Abstract
The redox protein flavodoxin has been shown earlier to be reduced by the pyruvate-oxidoreductase (POR) enzyme complex of Helicobacter pylori, and also was proposed to be involved in the pathogenesis of gastric mucosa-associated lymphoid-tissue lymphoma (MALToma). Here, we report its X-ray structure, which is similar to flavodoxins of other bacteria and cyanobacteria. However, H. pylori flavodoxin has an alanine residue near the isoalloxazine ring of its cofactor flavin mononucleotide (FMN), while the other previously crystallized flavodoxins have a larger hydrophobic residue at this position. This creates a solute filled hole near the FMN cofactor of H. pylori flavodoxin. We also show that flavodoxin is essential for the survival of H. pylori, and conclude that its structure can be used as a starting point for the modeling of an inhibitor for the interaction between the POR-enzyme complex and flavodoxin.
Up to 50% of the world's human population is chronically infected with Helicobacter pylori. The pathogen is regarded as responsible for type-B gastritis and peptic-ulcer diseases (Blaser 1992), gastric carcinoma (Nomura et al. 1991; Parsonnet et al. 1991) and mucosa-associated lymphoid-tissue lymphoma (MALToma) (Wotherspoon et al. 1993). The increasing occurrence of antibiotic-resistant H. pylori strains poses a threat to therapeutic regimes. New molecular targets suited for the eradication of H. pylori should fulfill the following criteria: they should be essential for the survival of H. pylori and absent in the human host to avoid strong side effects of medication.
The H. pylori genome encodes the small acidic redox protein flavodoxin (Tomb et al. 1997; Alm et al. 1999) with the fldA gene. Flavodoxins are flavin mononucleotide (FMN) containing proteins involved in a variety of electron transfer reactions. They have been identified in both prokaryotes and eukaryotes but not in mammals (Osborne et al. 1991; Romero et al. 1996), where flavodoxin-like domains are found as part of larger proteins, as for example cytochrome P450 reductase (Wang et al. 1997). Flavodoxins exist in three redox states: in an oxidized form (OX), as a semiquinone (SQ), or as a hydroquinone (HQ). The SQ to HQ reduction step is shifted by the apoprotein from −172 mV for free FMN to ∼−400 mV upon complex formation, and thought to be of physiological relevance, as most electron transfer reactions involving flavodoxins occur at a low redox potential (Mayhew and Ludwig 1975; Vervoort et al. 1994). Flavodoxins can be divided into two structural classes: short-chain flavodoxins with ∼150 residues and long-chain flavodoxins with an insertion of ∼20 amino-acid residues interrupting the final strand of β-sheet (Mayhew and Ludwig 1975).
The long-chain flavodoxin of H. pylori functions as electron acceptor to the pyruvate-oxidoreductase (POR) enzyme complex, which catalyzes the oxidative decarboxylation of pyruvate (Hughes et al. 1995; Tomb et al. 1997; Kaihovaara et al. 1998). Additionally, it was discovered recently that sera from patients with gastric MALToma contained antibodies against a 19 kD protein from H. pylori (Chang et al. 1999; Shiesh et al. 2000). The identification of this protein as flavodoxin inspired the sequencing of the fldA genes from 26 H. pylori strains isolated from patients with MALToma or other H. pylori-related diseases (Chang et al. 1999). In seven out of seven strains from patients with MALToma, but only in five out of 17 strains from patients with the other diseases, fldA encoded 164 amino acid residues, while the remaining 12 (out of 17) strains had a gene encoding 175 amino acids. The two forms of the gene differed by an additional guanine base at position 481 of the DNA sequence of the elongated form. This insertion altered the carboxy-terminal sequence 160G-S-F-A164 of the short-form protein to 160V-S-L-L164 in the long form, and also resulted in the addition of an extra 11 residues at the C-terminus. From the correlation between the occurrence of the short-form flavodoxin and the diagnosis of gastric MALToma, a potential role of the redox protein in the pathogenesis of H. pylori-associated MALToma was proposed (Chang et al. 1999).
Results and Discussion
Insertion mutagenesis of H. pylori fldA and por α
To investigate the role of flavodoxin in the metabolism of H. pylori, we tried to inactivate the fldA gene through insertion of an antibiotic resistance marker. Transformation of five different H. pylori strains (26695, 503, 504, 69A, and 888–0) with a fldA::cat construct yielded only a limited number of chloramphenicol-resistant colonies. In contrast, transformation with a plasmid containing an interrupted gene coding for a structural flagellin subunit (flaA::cat), shown to be nonessential under laboratory conditions (Haas et al. 1993), rendered a large number of chloramphenicol-resistant colonies (Table 1). To investigate if the fldA gene was indeed inactivated through homologous recombination in the chloramphenicol-resistant clones, we isolated genomic DNAs and performed PCR and Southern-blot analysis. In each case, an apparently intact fldA gene was found (Fig. 1 ▶). When examining the expression of flavodoxin in the putative knock-out clones by Western-blot analysis, in each case flavodoxin was present in the same amount as in the respective wild-type strain. We conclude that flavodoxin is essential for the five investigated H. pylori strains. This result is not unexpected, as flavodoxin is the electron acceptor of the pyruvate-oxidoreductase enzyme complex (POR; Hughes et al. 1995; Kaihovaara et al. 1998). POR is the only known pyruvate-decarboxylating enzyme in H. pylori (Tomb et al. 1997), and its β subunit was shown to be essential for the H. pylori strain NCTC11637 by insertional mutagenesis (Hughes et al. 1998). To confirm that the POR-enzyme complex is essential not only for H. pylori NCTC11637, we tried to inactivate the gene encoding its α subunit in five additional strains (26695, 503, 504, 69A, and 888–0). Similar to the results obtained for the fldA gene, only a small number of chloramphenicol-resistant colonies were obtained after transformation with a porα::cat construct (Table 1). All of these had a wild-type version of the porα gene and contained normal amounts of Porα protein, as was shown by PCR, Southern- and Western-blot analyses (data not shown). Therefore, both the POR enzyme complex and flavodoxin are essential for the five investigated H. pylori strains. This is in contrast, for example, to the cadA gene encoding a P-type ATPase, which was shown to be essential for only some H. pylori strains, while other strains were able to survive without an intact copy of this gene (Herrmann et al. 1999). Because neither flavodoxin nor POR are present in the human host, a substance interfering with their interaction might be well suited for eradication therapies of H. pylori.
Table 1.
Chloramphenicol-resistant (catR) colonies obtained after transformation with the flaA::cat, fldA::cat or porα::cat constructs
| Number of colonies after transformation with | |||
| H. pyloristrain | flaA::cat | fldA::cat | porα::cat |
| 26695 | 500 | 2 | 1 |
| 26695 | 500–1000 | 0 | 1 |
| 26695 | 500 | 0 | 0 |
| 26695 | 1000–5000 | 0 | 0 |
| 503 | 100 | 2 | 0 |
| 503 | 85 | 5 | 0 |
| 504 | 100 | 0 | 0 |
| 504 | 128 | 0 | 0 |
| 69A | 200 | 0 | 0 |
| 69A | 500–1000 | 0 | 0 |
| 69A | 500 | 0 | 0 |
| 888-0 | >5000 | 2 | 30 |
| 888-0 | 1000–5000 | 8 | 7 |
| 888-0 | 1000–5000 | 1 | 0 |
Each line represents an independent transformation experiment.
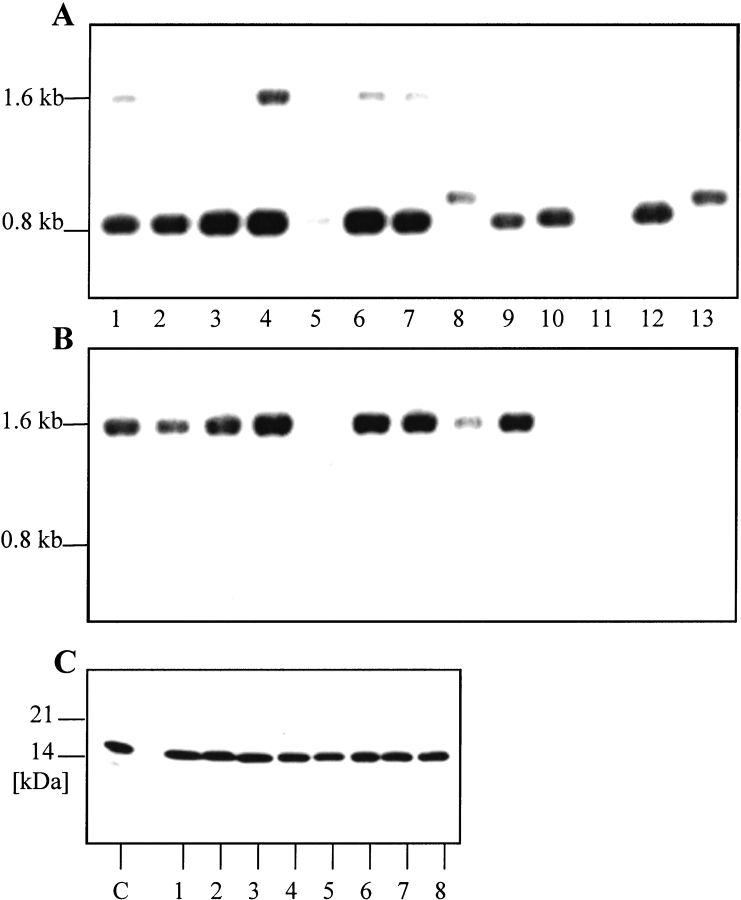
Fig. 1.
Knockout-mutagenesis of the fldA gene. (A, B) Southern-blot analysis of catR clones. Genomic DNA (∼2 μg) was digested with HindIII, separated by gel electrophoresis on a 1% agarose gel and blotted by capillary transfer onto a nylon membrane (Amersham Hybond-N; Southern 1975). Probes for the nonradioactive detection of DNA were generated with the PCR DIG Probe Synthesis Kit (Roche) according to the manufacture's protocol and covered the complete open reading frames. (A) Hybridization with a fldA-specific probe. (B) Hybridization with a cat-specific probe. (C) Western-blot analysis. 1 μg of total bacterial protein were separated under nonreducing conditions on a 10% PA gel. The proteins were transferred on a nitrocellulose membrane, and flavodoxin was detected with flavodoxin antiserum. 1–8: CatR Helicobacter pylori. 1, 2: 26695-fldA::cat; 3, 4: 503-fldA::cat; 5, 6, 7: 69A-fldA::cat; 8: 888–0-fldA::cat; 9–12: H. pylori wild types; 9: 26695; 10: 503; 11: 504; 12: 69A; 13: 888–0. (C) C, control. Recombinant flavodoxin, 25ng.
The overall structure of H. pylori flavodoxin
The structure of oxidized H. pylori flavodoxin (strain 69A) was solved by molecular replacement to 2.4 Å resolution (Table 2). The protein has an α/β-fold, and shares a high degree of similarity with the other structurally known flavodoxins, as can be inferred from a structure-based alignment (Fig. 2 ▶). Five parallel β strands (β1–β5) form a central β-sheet that is flanked on both sides by a total of five α helices (α1–α5). Helices α1 and α5 are on one side of the β-sheet, and helices α2, α3, and α4 are located on the opposing side (Fig. 3 ▶). Strand β5 is interrupted by an insertion of 21 residues, which makes the H. pylori protein a long-chain flavodoxin. The binding site for the cofactor FMN is located at the carboxy-terminal end of the β-sheet. Currently, the coordinates for the oxidized forms of the following flavodoxins are available from the protein data bank: The long-chain flavodoxins from Anabaena PCC 7120 (Rao et al. 1992), Anacystis nidulans (Drennan et al. 1999), Chondrus crispus (Fukuyama et al. 1992), and E. coli (Hoover and Ludwig 1997), and the short-chain flavodoxins from Clostridium beijerinckii (Ludwig et al. 1997) and Desulfovibrio vulgaris (Watenpaugh et al. 1973). A superposition of these structures revealed that H. pylori flavodoxin most closely resembles the proteins from Anabaena PCC 7120 and A. nidulans. The Cα-trace of the H. pylori protein deviates only in three loop regions significantly from the Cα-traces of these two proteins. The α2–β3 loop contains two residues less than the Anabaena and Anacystis proteins (Fig. 2 ▶). Similarly, the α4–β5 loop contains two residues less than the equivalent loops of the other flavodoxins. This different architecture affects the neighboring α3–β4 loop, which adopts a conformation significantly different from those found in other species. However, all described differences in the α2–β3, α3–β4, and α4–β5 loops are unlikely to affect the active site, as the distance from the loops at the amino-terminal end of the central β-sheet to the cofactor is larger than 15 Å.
Table 2.
Crystallographic data collection and refinement statistics
| Space group | P21 |
| Unit cell constants | a = 31.1 Å b = 47.2 Å, c = 48.3 Å, β = 97.1° |
| Resolution limitsa | 20.00–2.40 Å (2.49–2.40 Å) |
| Number of measured reflections | 13,654 |
| Number of unique reflections | 5346 |
| Completeness of dataa | 96.6% (99.2%) |
| Rsyma | 8.5% (32.5%) |
| I/σa | 12.2 (2.7) |
| Rwork | 18.2% |
| Rfree | 26.7% |
a The values for the highest resolution shell are given in parentheses.
Fig. 2.
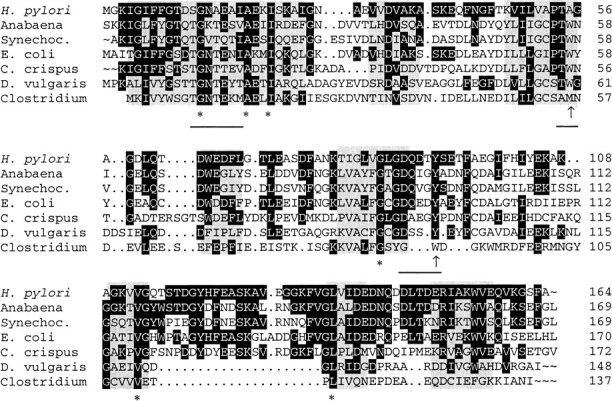
Structural alignment of flavodoxins with resolved three-dimensional structure. The alignments were performed according to regions of structural similarity as determined by Multiple Alignment of Proteins Structures (MAPS) (Guoguang 1998). Structural identity between all crystal structures is indicated by gray boxes. The 6 amino-acid residues conserved between these proteins are marked by asterisks, while amino acids identical to the Helicobacter pylori flavodoxin are shown with a black background. Hydrophobic amino acids in proximity to the isoalloxazine ring are indicated by an arrow, and residues involved in binding of the FMN cofactor are underlined.
Fig. 3.
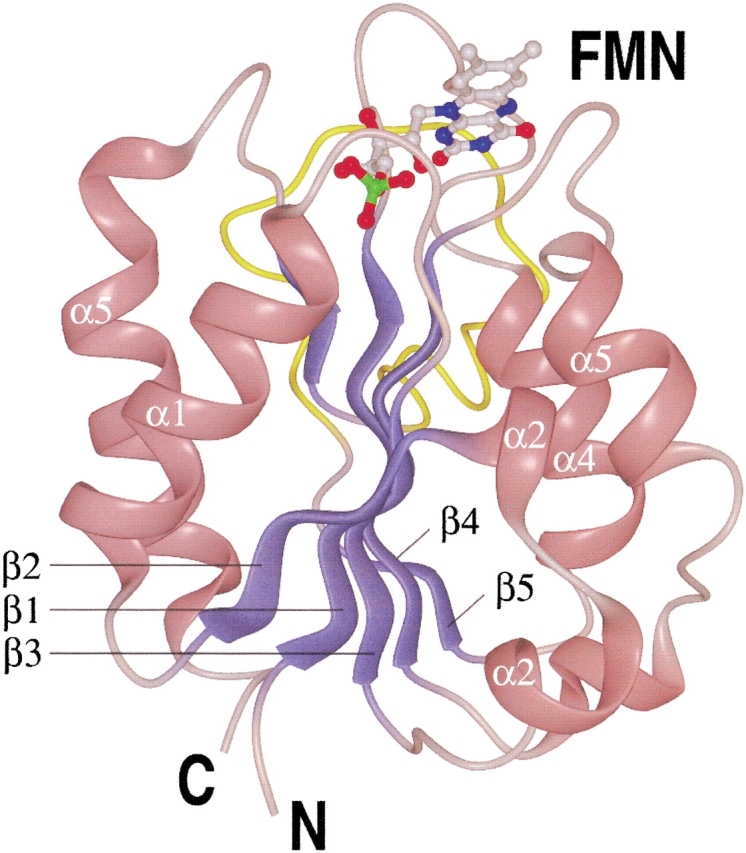
Overview of the Helicobacter pylori flavodoxin structure. Helices are shown in pink, β strands are shown in blue, and the insertion of 21 amino acids interrupting β-strand 5, making the protein a long-chain flavodoxin, is shown in yellow. The FMN cofactor is shown in a "ball and stick" representation.
The shorter α3–β4 and α4–β5 loops result in a more compact shape of the H. pylori protein, with only 7278Å2 water-accessible surface, while the other long-chain flavodoxins possess between 7665 and 8033 Å2. Even the short-chain flavodoxin from Desulfovibrio vulgaris has 7162Å2 water-accessible surface, although having 19 residues less than the H. pylori protein. Beside shorter loops, H. pylori flavodoxin has relative small amino-acid side chains on its surface. For example, the H. pylori protein has 18 alanine residues (11.0% of all residues) compared to the Anabaena flavodoxin with only eight alanine residues (4.7% of all residues). We can only speculate that the compact structure of H. pylori flavodoxin is complementary to the shapes of its interaction partners, and might thus be the result of functional constraints.
Phosphoribityl binding
The complexion of the cofactor FMN involves two distinct mechanisms in flavodoxins. The phosphoribityl part of the molecule is bound by hydrogen bonds, while the isoalloxazine ring is kept in place mainly by hydrophobic interactions. The phosphate group is bound by a loop that contains the key fingerprint motif for flavodoxins, T/S-X-T-G-X-T (Drennan et al. 1999). While the other flavodoxins of known structure fit exactly into this pattern, the H. pylori protein (10T-D-S-G-N-A15) does not (Fig. 2 ▶): Ala15 is at a position where in other flavodoxins a threonine forms a hydrogen bond with the phosphate. In Anabaena, the mutation of Thr15 to the isosteric valine results in a destabilization of the complex by ∼2 kcal/mol (Lostao et al. 2000). Additionally, a commonly found hydrogen bond from a tryptophan side chain to a phosphate oxygen is absent in the H. pylori flavodoxin, where an alanine residue is found at the equivalent position. Despite these differences involving side-chain atoms, the complexion of the phosphate is similar in all structurally known flavodoxins, as it mainly involves hydrogen bonds to main chain atoms. In contrast, the ribityl part of the cofactor mainly interacts with side-chain atoms. In the H. pylori structure, side-chain atoms from Asn14 and Asp142 form hydrogen bonds to the ribityl part of FMN. In most other flavodoxin structures, identical residues are found at equivalent positions.
Isoalloxazine binding
The isoalloxazine ring of FMN is embedded into the flavodoxin structure by stacking between two hydrophobic residues. In the case of flavodoxin from C. beijerinckii, these are a methionine at the re-, and a tryptophan at the si-face of the ring system. All other structurally known flavodoxins have a tyrosine at the re-, and a tryptophan at the si-face. This well-conserved architecture is not found in the H. pylori flavodoxin, where the tryptophan at the si-face is replaced by an alanine (Fig. 4 ▶). As a consequence, the isoalloxazine ring is rotated by ∼20° around an axis through the cofactor atoms N(1) and O(4). This rotation does not affect the hydrogen-bonding pattern of the cofactor atoms N(1), O(2), N(3), and O(4) to protein backbone atoms, which is highly conserved among flavodoxins. The absence of the indole group of tryptophan creates a cavity in the protein, which is only partially closed by the rotation of the cofactor. This makes the FMN cofactor more accessible to the solvent. No electron density for ordered water molecules were observed in this cavity (Fig. 5 ▶). We want to stress that the presence of an alanine at this key position is not restricted to flavodoxin from strain 69A, as all other published sequences of H. pylori fldA encode an alanine at this position (data not shown).
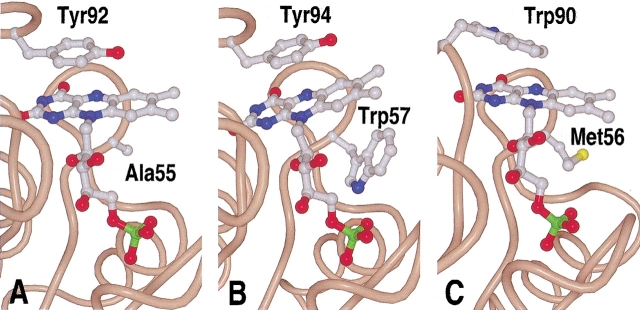
Fig. 4.
Complexion of the FMN cofactor by hydrophobic residues. Most so-far crystallized flavodoxins have a tyrosine at the si-face of the isoalloxazine ring, and a tryptophan residue at the re-face, as for example (B) the Anabaena flavodoxin. (A) In the Helicobacter pylori protein, Tyr92 is found at the si-face of the isoalloxazine ring, and an alanine residue at the re-face. (C) The Clostridium beijerinckii flavodoxin has an unusual structure in regard that Met56 is found at the si-face.
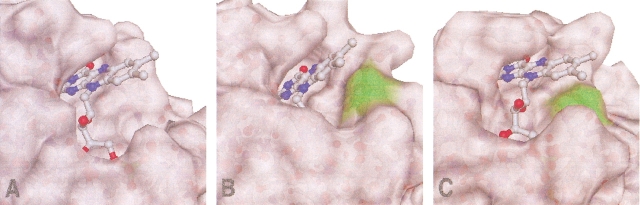
Fig. 5.
Solvent accessibility of the cofactor in the flavodoxins from (B) Anabaena, (A) Helicobacter pylori, and (C) Clostridium beijerinckii. The presence of an alanine residue at the si-face of H. pylori flavodoxin creates a cavity that renders the cofactor more accessible to the solvent.
The mutation of tryptophan to alanine at the equivalent position in the Anabaena flavodoxin leads to a destabilization of the complex by 3 kcal/mol, and to a 100-fold increase in the dissociation constant (Lostao et al. 1997). In H. pylori, the missing aromatic side chain combined with the above-mentioned effect of an alanine at position 15 of the phosphate-binding loop might result in a relative weak binding of the FMN cofactor. The fact that the residues in the phosphate-binding loop and the hydrophobic residues on both sides of the isoalloxazine ring are highly conserved among flavodoxins of different species indicates a general requirement for a tight FMN binding, with observed dissociation constants in the nanomolar range (Hoover and Ludwig 1997). We can currently offer no explanation for the comparatively loose binding of the FMN cofactor, which was inferred from the described structural details.
The solvent accessibility of FMN is correlated to the redox potential of flavodoxin, because solvent access has a stabilizing effect on the anionic hydroquinone (Swenson and Krey 1994). While the water-accessible surface of the FMN is between 99 and 109 Å2 in the other long-chain flavodoxins, it is 160A2 in H. pylori. The missing tryptophan on the si-face of FMN might result in a more positive redox potential of H. pylori flavodoxin. In D. vulgaris flavodoxin, the mutation W60A shifts the midpoint redox potential of the SQ/HQ step by 86 mV to a more positive value (Mayhew et al. 1996). However, in Anabaena, the equivalent mutation W57A produces only a minor shift of 19 mV to a more positive value (Lostao et al. 1997). It is not known why the observed effect differs so significantly between the two proteins despite their similarities at the active site. We therefore cannot rule out the possibility that the tryptophan residue at this position has an additional, still unidentified role in the function of flavodoxins.
Main chain conformation of the Gly56-Ala57 peptide
A crystallographic analysis of flavodoxin from C. beijerinckii in oxidized, semireduced, and fully reduced form revealed that reduction of the cofactor is accompanied by a conformational change of a peptide bond near the isoalloxazine ring. While in oxidized C. beijerinckii flavodoxin the Gly57-Asp58 peptide bond is in the cis-conformation, the trans-conformation is found in the reduced semiquinone and hydroquinone forms (Ludwig et al. 1997). In the latter case, the carbonyl oxygen of the peptide bond points towards the isoalloxazine and has therefore been termed trans O-up conformation. The importance of the side chain residue for the main chain conformation has been confirmed by structural analysis of the oxidized G57N mutant, which adopts a trans O-down conformation with the carbonyl group pointing away from the cofactor (Ludwig et al. 1997). Flavodoxin from Anabaena, which has an asparagine residue at the position equivalent to Gly57 from C. beijerinckii, also has a trans O-down conformation (Rao et al. 1992). H. pylori flavodoxin contains Gly56 at the position equivalent to Gly57 of the C. beijerinckii protein. The observed electron density is in good agreement with a trans O-down conformation, the same conformation that is found in the crystal structure of an Asn58Gly mutant of A. nidulans flavodoxin (Hoover et al. 1999). This mutation stabilizes the trans O-down conformation in the semiquinone oxidation state and raises the OX/SQ potential by 46 mV, while it lowers the SQ/HQ potential by 26 mV. The glycine found in the H. pylori protein might therefore to some extent compensate for the redox shift caused by the missing tryptophan side chain at the si-face of the FMN cofactor.
The structural role of the carboxy-terminal residues
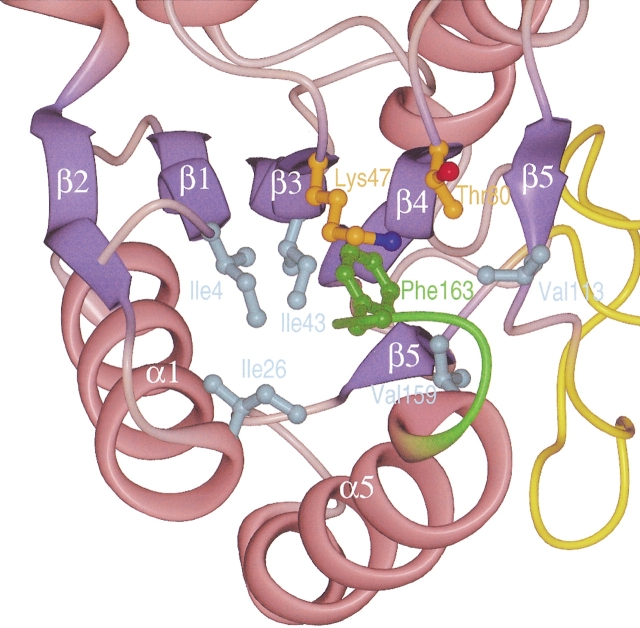
The suspected role of flavodoxin from H. pylori in the pathogenesis of gastric MALToma induced the sequencing of fldA genes from several clinical H. pylori isolates (Chang et al. 1999). Two different forms of flavodoxins were found: a long form with 175 residues, and a short form containing 164 amino-acid residues, the latter more frequently found in strains from patients with gastric MALToma. It was concluded that the short form of the protein represents a truncated version (Chang et al. 1999). If this were the case, the structure of the last four residues in the short form of the protein should differ significantly from other flavodoxins and also might be structurally disordered. The protein used in our study had 164 amino acids and represented the short form of H. pylori flavodoxin. The crystal structure of the carboxy-terminal part closely resembles the corresponding parts of the Anabaena sp. PCC 7120 and A. nidulans proteins. These two proteins also contain a phenylalanine at the position equivalent to Phe163 in H. pylori. This residue has an important role in the stabilization of the protein, as its aromatic side chain is an integral part of the hydrophobic core. In the H. pylori protein, Phe163 stabilizes the orientation of the helices α1 and α5 relative to the central β-sheet, as it forms hydrophobic interactions to Ile4 of strand β1, Ile26 of helix α1, Ile49 of strand β3, Val113 of strand β5, and Val159 of helix α5 (Fig. 6 ▶). In the elongated form of H. pylori, flavodoxin Phe163 is replaced by a serine. Because its short polar side chain can not fulfill a similar structural role, this difference would probably destabilize the protein fold. The elongated protein also contains 11 additional residues at its C-terminus. Of the structurally known flavodoxins, only the E. coli protein contains extra carboxy-terminal residues when compared to the H. pylori protein. These residues form a short helix of unknown function (Hoover and Ludwig 1997). However, the carboxy-terminal part of the E. coli protein does not show any sequence similarity to the elongated H. pylori protein, despite the high percentage of identical residues in the rest of the protein (Fig. 2 ▶).
Fig. 6.
Stabilization of the overall structure of Helicobacter pylori flavodoxin by the carboxy-terminal residues. The part of the protein that is altered in the elongated form is shown in green. Side-chain atoms are only shown for residue Phe163 of the short form of H. pylori flavodoxin, and of contacting residues. Amino acids belonging to the hydrophobic core of the protein are shown in cyan, and amphipatic residues shielding Phe163 from the solvent are shown in orange.
These considerations suggest that the short form of H. pylori flavodoxin is a fully functional protein, in contrast to a previous proposition (Chang et al. 1999). We conclude that the elongated form found in some strains most likely resulted from a deletion of one nucleotide in the fldA gene, which caused a random amino-acid sequence at the C-terminus. The important structural role of the carboxy-terminal residues in the short form of H. pylori flavodoxin imply a severe impairment of the stability and probably the function of the elongated form. Because we showed that flavodoxin is an essential protein in H. pylori, strains with an elongated version of flavodoxin might have a decreased metabolic capacity. There is, however, no obvious correlation between the structural differences of flavodoxins found in different H. pylori strains and the potential role of the protein in the pathogenesis of MALToma and other H. pylori-related diseases.
Conclusions
We reported the crystal structure of oxidized flavodoxin from H. pylori (strain 69A) and compared it with the structures of flavodoxins from other species. H. pylori flavodoxin has marked structural differences compared to other flavodoxins, e.g., differences in FMN binding and a more compact shape. We also demonstrated that flavodoxin and its electron donor, the POR enzyme complex, are essential for H. pylori. In regard to the recently proposed involvement of H. pylori flavodoxin in the pathogenesis of MALToma (Chang et al. 1999), our data suggest that the H. pylori flavodoxin with 164 amino acids is a stable and fully functional protein, while the H. pylori flavodoxin version with 175 amino acids is impaired both in stability and functionality.
Materials and methods
Bacterial strains and culture conditions
The H. pylori strains 26695 (Tomb et al. 1997), 503 (ATCC 43503), 504 (ATCC 49504), 69A, and 888–0 (clinical isolates obtained from R. Haas, Max-von Pettenkofer-Institut, Munich, Germany) were cultivated at 37°C in brain-heart infusion medium (BHI; Difco-BD Biosciences), supplemented with 6% fetal-calf serum (FCS; Eurobio). Microaerobic conditions were generated by an activated Anaerocult C pack (Merck & Co., Inc.) in an anaerobic jar (Merck & Co., Inc.). Liquid cultures were grown in a shaker incubator at 90 rpm. For transformation, H. pylori was cultivated until late logarithmic phase in BHI liquid medium and then diluted to an OD578 of 0.1 (Haas et al. 1993). Aliquots of 1 mL were incubated for 6 h in 24-well tissue culture plates (Greiner) under microaerobic conditions at 37°C. Then, 250–500 ng of plasmid DNA were added. After 16 h incubation, the bacteria were plated on GC agar plates (Schmitt and Haas 1994) containing 6 μg/mL chloramphenicol. The plates were incubated for 5 to 7 d, until bacterial colonies became visible. Plates without visible growth were incubated for at least 14 days. For conservation purposes, H. pylori was stored in BHI/20% glycerol at −80°C. Escherichia coli strains were propagated at 37°C, and liquid cultures were incubated on a shaker incubator at 200 rpm.
DNA isolation, PCR, and DNA sequencing
Genomic DNA was prepared according to the protocol for gram-negative bacteria from the QIAamp Tissue Kit (Qiagen). The standard PCR reaction contained 25 ng genomic DNA, 25 pmol of each primer (Interactiva, Ulm, Germany), 1 × PCR-reaction buffer, 200 nM dNTP-mix, and 2.5 U Expand High Fidelity DNA-polymerase mix (all from Roche) in a total volume of 50 μl. All PCR reactions were overlaid with mineral oil (Perkin Elmer) and performed in a Robocycler Gradient 40 (Stratagene). After initial denaturation for 2 min at 92°C, the DNA was amplified in 30 cycles (1 min denaturation at 92°C, 1 min annealing at 60°C, and 1–4 min extension at 72°C), followed by a final extension step of 7–15 min at 72°C. PCR products were purified with the PCR Purification Kit from Qiagen. Aliquots were analyzed by agarose gel electrophoresis. DNA cycle sequencing was performed by GATC (Konstanz, Germany).
Construction of suicide plasmids containing interrupted fldA or por α genes
A chloramphenicol resistance cassette (cat) was inserted by SOE-PCR (Ho et al. 1989; Horton et al. 1989) into the H. pylori fldA gene, under concomitant deletion of the amino acids 51–104 of the fldA open reading frame. The cat-cassette (Haas et al. 1993) was amplified with the primers I786 (GCACACTAGAAGCGAGC GATTTCCGGTTTTTGTTAATCCGCC) and I803 (AGTTTCGC TGTAAGTGTCTTGATTTACGCCCCGCCCTGCCACTC). The resulting PCR-product was ligated to the 5` and 3` region of the fldA gene in separate PCRs using the primer pairs I786 and I728 (CTCGAATTCGGTTTGGTTGTCTATTTCTAGCAT), and I803 and I727 (CTCGAATTCGTTCTTATAGCGCTTTTAATATGG), respectively. The products of these two PCR reactions were combined and amplified with the primers I727 and I728 to generate a fldA gene interrupted by a cat-antibiotic resistance cassette. The construct was cloned into the TOPO-TA vector (Invitrogen), which is unable to replicate in H. pylori. At the same time, a suicide vector containing an interrupted porα gene, under concomitant deletion of the amino acids 151–269 of the porα open reading frame, was constructed using the primers I722 (CTCGA ATTCCATATGAAGATTGGAATGAGTTTGAAATGG), I745 ( C G A A T T C G C T C T T C C G C A A A A A A A G C T C A T T T T A G G G C CGTG), I784 (GATTCTGGTTGGATAAGTTTATTCCGGTTT TTGTTAATCCGCC), and I785 (GGTTAAATGGTATTGCCGC CTTTACGCCCCGCCCTGCCACTC).
Recombinant expression of H. pylori flavodoxin
Recombinant H. pylori flavodoxin was purified as described (Paul et al. 2001). Shortly, the fldA gene of the H. pylori strain 69A was PCR-amplified and cloned into the NdeI and SapI restriction sites of the pTYB1 expression vector (New England Biolabs [NEB]). E. coli Top10 transformed with the pTYB1/fldA expression plasmid was induced 4 h 30 min with 1 mM IPTG at 30°C, harvested by centrifugation and lysed with a high-pressure homogenizer (APV-Gaulin). After 1 h centrifugation at 30,000g, the supernatant was applied to a chitin column (NEB). After extensive washing (with 5–500 mM NaCl; 20mM Tris-HCl pH 8.0; 0.1 mM EDTA), the cleavage between the flavodoxin and the intein-CBD part of the fusion protein was induced with 30 mM β-mercaptoethanol. After 16 h incubation at 4°C, flavodoxin was eluted from the chitin column and further purified on a MonoQ anion exchange column.
Analysis of proteins
The concentration of protein in solution was determined according to Bradford (1976), and PA gel electrophoresis was performed according to Schägger and von Jagow (1987). Proteins were visualized with either Coomassie blue or silver staining (Heukeshoven and Dernick 1988). For immunostaining, proteins were transferred onto nitrocellulose membranes (Protran BA85 Schleicher & Schüll; Towbin et al. 1979). The antiserum against flavodoxin was used in a 1:25,000 dilution (Paul et al. 2001). The secondary, antirabbit antibody (Diagen), which was conjugated to horseradish-peroxidase, was detected by ECL (Amersham).
Crystallization and data collection
For crystallization, the sitting-drop vapor diffusion method was used. Equal volumes of protein solution (5 mg/mL) and reservoir solution were mixed and equilibrated against a 20-fold volume of reservoir solution. To identify suitable crystallization conditions, the sparse matrix approach was used (Jancarik et al. 1991). The best crystals were obtained from condition 43 containing 30% (w/w) PEG 1500 at 17°C. They took 6 wk to grow to a final size of 200 × 200 × 200 μm and were of a bright yellow color. A single crystal was mounted in a quartz capillary and measured at room temperature with a conventional rotating anode generator as x-ray source. Diffraction data were recorded on an MAR345 image plate detector and reduced with Denzo/Scalepack (Minor et al. 1996). The crystals belonged to space group P21 with a = 31.1 Å; b = 47.3 Å; c = 48.3 Å; β = 97.1°; and contained one molecule per asymmetric unit. This resulted in a Matthews coefficient of 2.03 Å3/Da.
Molecular replacement and model refinement
Molecular replacement and refinement of the model were carried out with CNS version 0.4 (Brunger et al. 1998). The oxidized flavodoxin from E. coli (Hoover and Ludwig 1997) was used as a search model. Data from 15 to 4 Å were included in the rotation search, which yielded an unambiguous solution. Only the best solution was used for the subsequent translation search. The correct solution had an R-factor of 47.2% using all reflections between 15 and 4 Å resolution. During the energy minimization refinement, simulated annealing and individual temperature-factor refinement were used, alternating with manual model rebuilding. For all refinement procedures, the maximum likelihood target function was used. In addition, an overall anisotropic temperature factor and a bulk solvent mask correction were applied. A total of 32 water molecules were included in the final model.
Model analysis
Stereochemical parameters were analyzed with PROCHECK (Laskowski et al. 1996). Secondary structure assignment was calculated with DSSP (Kabsch and Sander 1984). For the optimized superposition of various flavodoxin structures from different organisms, the program SUPERIMPOSE (Diederichs 1995) was used. Calculation of root mean squared deviations of Cα-positions and the structure-based sequence alignments were calculated with MAPS (Guoguang 1998). Calculations of solvent-accessible surface areas were carried out with CNS version 0.4 (Brunger et al. 1998), using a probe radius of 1.4 Å.
Acknowledgments
The work described here was supported by a grant from the German Ministry of Education and Research (BMBF; BEO/ 22 031 0777). We thank Klaus Bensch, Uwe Bosch, Marion Eisenhauer, Sandro Ghisla, Imme Hartig-Beeken, and Klaus Melchers for discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.28602.
References
- Alm, R.A., Ling, L.S., Moir, D.T., King, B.L., Brown, E.D., Doig, P.C., Smith, D.R., Noonan, B., Guild, B.C., deJonge, B.L., Carmel, G., Tummino, P.J., Caruso, A., Uria-Nickelsen, M., Mills, D.M., Ives, C., Gibson, R., Merberg, D., Mills, S.D., Jiang, Q., Taylor, D.E., Vovis, G.F. and Trust, T.J. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397 176–80. [DOI] [PubMed] [Google Scholar]
- Blaser, M.J. 1992. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology 102 720–7. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–54. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T., and Warren, G.L. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D. Biol. Crystallogr. 54 905–21. [DOI] [PubMed] [Google Scholar]
- Chang, C.S., Chen, L.T., Yang, J.C., Lin, J.T., Chang, K.C., and Wang, J.T. 1999. Isolation of a Helicobacter pylori protein, FldA, associated with mucosa-associated lymphoid tissue lymphoma of the stomach. Gastroenterology 117 82–8. [DOI] [PubMed] [Google Scholar]
- Diederichs, K. 1995. Structural superposition of proteins with unknown alignment and detection of topological similarity using a six-dimensional search algorithm. Proteins 23 187–95. [DOI] [PubMed] [Google Scholar]
- Drennan, C.L., Pattridge, K.A., Weber, C.H., Metzger, A.L., Hoover, D.M. and Ludwig, M.L. 1999. Refined structures of oxidized flavodoxin from Anacystis nidulans. J. Mol. Biol. 294 711–24. [DOI] [PubMed] [Google Scholar]
- Fukuyama, K., Matsubara, H., and Rogers, L.J. 1992. Crystal structure of oxidized flavodoxin from a red alga Chondrus crispus refined at 1.8 A resolution. Description of the flavin mononucleotide binding site. J. Mol. Biol. 225 775–89. [DOI] [PubMed] [Google Scholar]
- Guoguang, L. 1998. MAPS (Multiple Alignment of Protein Structures), Version 1.0. http://gamma.mbb.ki.se/guoguang.maps.html.
- Haas, R., Meyer, T.F., and van Putten, J.P. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8 753–60. [DOI] [PubMed] [Google Scholar]
- Herrmann, L., Schwan, D., Garner, R., Mobley, H.L., Haas, R., Schafer, K.P., and Melchers, K. 1999. Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol. Microbiol. 33 524–36. [DOI] [PubMed] [Google Scholar]
- Heukeshoven, J. and Dernick, R. 1988. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9 28–32. [DOI] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 51–9. [DOI] [PubMed] [Google Scholar]
- Hoover, D.M. and Ludwig, M.L. 1997. A flavodoxin that is required for enzyme activation: The structure of oxidized flavodoxin from Escherichia coli at 1.8 A resolution. Protein Sci. 6 2525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, D.M., Drennan C.L., Metzger, A.L., Osborne, C., Weber, C.H., Pattridge, K.A., and Ludwig, M.L. 1999. Comparisons of wild-type and mutant flavodoxins from anacystic nidulans. Structural determinants of the redox potentials. J. Mol. Biol. 294 725–43. [DOI] [PubMed] [Google Scholar]
- Horton, R.M., Hunt, H.D., Ho, S.N., Pullen, J.K., and Pease, L.R. 1989. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 77 61–8. [DOI] [PubMed] [Google Scholar]
- Hughes, N.J., Chalk, P.A., Clayton, C.L., and Kelly, D.J. 1995. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J. Bacteriol. 177 3953–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, N.J., Clayton, C.L., Chalk, P.A. & Kelly, D.J. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180 1119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J., Scott, W.G., Milligan, D.L., Koshland, D.E., Jr., and Kim, S.H. 1991. Crystallization and preliminary x-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J. Mol. Biol. 221 31–4. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1984. On the use of sequence homologies to predict protein structure: Identical pentapeptides can have completely different conformations. Proc. Natl. Acad. Sci. 81 1075–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaihovaara, P., Hook-Nikanne, J., Uusi-Oukari, M., Kosunen, T.U., and Salaspuro, M. 1998. Flavodoxin-dependent pyruvate oxidation, acetate production and metronidazole reduction by Helicobacter pylori. J. Antimicrob. Chemother. 41 171–7. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–86. [DOI] [PubMed] [Google Scholar]
- Lostao, A., El Harrous, M., Daoudi, F., Romero, A., Parody-Morreale, A., and Sancho, J. 2000. Dissecting the energetics of the apoflavodoxin-FMN complex. J. Biol. Chem. 275 9518–26. [DOI] [PubMed] [Google Scholar]
- Lostao, A., Gomez-Moreno, C., Mayhew, S.G., and Sancho, J. 1997. Differential stabilization of the three FMN redox forms by tyrosine 94 and tryptophan 57 in flavodoxin from Anabaena and its influence on the redox potentials. Biochemistry 36 14334–44. [DOI] [PubMed] [Google Scholar]
- Ludwig, M.L., Pattridge, K.A., Metzger, A.L., Dixon, M.M., Eren, M., Feng, Y., and Swenson, R.P. 1997. Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: The role of conformation changes. Biochemistry 36 1259–80. [DOI] [PubMed] [Google Scholar]
- Mayhew, S.G. and Ludwig, M.L. 1975. Flavodoxins and electron-transferring flavoproteins. Academic Press, New York, NY.
- Mayhew, S.G., O'Connell, D.P., O'Farrell, P.A., Yalloway, G.N., and Geoghegan, S.M. 1996. Regulation of the redox potentials of flavodoxins: Modification of the flavin binding. Biochem. Soc. Trans. 24 122–7. [DOI] [PubMed] [Google Scholar]
- Minor, W., Steczko, J., Stec, B., Otwinowski, Z., Bolin, J.T., Walter, R., and Axelrod, B. 1996. Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution. Biochemistry 35 10687–701. [DOI] [PubMed] [Google Scholar]
- Nomura, A., Stemmermann, G.N., Chyou, P.H., Kato, I., Perez-Perez, G.I., and Blaser, M.J. 1991. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 325 1132–6. [DOI] [PubMed] [Google Scholar]
- Osborne, C., Chen, L.M., and Matthews, R.G. 1991. Isolation, cloning, mapping, and nucleotide sequencing of the gene encoding flavodoxin in Escherichia coli. J Bacteriol 173 1729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet, J., Friedman, G.D., Vandersteen, D.P., Chang, Y., Vogelman, J.H., Orentreich, N., and Sibley, R.K. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325 1127–31. [DOI] [PubMed] [Google Scholar]
- Paul, R., Bosch, U., and Schaefer, K.P. 2001. Overexpression and purification of Helicobacter pylori flavodoxin and induction of a specific antiserum in rabbits. Protein Expr. Purif. 22 399–405. [DOI] [PubMed] [Google Scholar]
- Rao, S.T., Shaffie, F., Yu, C., Satyshur, K.A., Stockman, B.J., Markley, J.L., and Sundarlingam, M. 1992. Structure of the oxidized long-chain flavodoxin from Anabaena 7120 at 2 A resolution. Protein Sci. 1 1413–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, A., Caldeira, J., Legall, J., Moura, I., Moura, J.J., and Romao, M.J. 1996. Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states. Eur. J. Biochem. 239 190–6. [DOI] [PubMed] [Google Scholar]
- Schägger, H. and von Jagow, G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–79. [DOI] [PubMed] [Google Scholar]
- Schmitt, W. and Haas, R. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: Structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 12 307–19. [DOI] [PubMed] [Google Scholar]
- Shiesh, S.C., Sheu, B.S., Yang, H.B., Tsao, H.J., and Lin, X.Z. 2000. Serologic response to lower-molecular-weight proteins of H. pylori is related to clinical outcome of H. pylori infection in Taiwan. Dig. Dis. Sci. 45 781–8. [DOI] [PubMed] [Google Scholar]
- Southern, E.M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98 503–17. [DOI] [PubMed] [Google Scholar]
- Swenson, R.P. and Krey, G.D. 1994. Site-directed mutagenesis of tyrosine-98 in the flavodoxin from Desulfovibrio vulgaris (Hildenborough): Regulation of oxidation-reduction properties of the bound FMN cofactor by aromatic, solvent, and electrostatic interactions. Biochemistry 33 8505–14. [DOI] [PubMed] [Google Scholar]
- Tomb, J.F., White, O., Kerlavage, A.R., Clayton, R.A., Sutton, G.G., Fleischmann, R.D., Ketchum, K.A., Klenk, H.P., Gill, S., Dougherty, B.A., Nelson, K., Quackenbush, J., Zhou, L., Kirkness, E.F., Peterson, S., Loftus, B., Richardson, D., Dodson, R., Khalak, H.G., Glodek, A., McKenney, K., Fitzegerald, L.M., Lee, N., Adams, M.D., Venter, J.C., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388 539–47. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. 76 4350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort, J., Heering, D., Peelen, S., and van Berkel, W. 1994. Flavodoxins. Methods Enzymol. 243 188–203. [DOI] [PubMed] [Google Scholar]
- Wang, M., Roberts, D.L., Paschke, R., Shea, T.M., Masters, B.S., and Kim, J.J. 1997. Three-dimensional structure of NADPH-cytochrome P450 reductase: Prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. 94 8411–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watenpaugh, K.D., Sieker, L.C., and Jensen, L.H. 1973. The binding of riboflavin-5`-phosphate in a flavoprotein: Flavodoxin at 2.0-Angstrom resolution. Proc. Natl. Acad. Sci. 70 3857–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotherspoon, A.C., Doglioni, C., Diss, T.C., Pan, L., Moschini, A., de Boni, M., and Isaacson, P.G. 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342 575–7. [DOI] [PubMed] [Google Scholar]