolabeled mouse SSI-1 cDNA probe, and mouse β-actin probe was used as an internal control.
RESULTS
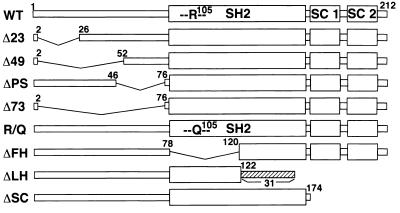
To determine which domain of the SSI-1 protein is required for its function, we constructed a series of expression vectors coding for various SSI-1 deletion and point mutants, including N-terminal mutants (Δ23, Δ49, ΔPS, and Δ73), SH2 mutants (R/Q, ΔFH, and ΔLH), and a C-terminal mutant (ΔSC), as shown schematically in Fig. 1.
Figure 1.
Schematic representation of WT and mutant SSI-1 used in this study. The SC-motif is composed of two conserved motifs, SC-motifs 1 and 2. The figure (Δ23, Δ49, or Δ73) shows the number of deleted amino acids. ΔPS lacks 29 aa in front of the SH2 domain. R/Q features a point mutation at the base of a putative phosphotyrosine binding pocket of the SH2 domain. ΔFH lacks the first half of the SH2 domain. ΔLH lacks both the latter half of the SH2 domain and the SC-motif and has 31 extra aa. ΔSC lacks the SC-motif.
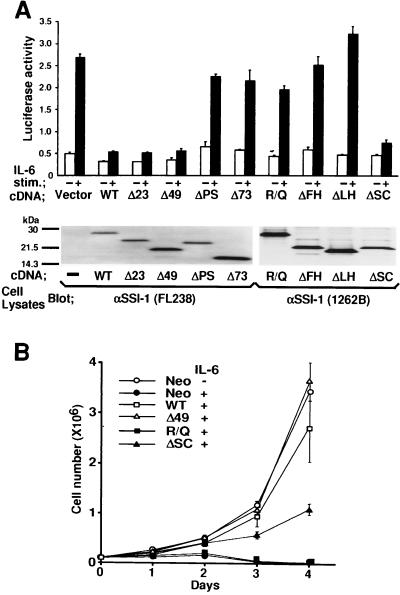
We transiently transfected the plasmid carrying either WT or mutated forms of SSI-1 together with the IRF-1 promoter-reporter gene into COS7 cells that express IL-6-signal transducer gp130 endogenously, and examined the biological effect of SSI-1 on IL-6 signaling. As shown in Fig. 2A, Δ23, Δ49, and ΔSC suppressed IL-6-induced luciferase activity, as did WT, but when the pre-SH2 domain (24 aa in front of the SH2 domain) was missing (ΔPS or Δ73) or the SH2 domain was mutated (R/Q, ΔFH, and ΔLH), IL-6-induced luciferase activity was not suppressed. Levels of expression of the WT and each of the mutant forms of SSI-1 were shown by blotting cell lysates with anti-SSI-1 mAbs (Fig. 2A, Lower). Thus, in COS7 cells, both the pre-SH2 and SH2 domains contributed to the function of SSI-1, but the N-terminal 49 aa and the SC-motif were not required for the function of SSI-1. To confirm that the N-terminal 49 aa and the SC-motif are not required for the function of SSI-1, we used M1 cells in which apoptosis was induced by IL-6. cDNAs encoding SSI-1/Δ49, R/Q, and ΔSC were transfected stably in M1 cells, and viable cells were counted after IL-6 stimulation. SSI-1/WT and Δ49 almost entirely suppressed IL-6-induced growth arrest in M1 cells, but R/Q did not (Fig. 2B). These findings were consistent with those obtained in COS7 cells. However, compared with the effect of SSI-1/ΔSC on IL-6 signaling in COS7 cells, suppression of IL-6 signaling by ΔSC was only partial in M1 cells.
Figure 2.
Effect of WT and mutant SSI-1 on IL-6 signaling. (A) COS7 cells were cotransfected with the reporter gene for the IRF-1 promoter and pEF-BOS carrying either WT, mutant SSI-1, or the vector alone. Luciferase activity (AU, arbitrary units) was measured after stimulation without (−) or with (+) IL-6 (100 ng/ml) plus soluble IL-6 receptor (50 ng/ml). Each bar represents the mean ± SD of three experiments. Levels of expression of the SSI-1 mutants were visualized by blotting cell lysates with anti-SSI-1 mAbs. FL238 recognizes the SH2 domain of SSI-1, and 1262B recognizes an N-terminal portion of SSI-1. (B) M1 cells stably transfected with cDNA for WT, Δ49, R/Q, ΔSC, or Neo gene alone were seeded with or without IL-6 (100 ng/ml) plus soluble IL-6R (50 ng/ml) at day 0. The numbers of viable cells were determined by trypan blue exclusion assay and counted in a hemocytometer on days 1–4. Averages of three independent clones of M1/SSI-1 transfectants are shown. Vertical bars indicate SD.
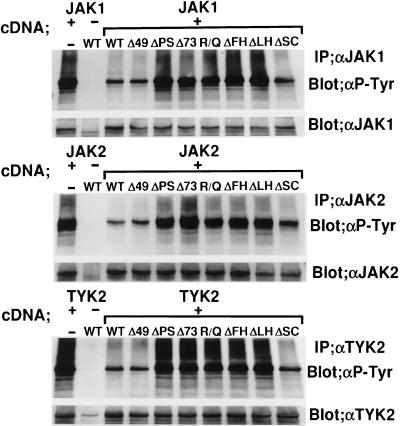
Having established the importance of the pre-SH2 and SH2 domains for the suppression of IL-6 signaling by SSI-1, we examined whether these domains were required for the suppression of JAK. Because it is known that JAKs, when highly expressed, are activated in the absence of cytokine stimulation and autophosphorylated on tyrosine residues (ref. 17 and M.N., unpublished data), we examined the effect of SSI-1 on autophosphorylation of three JAKs (JAK1, JAK2, and TYK2), which are known to be activated by IL-6 stimulation (3). JAK1, JAK2, or TYK2 were coexpressed with WT or mutant SSI-1 in COS7 cells. Cell lysates were immunoprecipitated with antirespective JAK antibodies, and the precipitates were immunoblotted with antiphosphotyrosine mAb. As shown in Fig. 3, Δ49 and ΔSC suppressed autophosphorylation of the three JAKs as well as WT, whereas ΔPS, Δ73, R/Q, ΔFH, and ΔLH did not. These findings demonstrated that the structural requirements of SSI-1 for inhibition of IL-6 signaling were consistent with those for inhibition of JAKs.
Figure 3.
The pre-SH2 and SH2 domains of SSI-1 are required for inhibition of JAK1, JAK2, and TYK2. COS7 cells were transfected with plasmids carrying JAK1, JAK2, or TYK2, together with SSI-1/WT or mutant SSI-1 as indicated at the top. After immunoprecipitation with anti-JAK1 (Top), anti-JAK2 (Middle), or anti-TYK2 (Bottom) antibody, the respective immunoprecipitates were blotted with antiphosphotyrosine mAb. The membranes were reprobed with anti-JAK antibody after the first blots were stripped.
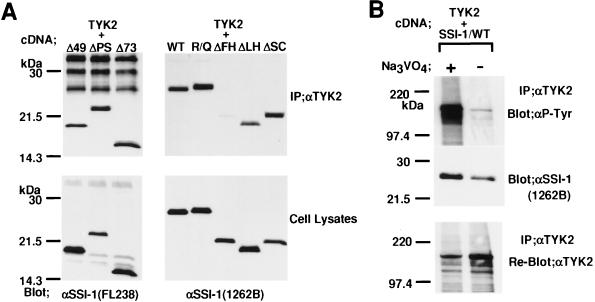
Next, we examined which domain of SSI-1 is required for association with JAK. After TYK2 together with mutant SSI-1 was expressed in COS7 cells, the immunoprecipitates obtained with anti-TYK2 antibody were immunoblotted with anti-SSI-1 mAbs. N-terminal mutants, R/Q and ΔSC mutants, were coprecipitated with TYK2 as well as WT, whereas ΔFH was not coprecipitated and ΔLH was weakly (Fig. 4A). The levels of expression of each SSI-1 were shown by blotting cell lysates with anti-SSI-1 mAbs. These findings indicated that SSI-1 associated with TYK2 via the entire structure of the SH2 domain. Furthermore, we also examined whether SSI-1 associates with tyrosine-phosphorylated form of TYK2. After TYK2 and SSI-1/WT were expressed in COS7 cells, the cells were lysed with RIPA lysis buffer in the presence or absence of tyrosine-phosphatase inhibitor Na3VO4. The lysates were immunoprecipitated with anti-TYK2 antibody, and the precipitates were blotted with antiphosphotyrosine mAb or anti-SSI-1 mAb. As shown in Fig. 4B, compared with the lysis in the presence of Na3VO4, the lysis in its absence reduced both the level of tyrosine-phosphorylation of TYK2 (Top) and the level of association of SSI-1 with TYK2 (Middle). The levels of immunoprecipitated TYK2 were shown by reblotting the membrane with anti-TYK2 antibody (Bottom). These results indicated that SSI-1 associated with the tyrosine-phosphorylated form of TYK2.
Figure 4.
The SH2 domain of SSI-1 is required for the association of SSI-1 with tyrosine-phosphorylated TYK2. (A) COS7 cells were transfected with plasmids carrying TYK2 together with SSI-1/WT or mutant SSI-1 as indicated at the top. After immunoprecipitation with anti-TYK2 antibody, the resultant immunoprecipitates were blotted with anti-SSI-1 mAb FL238 (Left) or anti-SSI-1 mAb 1262B (Right). Cell lysates were immunoblotted with anti-SSI-1 mAbs to show the levels of expression of the proteins. (B) SSI-1 associates with tyrosine-phosphorylated TYK2. After TYK2 together with SSI-1/WT were expressed in COS7 cells, the cells were lysed with RIPA buffer in the presence or absence of 2 mM Na3VO4. The lysates were immunoprecipitated with anti-TYK2, and the precipitates were separated by SDS/PAGE and transferred to a membrane. The upper half of the membrane was blotted with antiphosphotyrosine mAb (Top), and the lower half was blotted with anti-SSI-1 mAb 1262B (Middle). The former was reblotted with anti-TYK2 antibody after the first blots were stripped (Bottom).
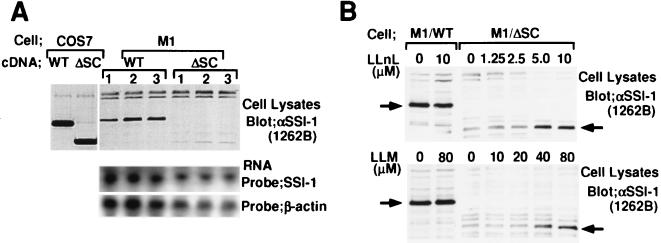
As described in Fig. 2, SSI-1/ΔSC suppressed IL-6 signaling in COS7 cells but only partially in M1 cells. To explore the difference in function of SSI-1/ΔSC between COS7 and M1 cells, we examined the levels of expression of SSI-1/WT and ΔSC proteins in both types of cells. Whole-cell extracts from COS7 or M1 cells transfected with cDNAs encoding SSI-1/WT or ΔSC were immunoblotted with anti-SSI-1 mAb. As shown in Fig. 5A, the expression of ΔSC protein was much less than that of WT protein in M1 cells, whereas in COS7 cells the levels of expression of these two proteins were almost the same. mRNAs corresponding to WT and ΔSC in M1 cells were detected equally by Northern blotting. Δ49 and R/Q mutants possessing the SC-motif were expressed at levels comparable to WT in M1 cells (data not shown). These findings suggested that ΔSC protein lost its stability in M1 cells. We next examined whether SSI-1/ΔSC is proteolytically degraded. M1 cells were treated with a proteasome-specific inhibitor, LLnL or LLM. As shown in Fig. 5B, although the expression levels of WT were not affected by proteasome inhibitors, the levels of expression of ΔSC protein were increased by inhibitors and reached maximum with treatment of 5 μM LLnL or 80 μM LLM. These findings demonstrated that SSI-1 was protected from proteolytic degradation by the SC-motif.
Figure 5.
The SC-motif is required for stable expression of SSI-1 in M1 cells. (A) Cell lysates from COS7 and M1 cells transfected with cDNA for SSI-1/WT and ΔSC were immunoblotted with anti-SSI-1 mAb 1262B. Three independent clones of M1 transfectant were used for this experiment. Northern analysis of the mRNAs from M1/SSI-1/WT and ΔSC revealed identical mRNA levels for the respective clones. (B) The effect of LLnL and LLM on SSI-1/ΔSC. M1 cells transfected with SSI-1/WT or ΔSC were treated with LLnL or LLM for 12 hr. Each treatment included 1% dimethyl sulfoxide. Cell lysates were prepared by lysis in RIPA buffer containing 10 μM LLnL and 10 μM LLM, separated by SDS/PAGE, transferred, and immunoblotted with anti-SSI-1 mAb 1262B. The positions of SSI-1/WT and SSI-1/ΔSC are indicated.
DISCUSSION
A family of proteins, SSI/SOCS/CIS, has been shown to be negative feedback molecules that are induced by various cytokines and then suppress cytokine signals. The proteins are structurally characterized by an SH2 domain and a C-terminal homologous domain (SC-motif/SOCS box/CH domain) (7–14). The first member of the SSI-family, CIS1, is reported to associate with the tyrosine-phosphorylated erythropoietin receptor and the IL-3 receptor β chain, and inhibits activation of STAT5. It is thought that CIS1 competes with STAT5 to reduce interaction between STAT5 and the tyrosine-phosphorylated cytokine receptors (7, 18), although interactions between the SH2 domain of CIS1 and these receptors have not yet been demonstrated.
In this study, to determine the function of the SH2 domain of SSI-1, we introduced point mutation or partial deletions into this domain of SSI-1. Arg-105 to Gln (R/Q) mutation at the base of a putative phosphotyrosine binding pocket of the SH2 domain resulted in loss of both the biological function of SSI-1 and its inhibition of three JAKs (JAK1, JAK2, and TYK2). However, the R/Q mutation did not affect the association of SSI-1 with TYK2, and this association was eliminated by lack of the first half of the SH2 domain including the Arg-105. It has been found on crystallographic analyses of Src kinase that several amino acids in the SH2 domain interact with amino acids of tyrosine-phosphorylated proteins (19, 20). Therefore, in the case of SSI-1, not a single Arg residue, but in fact several amino acids in the SH2 domain, may play roles in the binding of SSI-1 to TYK2, and the high expression system developed in our study may permit association between the R/Q mutant and TYK2. However, the R/Q mutant may lose stable interaction between SSI-1 and TYK2, resulting in the loss of function of SSI-1, probably because the Arg residue participates in the firm binding of SSI-1 to TYK2.
Both the SH2 and pre-SH2 domains were required for the function of SSI-1. We noted that the pre-SH2 domain of SSI-1 exhibits sequence homology with the other three members of the SSI family, SSI-3, SOCS-4, and SOCS-5. As shown in Fig. 6, a negatively charged amino acid-tyrosine and two leucine/isoleucines are conserved in this domain. Particularly significant homology was observed between SSI-1 and SSI-3 and between SOCS-4 and SOCS-5. Interestingly, both SSI-1 and SSI-3 have been found to suppress leukemia inhibitory factor-induced apoptosis in M1 cells and growth hormone-induced Sp 2.1 promoter activation and can associate with JAK2 (11, 12, 15). These findings suggest the possibility that the pre-SH2 domain may contribute to interaction with JAK and inhibit JAK specifically. And there probably might be a similarity of function between SOCS-4 and SOCS-5.
Figure 6.
Sequence alignment of the pre-SH2 domains of SSI-1, SSI-3, SOCS-4, and SOCS-5. Defined as AILMFPVW (nonpolar), NCQGSTY (polar but uncharged), RHK (positively charged), and DE (negatively charged) according to the chemical characteristics of their side chains. Residues conserved among three of the four proteins are boxed. Identical (∗) or similar (:) amino acids are indicated between SSI-1 and SSI-3 and between SOCS-4 and SOCS-5.
The SC-motif was found not to be required for the function of SSI-1, at least in COS7 cells. We detected a mutant SSI-1 protein lacking the SC-motif (ΔSC) in COS7 cells, but this mutant was barely detected in M1 cells. The proteasome inhibitors LLnL and LLM restored the levels of expression of ΔSC protein in M1 cells, thus the SC-motif functions to protect the SSI-1 molecule from proteolytic degradation. Because proteolytic degradation is one of the mechanisms down-regulating cytokine signal, it seems reasonable that negative feedback molecules are protected from degradation to function effectively. One possible reason for the difference in levels of expression of ΔSC protein between COS7 and M1 cells is that ΔSC may be continuously highly expressed in COS7 cells to overcome protein degradation, because COS7 cells express the simian virus 40 (SV40) large tumor (T) antigen, which can amplify the copy number of the pEF-BOS vector including the SV40 origin. Alternatively, COS7 cells may lack mechanisms that degrade the SSI protein.
In this study, we found that SSI-1 binds to JAK via the entire structure of the SH2 domain, and that both the pre-SH2 and the SH2 domains were required for the suppression of IL-6 signaling and inhibition of JAKs. We also found that SSI-1 was protected from proteolytic degradation by the SC-motif. Based on our findings, we propose mechanisms to explain the function of SSI-1. Ligand-induced dimerization or oligomerization of cytokine receptors and high expression of JAK protein are known to induce activation of JAK. These conditions allow JAK to approach its substrates, such as cytokine receptors, STAT, or JAK itself. Therefore, after binding of the SH2 domain of SSI-1 to tyrosine-phosphorylated JAK, the pre-SH2 domain may interfere with approach of JAK substrate to JAK. Alternatively, the pre-SH2 domain may induce conformational change resulting in inactivation of JAK.
Acknowledgments
We thank Dr. S. Nagata for the pEF-BOS vector, Dr. R. Fukunaga for JAK1 and JAK2 cDNAs, Dr. J. Krolewski for TYK2 cDNA, Dr. M. Revel for the IRF-1 promoter-luciferase gene, Dr. K. Yasukawa for recombinant IL-6 and soluble IL-6R, and Mss. N. Kameoka and M. Tagami for their secretarial assistance. This study was supported by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan.
ABBREVIATIONS
- IL
interleukin
- JAK
Janus kinase
- STAT
signal transducers and activators of transcription
- SSI
STAT-induced STAT inhibitor
- SOCS
suppressor of cytokine signaling
- CIS
cytokine-inducible SH2-containing protein
- JAB
JAK-binding protein
- SC-motif
SSI COOH-terminal motif
- LLnL
N-acetyl-l-leucinyl-l-leucinyl-norleucinal
- LLM
N-acetyl-l-leucinyl-l-leucinyl-methioninal
- WT
wild type
References
- 1. Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Ihle J N, Kerr I M. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 3.Kishimoto T, Akira S, Narazaki M, Taga T. Blood. 1995;86:1243–1254. [PubMed] [Google Scholar]
- 4.Rodig S J, Meraz M A, White J M, Lampe P A, Riley J K, Arthur C D, King K L, Sheehan K C F, Yin L, Pennica D, et al. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 5.Parganas E, Wang D, Stravopodis D, Topham D J, Marine J-C, Teglund S, Vanin E F, Bodner S, Colamonici O R, van Deursen J M, et al. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 6.Neubauer H, Cumano A, Müller M, Wu H, Huffstadt U, Pfeffer K. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 9.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Mitsui K, Sakamoto H, Ohtsubo M, Misawa H, Kanekura Y, Yoshimura A. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 10.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 11.Minamoto S, Ikegame K, Ueno K, Narazaki M, Naka T, Yamamoto H, Matsumoto T, Saito H, Hosoe S, Kishimoto T. Biochem Biophys Res Commun. 1997;237:79–83. doi: 10.1006/bbrc.1997.7080. [DOI] [PubMed] [Google Scholar]
- 12.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, et al. Biochem Biophys Res Commun. 1997;239:439–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 13.Hilton D J, Richardson R T, Alexander W S, Viney E M, Willson T A, Sprigg N S, Starr R, Nicholson S E, Metcalf D, Nicola N A. Proc Natl Acad Sci USA. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aman M J, Leonard W J. Curr Biol. 1997;7:784–788. doi: 10.1016/s0960-9822(06)00405-2. [DOI] [PubMed] [Google Scholar]
- 15.Adams T E, Hansen J A, Starr R, Nicola N A, Hilton D J, Billestrup N. J Biol Chem. 1998;273:1285–1287. doi: 10.1074/jbc.273.3.1285. [DOI] [PubMed] [Google Scholar]
- 16.Harroch S, Revel M, Chebath J. EMBO J. 1994;13:1942–1949. doi: 10.1002/j.1460-2075.1994.tb06463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto K, Quelle F W, Thierfelder W E, Kreider B L, Gilbert D J, Jenkins N A, Copeland N G, Silvennoinen O, Ihle J N. Mol Cell Biol. 1994;14:4342–4349. doi: 10.1128/mcb.14.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 19.Marengere L E M, Pawson T. J Biol Chem. 1992;267:22779–22786. [PubMed] [Google Scholar]
- 20.Waksman G, Shoelson S E, Pant N, Cowburn D, Kuriyan J. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]