Abstract
An automatic sequence search and analysis protocol (DomainFinder) based on PSI-BLAST and IMPALA, and using conservative thresholds, has been developed for reliably integrating gene sequences from GenBank into their respective structural families within the CATH domain database (http://www.biochem.ucl.ac.uk/bsm/cath_new). DomainFinder assigns a new gene sequence to a CATH homologous superfamily provided that PSI-BLAST identifies a clear relationship to at least one other Protein Data Bank sequence within that superfamily. This has resulted in an expansion of the CATH protein family database (CATH-PFDB v1.6) from 19,563 domain structures to 176,597 domain sequences. A further 50,000 putative homologous relationships can be identified using less stringent cut-offs and these relationships are maintained within neighbour tables in the CATH Oracle database, pending further evidence of their suggested evolutionary relationship. Analysis of the CATH-PFDB has shown that only 15% of the sequence families are close enough to a known structure for reliable homology modeling. IMPALA/PSI-BLAST profiles have been generated for each of the sequence families in the expanded CATH-PFDB and a web server has been provided so that new sequences may be scanned against the profile library and be assigned to a structure and homologous superfamily.
Keywords: Structural genomics, fold assignment, homology, sequence profiles, CATH
The challenge for the post-genomic era will be to understand the functions and biological roles of the thousands of sequences being determined by the international genome initiatives. There are already more than 500,000 nonredundant sequences deposited in GenBank (Benson et al. 2000), and as the number increases it will become even more important to derive functional and structural information for these sequences. By integrating genomic sequences within the CATH database, we aim to facilitate the assignment of structural and functional properties to these newly determined sequences.
As proteins evolve their sequences diverge, but their structures are generally conserved. Consequently, homologous relationships between distantly related proteins may not be identified until their structures are resolved. The CATH database (Orengo et al. 1997; Pearl et al. 2001) contains structural domains derived from the Protein Data Bank (PDB, Berman et al. 2000), organized according to Class, Architecture, Topology (fold), and Homologous superfamily. Proteins are clustered into their evolutionary families if they have high sequence similarity or high structural similarity and some sequence/functional similarity. Pairs of proteins with the same fold but different sequence and function are classed as analogous unless there is other evidence to suggest the proteins are related by divergent evolution. One approach to address the sequence/structure gap is to look for common sequence features between uncharacterized genes with already resolved protein structures, so that structural and functional information can be inherited.
Numerous pairwise sequence comparison methods (e.g., BLAST, FASTA, see Brenner et al. 1998) can reliably and quickly detect similarities between proteins sharing at least 30% of their amino acid sequence. These proteins will adopt the same fold and will often exhibit similar function. For more distant homologs these pairwise methods can be insensitive. In the twilight zone of sequence comparison (20%–30% sequence identity) only about half of the relationships can be detected (Brenner et al. 1998), whereas in the midnight zone (<20%, Rost 1999) the proportion is even smaller. Sequence searching methods that use profile-based approaches, for example, PSI-BLAST (Altschul et al. 1997) and hidden Markov models (HMMs) (SAM-T98, Karplus et al. 1998) or intermediate sequences (ISS, Park et al. 1997) can detect more distant homologs with up to three times as much coverage and greater reliability than pairwise methods (Park et al. 1998; Salamov et al. 1999a).
Both PSI-BLAST and HMMs generate family-specific profiles. In PSI-BLAST, a database is scanned using BLASTPGP, a version of BLAST that gives gapped alignments. The program creates multiple alignments and produces a profile or position-specific scoring matrix (PSSM) that is used to search the database for further distant homologs. Multiple alignments reveal position-specific residue propensities particular to a family. This procedure is iterative and continues until no more homologs are found. In contrast, an HMM profile can be built from a pre-aligned set of related sequences (Eddy 1996), but also benefits from iteration (Karplus et al. 1998).
ISS is based on the phenomenon that when sequences of a pair of proteins have diverged beyond a point where their relationship can be detected by pairwise sequence comparison directly, a third intermediate sequence that matches each of the pair indicates the sequences are evolutionarily related. The original ISS method (Park et al. 1997) used FASTA libraries to detect homologous relationships, however, an intermediate library collated using PSI-BLAST intermediates gives better coverage (Salamov et al. 1999a). Several intermediate libraries have been established based on either CATH or SCOP homology assignment (Muller et al. 1999; Salamov et al. 1999a; Teichmann et al. 2000).
In conjunction with PSI-BLAST, the complementary IMPALA computer package (Schaffer et al. 1999) is now available. IMPALA allows a single query sequence to be compared against a database of PSSMs generated from PSI-BLAST searches. IMPALA's sensitivity is comparable to that of PSI-BLAST and it is much faster when screening small data sets. A better local alignment algorithm is employed using the Gotoh implementation of the Smith-Waterman alignment method.
Earlier work of Huynen et al. (1998) and Salamov et al. (1999a,b) based on the CATH database explored the performance of PSI-BLAST for providing structural annotation for genome sequences. Here, we have extended that approach and developed a protocol (DomainFinder) for using PSI-BLAST to reliably assign domain boundaries to protein sequences related to CATH structural families. In addition to increasing the CATH database tenfold from 19,563 to create an extended protein family database (CATH-PFDB) containing 176,597 domains, this has resulted in further validation of the homologous families in CATH by confirming evolutionary relationships.
IMPALA profiles generated from the PSI-BLAST searches have been established for all nonidentical structures in the CATH database. These can be used to assign domain boundaries to newly solved structures and sequences that are distant homologs to previously assigned domains. IMPALA profiles created for the extra 9560 sequence family representatives from the extended database have improved the recognition of distant homologs, increasing the coverage from 53% to 76% in benchmark tests using manually validated CATH superfamilies. A web server is available at http://www.biochem.ucl.ac.uk/bsm/cath_new/Impala/ that allows the user to search for a homologous relative within the extended CATH protein family database (CATH-PFDB) by scanning a sequence against these IMPALA profiles.
Results and Discussion
Benchmarking
PSI-BLAST was benchmarked to derive conservative thresholds to reliably predict sequence domains for inclusion as input for the DomainFinder algorithm. A data set of sequences was derived from the single segment domains in the CATH structural domain database. This contained 1351 representatives (CATH-35, see Materials and Methods section at end) from the majority of homologous superfamilies in CATH (773 families from the April 5, 2000 release of CATH). Only relatives from each superfamily exhibiting <35% sequence identity with any other selected relative were included, ensuring that the data set contained only remote homologs from each superfamily. This was to maximize the performance for recognizing distant relatives from different CATH-35 sequence families, because homologs with sequence identities >35% are easily identified by pairwise sequence comparison methods (Pearl et al. 2000). The 1351 single-segment homologs give a total of 911,925 (1351 × 1350/2) pairwise relationships (false + true). The optimal implementation of the PSI-BLAST algorithm should be able to detect all the true pairwise relationships within a homologous superfamily (H-family, 2478 in total) without detecting sequences from any other superfamily.
PSI-BLAST was run for each of the CATH-35 representatives for a range of E-values. When a CATH-35 matched another CATH-35 from the same homologous superfamily, a hit was recorded and scored accordingly. If it matched a CATH-35 from a different H-family that had the same fold (same T-level), this was not counted either as a true or a false match. The H-families in CATH have traditionally been assigned very conservatively, with only strong evidence of functional similarity allowing them to merge. Therefore, PSI-BLAST matches having the same fold group but with different homologous superfamily assignments suggest putative evolutionary relationships, for which no strong functional evidence currently exists or in which functional properties have diverged.
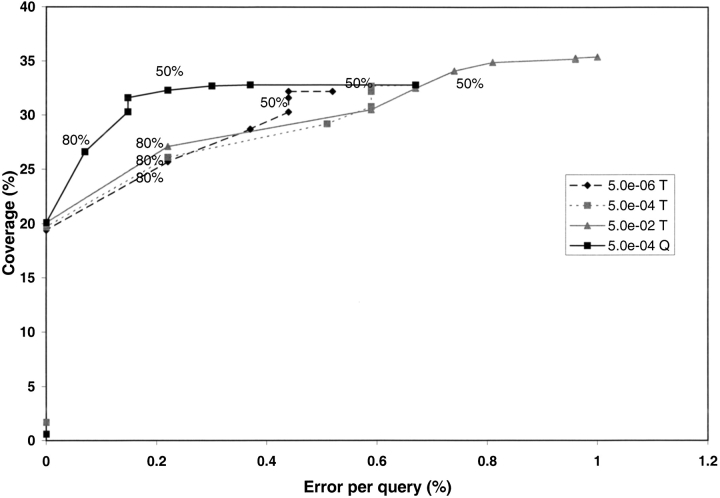
Park et al. (1998) and Teichmann et al. (2000) found that for PSI-BLAST an E-value of 5.0 × 10−4 gave an error per query (EPQ), calculated as the number of unrelated sequences matched as a percentage of the number of query sequences, of ∼1%. Muller et al. (1999) took this further and investigated the percentage of a target correctly identified by PSI-BLAST [overlap(target)]. Figure 1 ▶ shows coverage plotted against error per query, for the different overlap thresholds from 0%–100% in steps of 10%. For an E-value of 5.0 × 10−4 in a one-to-one relationship half (50%) of the target is identified in 32% of the cases, with an EPQ of 0.6%. However, when 80% of the target is identified, although the coverage drops to 26%, the EPQ is halved. Clearly, by having a more stringent overlap criterion the number of false positives can be considerably reduced, and the coverage remains more than twice that produced by a single BLAST run (Park et al. 1998).
Fig. 1.
Coverage obtained when detecting one-to-one relationships. The coverage is measured using the CATH-35 sequences for different PSI-BLAST parameters. The graph shows the percent coverage of true positive matches divided by the total number of possible assignments plotted against the numbers of errors per query. These values are plotted for the different percentages of the target domain (T) included in the alignment, at different E-values. These values are also plotted for the different percentages of the query domain (Q) included in the alignment.
Generating an extended CATH database (CATH-PFDB) containing clear gene relatives while maintaining the integrity of the data required a very low error rate for recruiting gene relatives. When populating a database with sequences, the percentage of target identified cannot be used as an overlap criterion as the answer is not known. Therefore overlap(query) was selected as the overlap criteria; this was the length of the alignment of the query sequence that matches a target, divided by the length of the query sequence. If the query aligned 80% of itself with a target gene sequence using an E-value of 5.0 × 10−4, the coverage remained the same (26.5%) as using the other measure of overlap, however a greatly reduced error rate was obtained with an EPQ of 0.07% (see Fig. 1 ▶). Consequently, the overlap(query) criteria for integrating gene domain sequences into the CATH-PFDB using the DomainFinder algorithm was set at 80%.
DomainFinder
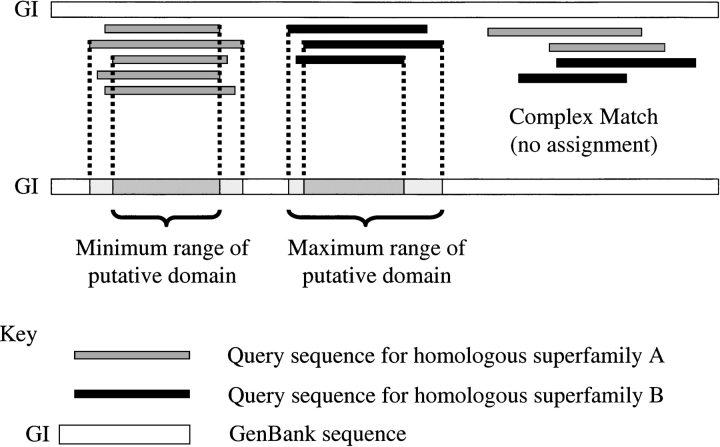
The DomainFinder algorithm exploits PSI-BLAST or IMPALA matches (see below) of a CATH sequence to a gene sequence to identify the residue range of a homologous domain within that gene sequence. Where a gene sequence is matched by a single CATH sequence, the domain region of the gene sequence to be integrated into CATH is inherited directly via the alignment. Where several CATH sequences from the same homologous superfamily are aligned to a similar region of a gene sequence, the minimum and maximum position of the overlap on the gene sequence is determined together with the best (relative with lowest E-value) and consensus overlap regions (see Fig. 2 ▶).
Fig. 2.
Assigning domain boundaries to GenBank sequences. Diagram of simple hits and cross-hits mapped onto a GenBank sequence. DomainFinder automatically assigns simple hits, calculating the ranges for the minimum and maximum, and consensus regions to the appropriate homologous superfamily. The domain range corresponding to the lowest E-value is also recorded. Cross-hits are stored separately and are validated manually. (Shaded) Query sequence for homologous superfamily A; (solid) query sequence for homologous superfamily B; (GI, open) GenBank sequence.
In some cases, the same region of a gene sequence is matched to CATH sequences from different homologous superfamilies or even to different fold groups in CATH. These are referred to as cross-hits to distinguish them from the simple case of a single superfamily matching (Fig. 2 ▶). The factors giving rise to these cross-hits and some methods for handling them are discussed below.
DomainFinder is designed to be capable of resolving multisegment domains. PSI-BLAST is run on all the segments from a domain. Only if an assignment to the largest segment is made are the assignments to the smaller segments searched for. Cross-hits are disregarded if they involve only smaller segments from multisegment domains. In domains where there is little difference in the size of the larger and smaller segments (i.e., the larger segment is <60% of the domain), both segments are weighted equally.
To validate the performance of DomainFinder, the domain assignments were compared with the domain boundary ranges identified for a set of 333 sequence representatives from multidomain sequences in CATH. These 333 sequence representatives (CATH-35) from the multidomain data set were scanned against the GenBankCATHnr library using PSI-BLAST and scanned against the profiles in the CATH-IMPALA library using IMPALA (see below). Domain ranges were then assigned automatically by DomainFinder using the structural representatives (CATH-95). Close homologs (<35% sequence identity) were discounted, so that we could test the performance of DomainFinder in assigning domain boundaries for distant homologs. Matches were identified using the optimal thresholds established by the benchmark trials (i.e., >80% overlap E-value <5 × 10−4). The predicted boundaries were then compared with those that had been previously assigned structurally and with manual validation in the CATH database. These boundary ranges had been identified using algorithms based on structural criteria (e.g., compactness of domain and hydrophobic clusters, Jones et al. 1998) and manually validated.
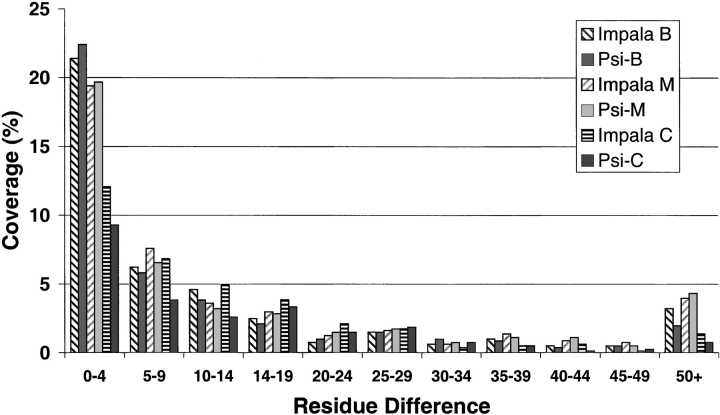
DomainFinder identified 41% of the domain boundaries using both IMPALA and PSI-BLAST assignments, where 80% of a structural domain sequence was aligned with the multidomain sequence (Fig. 3 ▶). Extracting the domain boundaries derived from the best PSI-BLAST match gave the optimal boundary assignment, with the majority of these assigned domains (71%) having boundary assignment within 10 residues of the manually validated boundary. Complete domain assignment was possible for 18% of the chains.
Fig. 3.
Accuracy of domain boundary identification. Histogram of the frequency (%) of the offset error in domain identification, where offset is the number of residues error in the delineation of a domain boundary. Both PSI-BLAST and IMPALA were used by DomainFinder to predict the domain boundaries, which were then compared with those determined by structural methods (DBS, Jones et al. 1998) and validated manually. (Hatched bars) IMPALA; (solid bars) PSI-BLAST. Three different domain boundary criteria were used: domain boundaries extracted from the match with the lowest E-value (B), domain boundaries extracted using the minimum-maximum regions (M), and the consensus regions (C) (see Fig. 2 ▶).
When updating CATH, the existing protocol for identifying domain boundaries is based on structural approaches (Jones et al. 1998). The Domain Boundary Suite (DBS) is a suite of three independent automatic domain recognition algorithms; boundary assignment is only possible when all three programs are in agreement. To calculate the number of chains that could have been automatically assigned by structural approaches, DBS was run on the structures of the 333 sequence representatives and 21% of the boundaries were identified. For 18% of the 333 sequence representatives complete domain assignment was possible by PSI-BLAST. However, only 5% of these could have been identified using structural approaches (DBS, Jones et al. 1998), that is, an extra 13% of chains could have been identified by sequence-based domain boundary assignment. Consequently, by implementing PSI-BLAST, the automatic assignment of domain boundaries of multidomain proteins in CATH was extended from 21% by using structural approaches alone (DBS) to 34% (DBS and DomainFinder).
Populating the CATH-PFDB using DomainFinder
The CATH-PFDB database was populated with homologs using DomainFinder. PSI-BLAST searches were run for each single segment CATH-95 sequences and then DomainFinder was used to assign domain sequences to their homologous superfamily using optimal thresholds (i.e., E-value <5 × 10−4 and 80% overlap criteria). Prior to integrating the domain gene sequences into an extended CATH-PFDB, their sequence similarity with all relatives in their assigned homologous superfamily was determined using a standard Needleman and Wunsch algorithm (HOMOL, Orengo et al. 1997). Single linkage clustering could then be applied to cluster the sequences into their respective sequence subfamilies at the different levels of identity employed within the CATH database (35%, 60%, 95%, 100%). Gene sequences matching a homologous superfamily with significant E-values but that had no significant sequence identities to structural relatives in the superfamily were integrated into the superfamily as separate sequence families. Using single-segment domain sequences as probes a further 144,107 single-segment domain gene sequences could be unambiguously assigned to a homologous superfamily (simple matches) and automatically integrated into the CATH-PFDB.
However, almost a quarter (23%) of the CATH-95 representative domains from the CATH database are discontiguous, multisegmented domains. Most (79%) of the discontiguous domains in the CATH database had two segments and for the majority (73%) >70% of the fold was found in the largest segment. Consequently, for discontiguous gene relatives to be recruited, a slightly less stringent overlap criteria was used in which only 70% of the domain has to be identified.
CATH-95 representatives for multisegmented discontiguous domains (751 domains) were added to the input for DomainFinder and an extra 12,938 gene sequences were assigned to a homologous superfamily. Almost half (6044) of the relatives recruited to the CATH-PFDB by matching these discontiguous domains could be identified on the basis of a single segment. A further 6686 gene relatives were recruited by identification of two sequence segments, and 208 sequences were identified on the basis of multiple segments. Multisegment domains were clustered on the basis of the concatenated sequence.
Generating the CATH-IMPALA and the CATH-PFDB IMPALA libraries
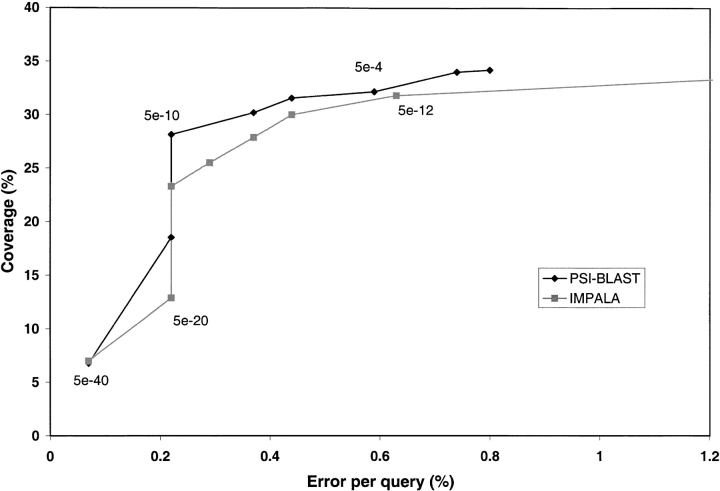
The matrices derived for each of the CATH-95 segment sequences generated to populate the CATH-PFDB were used to establish a CATH-IMPALA library. IMPALA was calibrated so that its performance was comparable to that of PSI-BLAST (see Fig. 4 ▶). In Figure 4 ▶, each point corresponds to an E-value threshold; an IMPALA E-value of 5 × 10−12 (when using the CATH-IMPALA library) was found to give the same error rate as a PSI-BLAST E-value of 5 × 10−4 (used in conjunction with the GenBankCATHnr).
Fig. 4.
Calibrating IMPALA using the CATH-35 sequences. Coverage corresponding to different PSI-BLAST (♦) and IMPALA (▪) E-values plotted against error per query, allowing one-to-one relationships and 50% overlap.
IMPALA was found to have slightly lower coverage for a set error rate (see Fig. 4 ▶), because in PSI-BLAST the PSSM alters at each iteration until convergence. For example, in 10 iterations of PSI-BLAST there will be 10 slightly different PSSMs, each probing slightly different areas of sequence space and each identifying a slightly different set of relatives. In our implementation of PSI-BLAST, the sequences from each iteration are kept rather than those just from the final iteration. IMPALA uses a single PSSM to represent all the members of a sequence family, which is equivalent to the hits being taken from the final iteration alone. Nevertheless, the difference in coverage was <1%, and IMPALA is considerably faster (Schaffer et al. 1999) when scanning small numbers of sequences against a relatively small profile database (e.g., the CATH IMPALA library), as opposed to using PSI-BLAST on the same sequences and scanning against the complete GenBankCATHnr. When updating CATH on a weekly basis, an increase in speed of identification of homology more than compensates for the minimal loss in coverage.
Once the extended CATH-PFDB database had been generated, gene sequence CATH-35 representatives were identified for sequence families without a structural representative. PSI-BLAST was used to generate IMPALA profiles for each of these extra representatives to include in the IMPALA library previously generated for the structural CATH-95 representatives. The expansion of the IMPALA profile library in this way considerably broadens the region of sequence space currently represented by the CATHIMPALA profiles and should improve the recognition of distant homologs.
Using the CATH-PFDB to detect distant evolutionary relationships
The recognition of evolutionary relatives is one of the most important stages in the classification of new structures and this level in the CATH hierarchy is of particular interest to biologists. When updating CATH, the assignment of a newly determined sequence to a protein family can be based on a single match to a relative within that family. To measure the relative performance of PSI-BLAST and IMPALA in the identification of homologs for the purpose of updating the CATH database, the coverage obtained when one-to-many relationships are allowed was calculated (Muller et al. 1999). Because we also wanted to assess the increase in sensitivity that could be obtained by using the CATH-PFDB, the special data set generated for benchmarking (CATH-35) was amended to include representative sequences from the CATH-PFDB rather than from the CATH database. However, the presence of extra intermediate sequences had caused some sequence families to merge so that the remaining data set contained a higher proportion of more diverse sequences and was therefore more stringent.
To measure the one-to-many success rate of PSI-BLAST, a given sequence representative (CATH-35) need only match one other nonidentical representative (CATH-95) from any other CATH-PFDB-35 sequence family within that homologous superfamily (rather than all) to be identified as belonging to that particular superfamily. Using PSI-BLAST and scanning CATH allowing a one-to-many relationship produces coverage of 51%, compared with 23% for a one-to-one relationship [using an E-value of 5.0 × 10−4, overlap(query) of 80%]. So although approximately 2 in 10 homologous relationships can be detected by PSI-BLAST, approximately 5 out of 10 domains with distant homology (<35% sequence identity) to a domain in the CATH database could have been reliably classified. By scanning the PSI-BLAST matches against the CATH-PFDB database, which had been populated by clear gene relatives to entries in CATH, a further substantial increase in coverage from 51% to 82% was obtained.
Similarly, using IMPALA a one-to-many relationship produces coverage of 53%, compared with 24% for a one-to-one relationship [using an E-value of 5 × 10−12, overlap(profile) of 80%]. IMPALA profiles created for the extra 9560 sequence family representatives from the extended database (CATH-PFDB IMPALA library) have improved the recognition of distant homologs, increasing the coverage from 53% to 76% in benchmark tests using manually validated CATH superfamilies.
Distant evolutionary relationships detected by DomainFinder
DomainFinder identified 16,000 significant regions in gene sequences that matched CATH-95 representatives from more than one homologous superfamily in CATH (cross-hits). This corresponded to 92 homologous superfamilies being linked via a match to a common gene sequence. These pairs were further analyzed to determine whether they represented very distant homologs, for which there had been insufficient evolutionary evidence at the time of classification in CATH. Alternatively, these matches could have been the result of the PSI-BLAST profiles drifting too far in sequence space. No homologous superfamilies were merged in CATH as a result of matching the same gene sequence, unless they shared the same fold and there was clear indication of common functional properties. When the evidence suggested that proteins exhibited distant evolutionary relationships, but either the fold or function had changed significantly, the homologous superfamilies were linked in a neighbor table and the homologous relationship stored within the CATH Oracle database (A.J. Shepherd and N. Martin, unpubl.). Where the evidence was insufficient or ambiguous the PSI-BLAST data describing the relationship between the CATH superfamilies was stored as putative.
One third of these homologous superfamilies matched a homologous superfamily from the same fold group in CATH. Manual analysis of these cases showed that 82% belonged to highly populated fold groups in CATH, which have been described as superfolds (Orengo et al. 1994) and within which the function is often observed to vary (Todd et al. 1999,Todd et al. 2001). In these cases it is often difficult to recognize very distant homologs without further evidence, such as a highly unusual structural feature that is conserved among the relatives. In some cases decisions were made to keep the families separated. There were six examples of TIM barrel folds that were linked through PSI-BLAST. Although functionally diverse (see Table 1) it has been well documented (Murzin et al. 1995; Copley and Bork 2000) that they share a phosphate-binding motif and there is evidence that they are distantly evolutionarily related. Although these superfamilies are currently not merged, these relationships are recorded as distant homologs in CATH neighbor tables and stored in the CATH Oracle database.
Table 1.
TIM barrel homologous superfamilies in CATH
| Family 1 | Family 2 | No. sequences in common |
| Dihydropteroate (DHP) synthetase (CATH:3.20.20.20) | Enolase superfamily (CATH:3.20.20.120) | 111 |
| Dihydropteroate (DHP) synthetase (CATH:3.20.20.20) | Quinolinic acid phosphoribosyl (QAPR) transferase1 (CATH:3.20.20.200) | 643 |
| Enolase superfamily (CATH:3.20.20.120) | FMN-dependent oxidoreductase and phosphate (PP) binding enzymes (CATH:3.20.20.130) | 156 |
| Enolase superfamily (CATH:3.20.20.120) | Quinolinic acid phosphoribosyl (QAPR) transferase1 (CATH:3.20.20.200) | 109 |
| Triose phosphate isomerase (TIM) (CATH:3.20.20.90) | FMN-dependent oxidoreductase and phosphate (PP) binding enzymes (CATH:3.20.20.130) | 11 |
| Triose phosphate isomerase (TIM) (CATH:3.20.20.90) | Quinolinic acid phosphoribosyl (QAPR) transferase1 (CATH:3.20.20.200) | 12 |
| FMN-dependent oxidoreductase and phosphate (PP) binding enzymes (CATH:3.20.20.130) | Aldolase class II (CATH:3.20.20.170) | 3 |
| FMN-dependent oxidoreductase and phosphate (PP) binding enzymes (CATH:3.20.20.130) | Quinolinic acid phosphoribosyl (QAPR) transferase1 (CATH:3.20.20.200) | 474 |
These families are all distant homologs, and although they have various functions they share a common phosphate binding site. Analysis of the cross-hits from DomainFinder confirms the distant evolutionary relationships between these families, and they have now been stored as homologs in neighbor tables in the CATH Oracle database. The column, No. sequences in common, indicates how many evolutionary relatives the families share that can be detected using PSI-BLAST.
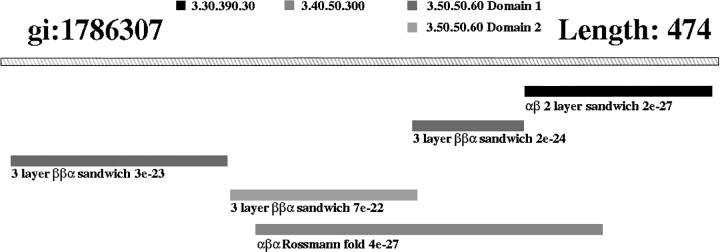
The remaining superfamilies had matched different fold groups in CATH. To check the possibility that these proteins were distant homologs, representative structures from the superfamilies were aligned using the structure comparison program SSAP (Taylor and Orengo 1989) and the results were manually validated. Several of the DomainFinder cross-hits were found to be the result of fold evolution. Lipoamide dehydogenase from Escherichia coli (SWISSPROT, P00391 GI: 1786307) belongs to the pyridine nucleotide-disulphide oxidoreductase (Class I) family. Although there is no structure for this individual protein, other members of this family comprise two three-layer αββ FAD/NAD(P) binding domains with a further C-terminal αβ domain (Todd et al. 2001). The PSI-BLAST data supports this structural assignment. However, in the analysis of cross-hits, sequences from the three-layer αβα nucleotide-binding Rossmann-like domains also match with this sequence (see Fig. 5 ▶) with significant E-values (E-values <4 × 10−22).
Fig. 5.
Diagram illustrating a cross-hit DomainFinder match. Domain assignment for lipoamide dehydogenase from the pyridine nucleotide-disulphide oxidoreductase family, which comprises a discontiguous αββ FAD/NAD(P) binding domain (3.50.50.60 domain 1) with a contiguous αββ FAD/NAD(P) binding domain inserted within it (3.50.50.60 domain 2), followed by a further αβ domain (3.30.390.30). Another significant match from a very distant homolog of different fold is also shown, the αβα nucleotide-binding domain (3.40.50.300) from which the αββ FAD/NAD(P) binding domains are thought to have evolved. (3.40.50.300 has moved to 3.40.50.720 in version 2.3 of CATH).
The three-layer αββ FAD/NAD(P) binding domain superfamily is thought to have evolved from the αβα nucleotide-binding Rossmann-like domain superfamily (Murzin et al. 1995; Vallon 2000). Members of the two superfamilies have different folds and architectures with an α-helix found between the third and fourth strand of the parallel β-sheet of the αβα nucleotide-binding Rossmann-like domains that is substituted by a small antiparallel β-sheet in the αββ FAD/NAD(P) domains. Analysis of the PSI-BLAST data suggests that these superfamilies are indeed distant evolutionary homologs. Further evidence (Vallon 2000) supports this view, including similarities in the nucleotide binding modes between the two proteins. These two superfamilies are not merged in the CATH database as they have different folds, however they are recorded as distant evolutionary homologs in the neighbor tables in the CATH Oracle database.
The majority of the remaining DomainFinder cross-matches were found to be a result of PSI-BLAST drift or motif matching; when small proteins matched large structures containing repetitive secondary structures, such as the six- and seven-bladed β-propellors, αβ and α-horseshoes and the β-solenoids. However, the analysis of the cross-hits from DomainFinder helped improve the quality of the superfamily assignments within the CATH database.
Automatic and manual procedures to speed up CATH homolog identification
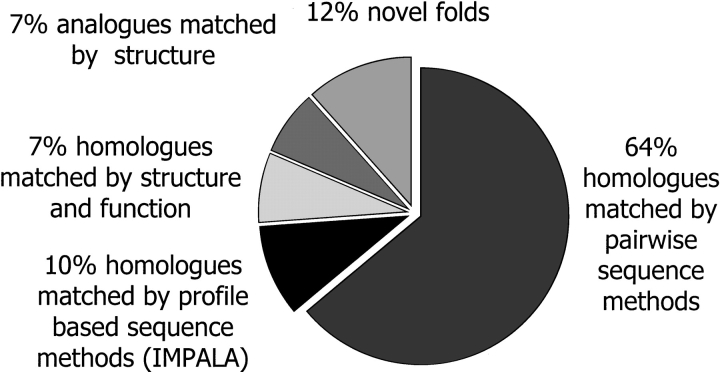
The development of the IMPALA profiles for the CATH structural domains means that a larger proportion of structural homologs can be rapidly classified in CATH using sequence-based approaches rather than the much slower structure comparison methods. To reflect these developments the CATH classification has been revised (Pearl et al. 2001). Preliminary sequence clustering using a Needleman and Wunsch algorithm is followed by scanning all the nonidentical structures against the CATH-IMPALA profiles. Any matches indicating putative homologs are subsequently checked by the structure comparison method SSAP (Taylor and Orengo 1989) and where validated added to their homologous superfamily. Figure 6 ▶ shows that for a subset of 2646 classified domains 64% could be classified by pairwise sequence methods, leaving 36% of the entries to be classified by structural comparison. However, 10% of these domains could be assigned to homologous superfamilies in CATH from matches to IMPALA profiles. This reduced the number of structures subjected to structural comparison against a large proportion of the CATH database, by over one quarter. Identification of homology using fast sequence comparison methods considerably reduces the number of structural comparisons that need to be performed in classifying newly determined protein structures and will allow CATH to keep pace with the structural genomic initiatives.
Fig. 6.
Pie-chart showing the classification of 2646 newly determined structures in the CATH database. The proportion matched by pairwise sequence methods (sequence identity ≥35%) and by IMPALA are indicated. The proportion of both homologs and analogs identified by structure comparison are also shown.
DomainFinder was also used to detect distant evolutionary relationships when assigning homologous superfamily in the CATH classification update procedure. Sequence relatives detected by PSI-BLAST/IMPALA using the DomainFinder algorithm are first validated using the structural comparison algorithm (SSAP). This is followed by manual validation of distant evolutionary relationship undertaken by visual inspection and consideration of available functional data. In a recent classification update, this procedure revealed that the N-terminal domain from folylpolyglutamate synthetase (1fgs01), a nucleotide-binding domain with a putative ATP binding P-loop, was a distant evolutionary relative of the N-terminal domain (1ffh02) of the signal sequence-binding protein (Ffh from E. coli), a GTPase that binds GTP via a P-loop. The SSAP algorithm confirmed that the structures were similar (SSAP score >74) and the P-loops were located in structurally identical sites.
Analysis of the CATH-PFDB
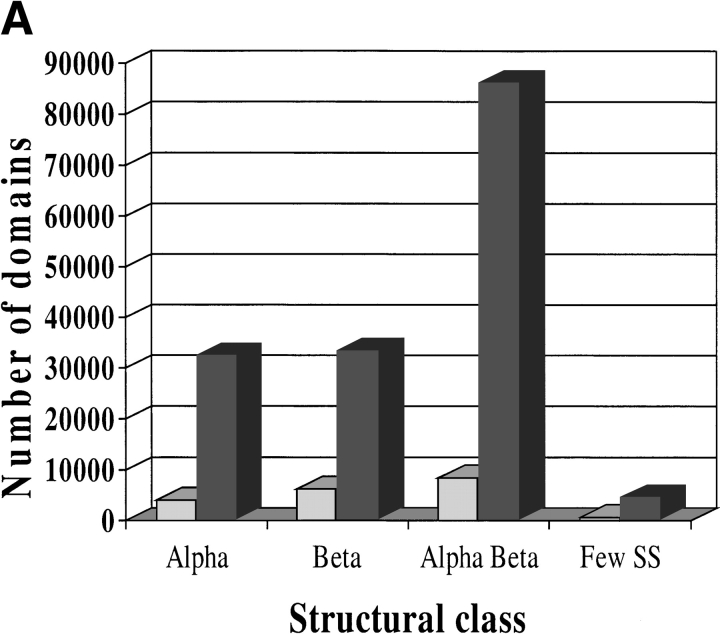
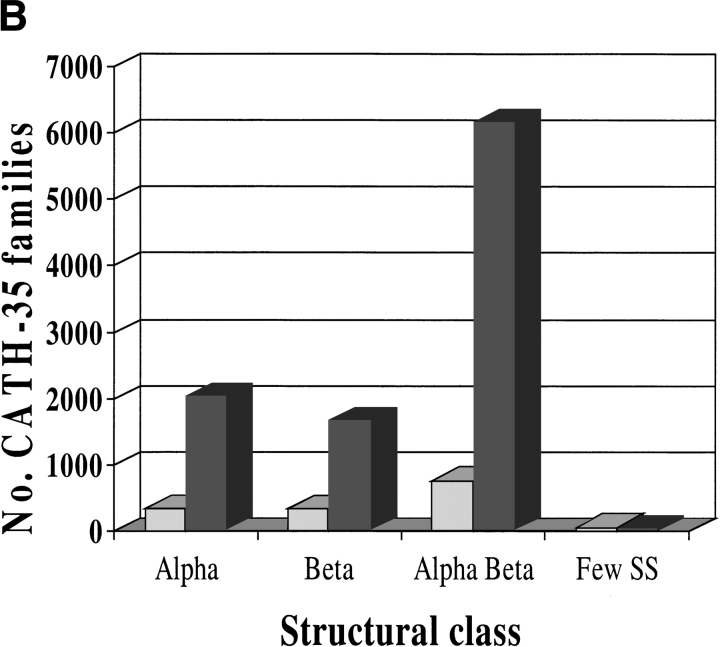
In total, 157,034 domain sequences were added to the extended CATH-PFDB from GenBank (Fig. 7A ▶). The ratio of α:β:α/β proteins is 1:1:3 where the proportion of the number of entries in CATH is 3:4:6. The sequences were then clustered into CATH-35 sequence families: Those sequences that clustered into CATH-35 families with a structural relative were termed "close" homologs. There were 1541 highly populated sequence families, comprising 73% of the sequences in the database, that had at least one close structural relative. Sequences that clustered into the sequence families without a structural relative were termed "distant" homologs. A total of 9961 extra distant sequence families have been assigned to homologous superfamilies in CATH. Figure 7B ▶ shows the distribution of the sequence families within each class.
Fig. 7.
Expansion in the number of (A) domains and (B) sequence families in the CATH database obtained by incorporating sequence relatives from GenBank. Relatives were identified using PSI-BLAST and the DomainFinder protocol. Populations are shown for the few secondary structures, mainly α, mainly β, and α/β classes in CATH, version 1.6. In A, shaded bars contain CATH domains and solid bars show domains recruited to the CATH-PFDB. In B, shaded bars contain sequences with structural relatives with ≥35% sequence identity. Solid bars contain sequences for which no close structural relative currently exists.
Therefore, of the 11,500 sequence families in the CATH-PFDB, all have a structural homolog but only 15% have a close structural homolog (>35% sequence identity) and can be accurately modeled. Thus, a structural genomic protocol that targets one model for each fold will not be sufficient to produce reliable models for functional analysis and drug design. Todd et al. (2001) also showed that below 35% sequence identity it is difficult to inherit function within an enzyme superfamily. Therefore, the CATH-PFDB can be used to extract targets for structural genomics for each sequence family within these large homologous superfamilies. The fact that the fold is already known would make these relatively easy targets that could be solved crystallographically using molecular replacement techniques with the structures available within the PDB.
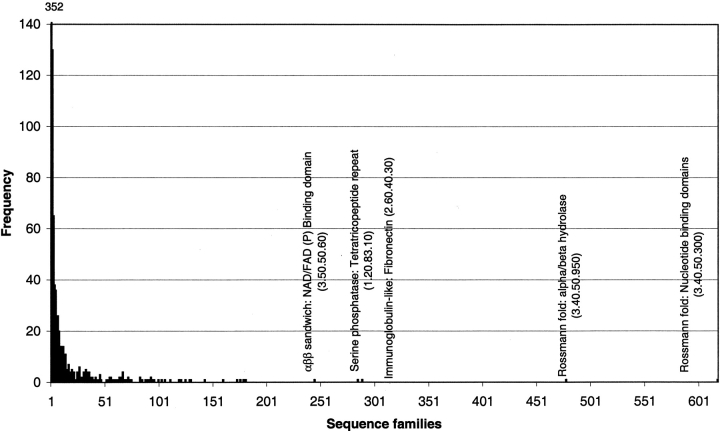
The distribution of sequence families (Fig. 8 ▶) shows that the majority (61%) of the homologous superfamilies in the CATH-PFDB are represented by more than a single sequence family. The superfamilies containing the most sequence families are the Rossmann nucleotide binding domains, which in CATH include the (P-loop) nucleotide binding proteins (618 sequence families), the αβ-hydrolases (478 sequence families), both of which are three-layer αβ-sandwiches, the αββ-FAD/NAD(P) binding domains (245 sequence families), and the two-layer β-sandwich immunoglobulins (289 sequence families).
Fig. 8.
Populations of sequence families within homologous superfamilies in the CATH-PFDB.
In the CATH-PFDB there were 593 folds distributed between 37 architectures, with 903 homologous superfamilies distributed within the fold groups. One fifth of the homologous superfamilies in the CATH-PFDB adopt one of five folds described as superfolds (Orengo et al. 1994): the mainly α-Arc-repressor, the TIM-barrel, the αβ-plait, the immunoglobulin-like β-sandwich, and the αβ-Rossmann fold. In contrast, 84% folds are just represented by one homologous superfamily. As with earlier analyses of the superfamilies and fold groups within CATH, the population of fold groups within the extended CATH-PFDB database shows a skewed distribution with at least a third of the superfamilies adopting fewer than 15 folds. This probably reflects some bias in the PDB, which contains a high proportion of structures that are enzymes. Alternatively, it may support previous hypotheses (Chothia 1992; Orengo et al. 1994) of some highly favored fold groups in nature. Whether these represent ancient ancestral folds that have been more extensively reused during evolution or favored folding arrangements that are more frequently adopted because of ease of folding, increased stability, or even extraordinary plasticity in evolving new functions is still unclear.
IMPALA Web Server
There are 176,597 entries in the CATH-PFDB of which 157,034 have been recruited from GenBank. They can be extracted by selecting to view putative sequence relatives for a given structural entry in CATH on the CATH-Gene3D website (http://www.biochem.ucl.ac.uk/bsm/cath_new/Gene3D, D.W.A. Buchan, A.J. Shepherd, D. Lee, F.M.G. Pearl, S.C.G. Rison, J.M. Thornton, and C.A. Orengo, in prep.).
A web server is available at http://www.biochem.ucl.ac.uk/bsm/cath_new/Impala/ that allows the user to search for a homologous relative within the extended CATH protein family database (CATH-PFDB). For any sequences matched, links are provided to the CATH homologous superfamily and to PDBsum (Laskowski 2001). There are also links to the CATH Dictionary of Homologous Superfamilies (Bray et al. 2000), which contains validated multiple structural alignments annotated with consensus functional information including corresponding SWISS-PROT keywords and Enzyme Commission (EC) classifications.
Conclusions
We have used PSI-BLAST to populate the CATH database with relatives from the GenBank sequence database to create the CATH protein family database (CATH-PFDB). This will enable reliable functional annotation to be added to the existing homologous superfamilies and allow extra family-specific sequences to be incorporated into sequence and sequence/structure profiles (e.g., HMMs) with confidence. The recruited sequences have been clustered with the CATH homologous superfamilies, resulting in a seven-fold expansion of the number of sequence families to which a structural assignment can be made. IMPALA profiles have been generated for each of the sequence families in the CATH-PFDB, giving a 23% increase in the number of homologs that can be detected using CATH-IMPALA profiles from 53% to 76% by allowing one-to-many relationships. New sequences can be scanned against the CATH-PFDB IMPALA library so that homology and structural data can be assigned to new sequences. As the extended protein family database contains 176,597 domain sequences from the nonredundant GenBank database, the associated IMPALA server should provide an important resource for structural genomics.
Materials and methods
Deriving a nonredundant sequence database
A nonredundant sequence database (GenBankCATHnr) was generated for PSI-BLAST searching, comprising all the sequences (478,079) in the GenBank nonedundant database (February 29, 2000) and sequences derived from structural data deposited in the PDB (Berman et al. 2000) that had been classified within the CATH database (version 1.6, April, 2000). All sequences were in FASTA format, which is the required input to PSI-BLAST.
CATH (version 1.6) is a hierarchical classification of 19,563 structural domains into evolutionary families and structural groupings comprising 903 homologous superfamilies. Version 1.6 contains 2807 near-identical protein families (CATH-95) in which the proteins in CATH with >95% sequence identity have been clustered together, and 1798 sequence families (CATH-35) in which proteins with >35% sequence identity have been clustered. A CATH-95 FASTA library was generated from one representative from each CATH-95 family. For discontiguous protein domains comprising more than a single segment, FASTA sequences for each segment were placed within the library. A total of 6710 domain and segment sequences were added to the CATH-95 library and used in the GenBankCATHnr data set, which was then masked for low complexity regions, membrane regions, and coiled coils using PFILT (D.T. Jones, pers. comm.).
Deriving sequences for CATH domains
To derive the most reliable sequence for a CATH domain, a FASTA pairwise alignment was performed between the sequence recorded in the atom records and the sequence recorded in the SEQ records within the PDB file. There are often discrepancies between the two entries when crystallographers have not been able to determine parts of the structure in disordered regions such as long loops. The SEQ records were used as the definitive sequence. However, if ATOM records were found to be missing, insertions from the SEQ records of up to 15 residues were allowed. Similarly, if either the N-terminal or C-terminal residues were missing from the structure, as long as no more than 15 were missing from either end, these residues were extracted from the SEQ records. Extensions larger than this were not allowed as they may encode a separate domain. Sequences were output in FASTA format with the CATH homology assignment added.
Generation of the IMPALA library
Each of the CATH-95 segment sequences was used as a query sequence for a PSI-BLAST (version 2.1.2) run against the GenBankCATHnr database. The initial matrix was BLOSUM62, the maximum number of iterations allowed was 20, the E-value for inclusion in the next pass was 5 × 10−4, a value recommended by Park et al. (1998) to minimize false positives, and the maximum E-value displayed was 0.1. The matrices derived for each of the CATH-95 segment sequences were used to establish a CATH-IMPALA library. When implementing IMPALA, Schaffer et al. (1999) suggested scaling the PSI-BLAST E-value cut-off by the difference in size between the IMPALA library and the original data set used to produce the library (i.e., size of IMPALA library/size of original nonredundant data set). However, we derived the IMPALA E-value by benchmarking (see Results).
Addressing PSI-BLAST drift
The sensitivity of PSI-BLAST relies on the PSSM, determining which sequences are matched. The profile moves away from the original sequence pattern as more distant relatives are pulled in and this enables it to probe more diverse regions of sequence space. However, if the profile moves too far, that is, "drift", the chances of spurious matches are increased and the profile may become corrupted (Muller et al. 1999; Park et al. 1998).
One way of measuring the extent of profile drift is to compare the structure of the query CATH-95 with the structures of any CATH-95 sequences from the GenBankCATHnr identified as homologs. This was done using the CATH classification. If a structure from a different fold group was identified as a homolog, the pair of structures (query and target) were then structurally compared using the SSAP algorithm (Taylor and Orengo 1989), which returns a normalized score in the range of 0 to 100 for identical proteins. The variation in SSAP scores with the proteins identified from each iteration can be used diagnostically to determine whether the profile has drifted too far from the original sequence. For example, if any matched sequence is found to belong to a different fold group from the query sequence, and the SSAP score is low, the PSI-BLAST algorithm can be rerun with stricter E-value constraints for inclusion in the database. SSAP score plots demonstrate when profiles have drifted. When the SSAP score between the query and structural targets is plotted, at each iteration, the SSAP score falls as the profile drifts. Only the sequences identified as relatives before the drifting occurs are allowed into the DomainFinder algorithm.
The identification by PSI-BLAST of a match between proteins with either different fold classifications or with low structure-comparison scores, and therefore significant differences in their folds, does not necessarily rule out common ancestry. There are increasing numbers of examples of fold evolution where proteins exhibiting different folds are proved to have evolved from a common ancestor (Grishin 2001). Although fold classification numbers and SSAP plots are used to identify drifting profiles and exclude them from the automatic procedure, manual interpretation of these data can reveal interesting biological discoveries.
Acknowledgments
We thank Farhana Abdulla for some preliminary analyses and David Jones for use of his computer program PFILT. We thank the MRC and the BBSRC for financial support and acknowledge support from the BSM and MRC computer services.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.16802.
References
- Altschul, S. F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D. 2000, GenBank. Nucleic Acids Res. 28 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, J.E., Todd, A.E., Pearl, F.M.G., Thornton, J.M., and Orengo, C.A. 2000. The CATH dictionary of homologous superfamilies (DHS): A consensus approach for identifying distant structural homologs. Protein Eng. 13 153–165. [DOI] [PubMed] [Google Scholar]
- Brenner, S.E., Chothia, C., and Hubbard, T.J. 1998. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl. Acad. Sci. 956073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. (1992) Proteins. One thousand families for the molecular biologist. Nature 357 543–544. [DOI] [PubMed] [Google Scholar]
- Copley, R.R. and Bork, P. 2000. Homology among (β/α)(8) barrels: Implications for the evolution of metabolic pathways. J. Mol. Biol. 303627–641. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6361–365. [DOI] [PubMed] [Google Scholar]
- Grishin, N.V. 2001. Fold change in evolution of protein structures. J. Struct. Biol. 134167–185. [DOI] [PubMed] [Google Scholar]
- Huynen, M., Doerks, T., Eisenhaber, F., Orengo, C., Sunyaev, S., Yuan, Y., and Bork, P. 1998. Homology-based fold predictions for Mycoplasma genitalium proteins. J. Mol. Biol. 280 323–326. [DOI] [PubMed] [Google Scholar]
- Jones, S., Stewart, M., Michie, A., Swindells, M.B., Orengo, C.A. and Thornton, J.M. 1998. Domain assignment for protein structures using a consensus approach: Characterisation and analysis. Protein Sci. 7233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, K., Barrett, C., and Hughey, R. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 14846–856. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A. 2001. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res. 29221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, A., MacCallum, R.M., and Sternberg, M. 1999. Benchmarking PSI-BLAST in genome annotation. J. Mol. Biol. 2931257–1271. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. 1995. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247536–540. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A, Jones, D.T., and Thornton, J.M. 1994. Protein superfamilies and domain superfolds. Nature 372631–634. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Michie, A.D., Jones, S., Jones, D.T., Swindells, M.B. and Thornton, J.M. 1997. CATH—A hierarchical classification of protein domain structures. Structure 5 1093–1108. [DOI] [PubMed] [Google Scholar]
- Park, J., Teichmann, S.A., Hubbard, T., and Chothia, C. 1997. Intermediate sequences increase the detection of homology between sequences. J. Mol. Biol. 273349–354 [DOI] [PubMed] [Google Scholar]
- Park, J., Karplus, K., Barrett, C., Hughey, R., Haussler, D., Hubbard, T., and Chothia, C. 1998. Sequence comparisons using multiple sequences detect three times as many remote homologs as pairwise methods. J. Mol. Biol. 2841201–1210. [DOI] [PubMed] [Google Scholar]
- Pearl, F.M.G., Lee, D., Bray, J.E., Sillitoe, I., Todd, A.E., Harrison, A.P., Thornton, J.M., and Orengo, C.A. 2000. Assigning genomic sequences to CATH. Nucleic Acids Res. 28277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, F.M.G., Martin, N.M., Bray, J.E., Buchan, D.W.A., Harrison, A.P., Lee, D., Reeves, G.A., Shepherd, A.J., Sillitoe, I., Todd, A.E., Thornton, J.M., and Orengo, C.A. 2001. A rapid classification protocol for the CATH domain database to support structural genomics. Nucleic Acids Res. 29223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12 85–94. [DOI] [PubMed] [Google Scholar]
- Salamov, A.A., Suwa, M., Orengo, C.A., and Swindells, M.B. 1999a. Combining sensitive database searches with multiple intermediates to detect distant homologues. Protein Eng. 1295–100. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Genome analysis: Assigning protein coding regions to three-dimensional structures. Protein Sci. 8 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, A.A., Wolf, Y.I., Ponting, C.P., Koonin, E.V., Aravind, L., and Altschul, S.F. 1999. IMPALA: Matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics. 151000–1011. [DOI] [PubMed] [Google Scholar]
- Taylor, W.R. and Orengo, C.A. 1989. Protein structure alignment. J. Mol. Biol. 208 1–22 [DOI] [PubMed] [Google Scholar]
- Teichmann, S.A., Chothia, C., Church, G.M., and Park, J. 2000. Fast assignment of protein structures to sequences using the intermediate sequence library PDB-ISL. Bioinformatics 16117–124. [DOI] [PubMed] [Google Scholar]
- Todd, A.E., Orengo, C.A., and Thornton, J.M. 1999. Evolution of protein function, from a structural perspective. Curr. Opin. Chem. Biol. 3 548–556. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307 1113–1143. [DOI] [PubMed] [Google Scholar]
- Vallon, O. 2000. New sequence motifs in Flavoproteins: Evidence for common ancestry and tools to predict structure. Proteins 3895–114. [DOI] [PubMed] [Google Scholar]