Abstract
A prerequisite for structural genomics and related projects is to standardize the process of gene overexpression and protein solubility screening to enable automation for higher throughput. We have tested a methodology to rapidly subclone a large number of human genes and screen these for expression and protein solubility in Escherichia coli. The methodology, which can be partly automated, was used to compare the effect of six different N-terminal fusion proteins and an N-terminal 6*His tag. As a realistic test set we selected 32 potentially interesting human proteins with unknown structures and sizes suitable for NMR studies. The genes were transferred from cDNA to expression vectors using subcloning by recombination. The subcloning yield was 100% for 27 (of 32) genes for which a PCR fragment of correct size could be obtained. Of these, 26 genes (96%) could be overexpressed at detectable levels and 23 (85%) are detected in the soluble fraction with at least one fusion tag. We find large differences in the effects of fusion protein or tag on expression and solubility. In short, four of seven fusions perform very well, and much better than the 6*His tag, but individual differences motivate the inclusion of several fusions in expression and solubility screening. We also conclude that our methodology and expression vectors can be used for screening of genes for structural studies, and that it should be possible to obtain a large fraction of all NMR-sized and nonmembrane human proteins as soluble fusion proteins in E. coli.
Keywords: Fusion proteins, solubility, recombination, genetic disorder, structural genomics
Structural biology, as most fields in molecular biology, is facing a massive increase in potential targets for structure determination due to the success of the large scale genome sequencing efforts. Structural biology on a genomic scale is referred to as structural genomics (Shapiro and Lima 1998). A prerequisite for structural genomics is to standardize the process of target screening and sample preparation and thus enable the automation needed for higher throughput (Edwards et al. 2000). This is in contrast to traditional structural biology for which purification protocols are optimized for each individual protein.
Regardless of approach, E. coli is still the expression host of choice due to its advantages in ease of use, high growth and production rates, cheapness, and availability. It has some disadvantages in that success rates for expressing eukaryotic proteins are low, most post-translational modifications are absent, and the product is often in the form of inclusion bodies (Baneyx 1999). The process used to refold proteins from inclusion bodies must be considered as difficult to set up in a high throughput format, and any improvements in the solubility of recombinant proteins in E. coli are therefore beneficial in a high throughput project. One commonly used strategy to increase solubility is to make a fusion to a protein that is known to have high solubility. There are several fusion proteins that have been shown to increase solubility in E. coli. For instance glutathion-S-transferase (GST) (Smith and Johnson 1988), the maltose binding protein (MBP) (Bedouelle and Duplay 1988; di Guan et al. 1988), and the Z-domain from protein A (Nilsson et al. 1987), which were developed as affinity fusion proteins, have been shown to increase solubility. The His tag (Gentz et al. 1988; Hochuli et al. 1988; Smith et al. 1988) was also developed as an affinity tag, but it is not commonly used to increase solubility. Later and more specialized fusion proteins are thioredoxin (LaVallie et al. 1993), NusA (Davis et al. 1999), and the Gb1-domain from protein G (Gb1) (Huth et al. 1997). There has not been a comprehensive comparative study of the effects of these different fusion protein and tag possibilities, and available comparisons have only been carried out with a limited set of genes (Sachdev and Chirgwin 1998b; Kapust and Waugh 1999; Wang et al. 1999). One objective with the present study is to compare the effects of many different fusions upon expression and solubility for a large set of human gene products in E. coli.
A second objective is to establish a methodology for gene expression screening that can be used in structural genomics. One of many hurdles in large-scale studies has been the cloning step for which numerous restriction and ligation reactions are needed. An alternative to restriction and ligation is to use recombination reactions. We have, therefore, adopted the Gateway cloning technology (Walhout et al. 2000) (Life technologies), which is a modification of the recombination system of phage lambda. This system allows for directional cloning in a two-step reaction. In the first reaction, called the BP reaction, the gene of interest is inserted into a donor vector to create what is referred to as an entry clone. In the second reaction, called the LR reaction, the gene of interest is moved from the entry clone into a destination vector to create an expression clone. One advantage of this system lies in the fact that a large number of different expression clones can easily be created from one entry clone. Thus, it is easy to test a number of different conditions affecting expression, like gene fusions and promoter. A second advantage is that the system is host independent, so it can be used for prokaryotic as well as for eukaryotic expression hosts.
We have converted four different expression vectors to be able to use them in addition to three destination vectors available from Invitrogen/Life Technologies. Thus, we have a set of seven different expression vectors that were used to screen for expression and solubility of proteins in E. coli. As a realistic set of genes for testing the methodology we have selected 32 different human proteins with unknown structure and that are associated with genetic disorders. We have developed a protocol for rapid cloning and expression, and adopted the methods to allow for automation. We have thus created an easily expanded platform that allows for the consistent comparisons needed to select optimal expression conditions. We also find that there are profound differences with regard to expression levels and solubility depending on the fusion partner used.
Results
Target selection
We combined information from different databases to find 126 interesting proteins that would be potential targets for structure determination within a structural genomics project. The targets were selected to fulfill our criteria of being human proteins without transmembrane regions, which are nonhomologous to any protein with known structure, and for which entries are linked to the OMIM database of genes associated with disease (Hamosh et al. 2000). Of the 126 selected targets, 32 could be obtained as full-length cDNA clones through the IMAGE consortium (Lennon et al. 1996). The proteins, which are listed in Table 1, have sizes in the range of 6 to 20 kD, and are therefore all potentially suitable for structural studies using NMR as well as X-ray crystallography.
Table 1.
Human protein targets and efficiency of the different cloning and expression stepsa
| Gene | SwissProt ID | Size (kD) | Cysteine content | Solubility probabilityb | PCR-product | Entry clone | Expr. clonec | Expressionc | Soluble productc | Commentd |
| 1 | MAR1_HUMAN | 13.2 | 4% | 12% | Y | Y | Y | Y | Y | |
| 2 | Q9UH52 | 8.0 | 0% | 11% | Y | Y | Y | Y | Y | |
| 3 | SMPX_HUMAN | 9.6 | 1% | 2% | Y | Y | Y | Y | Y | |
| 4 | Q9UMT3 | 12.1 | 3% | 16% | Y | Y | Y | Y | Y | |
| 5 | Q9Y260 | 11.4 | 6% | 6% | Y | Y | Y | Y | Y | Low solubility |
| 6 | SAA_HUMAN | 13.5 | 1% | 21% | Y | Y | Y | Y | Y | |
| 7 | Q9Y605 | 14.6 | 1% | 97% | Y | Y | Y | Y | Y | |
| 8 | BC10_HUMAN | 9.9 | 8% | 31% | Y | Y | Y | Y | N | Not soluble |
| 9 | STP2_HUMAN | 15.6 | 4% | 64% | N | Wrong size | ||||
| 10 | APR_HUMAN | 6.0 | 4% | 72% | Y | Y | Y | Y | Y | |
| 11 | Q9UI30 | 14.2 | 2% | 54% | Y | Y | Y | Y | Y | Low solubility |
| 12 | Q9NZA6 | 12.5 | 0% | 47% | Y | Y | Y | Y | Y | |
| 13 | HBP1_HUMAN | 8.5 | 0% | 98% | Y | Y | Y | Y | Y | |
| 14 | NCYM_HUMAN | 11.7 | 7% | 18% | Y | Y | Y | Y | Y | |
| 15 | Q9UI41 | 10.9 | 3% | 42% | Y | Y | Y | N | Not expressing | |
| 16 | IPKA_HUMAN | 8.0 | 0% | 74% | Y | Y | Y | Y | Y | |
| 17 | Q9UMZ1 | 11.0 | 0% | 100% | N | Wrong cDNA | ||||
| 18 | HSP1_HUMAN | 6.7 | 12% | 100% | N | No PCR product | ||||
| 19 | TCTP_HUMAN | 19.5 | 1% | 79% | Y | Y | Y | Y | Y | |
| 20 | MCS_HUMAN | 12.7 | 17% | 4% | Y | Y | Y | Y | Y | |
| 21 | O75394 | 7.6 | 2% | 98% | Y | Y | Y | Y | N | Not soluble |
| 22 | STP1_HUMAN | 6.2 | 0% | 78% | Y | Y | Y | Y | N | Not soluble |
| 23 | STHM_HUMAN | 17.2 | 0% | 71% | N | No PCR product | ||||
| 24 | DAP1_HUMAN | 11.2 | 0% | 5% | Y | Y | Y | Y | Y | |
| 25 | RP14_HUMAN | 13.7 | 3% | 29% | Y | Y | Y | Y | Y | Low solubility |
| 26 | WIT1_HUMAN | 10.4 | 2% | 42% | N | No PCR product | ||||
| 27 | ALR_HUMAN | 15 | 6% | 62% | Y | Y | Y | Y | Y | |
| 28 | BTG1_HUMAN | 19.2 | 2% | 15% | Y | Y | Y | Y | Y | |
| 29 | BIR5_HUMAN | 16.4 | 4% | 67% | Y | Y | Y | Y | Y | |
| 30 | MGN_HUMAN | 17.2 | 1% | 58% | Y | Y | Y | Y | Y | |
| 31 | PPR8_HUMAN | 13.3 | 0% | 14% | Y | Y | Y | Y | Y | |
| 32 | PFD3_HUMAN | 18.7 | 1% | 48% | Y | Y | Y | Y | Y | |
| Stepwise success | 84% | 100% | 100% | 96% | 88% | |||||
| Overall success | 84% | 84% | 84% | 81% | 72% | |||||
a Y denotes a successful outcome of the experiment, for example, a PCR product or expression in the expression test. N denotes an unsuccessful outcome of the experiment, for example, a PCR product of incorrect size or no expression. Blank denotes that the experiment could not be performed due to an earlier unsuccessful experiment.
b The probability that the protein will be soluble when expressed in E. coli calculated using the empirically derived "revised Wilkinson-Harrison solubility model" (Davis et al. 1999).
c In these cases Y denotes a successful outcome of the experiments with any of the seven different expression vectors.
d Comment on current status; see the Results section.
Success rates for subcloning, expression, and solubility screening
The overall outcome of the various steps in the screening procedure is summarized in Table 1. DNA fragments containing the genes of interest with flanking recombination sequences were obtained by PCR. Of the 32 ordered cDNA clones, 27 (84%) gave a PCR product of expected size in the amplification step. Of the five cDNA clones that failed in the amplification step, three yielded no PCR product, one yielded product of the wrong size, and one was not the correct cDNA clone. For the four with no product or product of the wrong size, variations in annealing temperature and substrate concentrations in the PCR reaction were tested without any positive effect, indicating that either their gene sequences are problematic to amplify or there is something wrong with these cDNA clones.
The PCR fragments were recombined with a Gateway donor vector to create a set of 27 entry clones. These were, in turn, recombined with three Gateway destination vectors containing an N-terminal 6*His tag, a GST fusion, and a thioredoxin fusion, respectively, and also with four additional Gateway-compatible vectors that we constructed. These four vectors code for NusA, Gb1, MBP, and a double Z domain (ZZ) as N-terminal fusion proteins. Thus, a total of 189 different expression clones were created. We found the efficiency of both the BP and the LR Gateway reactions to be very high: 95% of screened colonies were positive, and entry and expression clones could be established for all 27 (100%) genes for which a PCR product of correct size could be obtained.
Expression and solubility screening was then carried out in E. coli. Of the 27 successfully cloned genes, 26 (96%) showed expression in at least one of the seven tested expression vectors. Soluble products could be obtained for 23 out of 26 expressing genes (88%). The overall success rate of going from identified target to soluble protein product was, therefore, 72%. However, we note that the largest attrition occurs at the initial step of obtaining a PCR product from the cDNA clone. Hence, the success rate for obtaining a soluble fusion protein, given a PCR fragment with recombination overhangs, is as high as 85%.
Expression and solubility screening and differences between N-terminal fusions
We tested expression at the temperatures 20, 28, and 37°C, and could see only minor effects of temperature on the amount of soluble protein. As growing at suboptimal temperatures greatly increased the growth time but showed no consistent effect on solubility, we performed our tag comparisons at 37°C.
The effects of different fusion proteins upon expression and solubility levels are shown in Table 2. The relative number of genes that are expressed and the levels of expression are generally high. An example is shown in Figure 1 ▶. Overall, there is also a good consistency in the results for any given gene or any given fusion vector. Gene number 7, for instance, has high expression, and the product has high solubility in all different constructs; genes that have high expression in any vector generally have high expression also in the other vectors. However, a closer examination of Table 2 reveals several facts worth noting.
Table 2.
Expression and solubility levels of the 27 cloned targets when expressed as seven different gene fusions at 37°a,b
| 6*His | GST | NusA | ZZ | Gb1 | MBP | Thioredoxin | ||||||||
| Gene | Exp | Sol | Exp | Sol | Exp | Sol | Exp | Sol | Exp | Sol | Exp | Sol | Exp | Sol |
| 1 | +++ | 0 | +++ | + | +++ | + | + | + | +++ | + | ++ | + | +++ | ++ |
| 2 | +++ | ++ | +++ | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 3 | − | − | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 4 | ++ | 0 | * | * | ++ | 0 | ++ | 0 | + | ++ | + | + | +++ | + |
| 5 | − | − | − | − | − | − | − | − | − | − | + | + | ++ | 0 |
| 6 | − | − | − | − | +++ | +++ | − | − | * | * | * | * | ||
| 7 | +++ | ++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 8 | + | 0 | − | − | − | − | − | − | − | − | − | − | − | − |
| 10 | +++ | 0 | +++ | + | ++ | + | +++ | + | ++ | ++ | ++ | + | ++ | + |
| 11 | +++ | 0 | +++ | + | +++ | + | +++ | 0 | +++ | + | +++ | + | +++ | + |
| 12 | − | − | − | − | − | − | − | − | ++ | ++ | − | − | + | 0 |
| 13 | +++ | + | +++ | ++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 14 | +++ | 0 | ++ | + | − | − | + | + | +++ | + | ++ | ++ | +++ | ++ |
| 15 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 16 | +++ | + | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 19 | +++ | ++ | +++ | ++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 20 | + | 0 | ++ | + | +++ | +++ | +++ | ++ | ++ | ++ | +++ | +++ | ++ | +++ |
| 21 | − | − | * | * | ++ | 0 | +++ | 0 | − | − | + | 0 | ++ | 0 |
| 22 | − | − | +++ | 0 | ++ | 0 | +++ | 0 | + | 0 | ++ | 0 | +++ | 0 |
| 24 | − | − | +++ | ++ | +++ | ++ | +++ | +++ | +++ | +++ | ++ | ++ | +++ | +++ |
| 25 | − | − | − | − | +++ | 0 | +++ | 0 | − | − | ++ | 0 | ++ | + |
| 27 | +++ | + | +++ | ++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 28 | +++ | + | +++ | 0 | +++ | 0 | +++ | +++ | ++ | ++ | +++ | ++ | +++ | +++ |
| 29 | − | − | * | * | − | − | * | * | ++ | ++ | +++ | ++ | ||
| 30 | + | + | ++ | 0 | +++ | 0 | +++ | ++ | ++ | ++ | +++ | ++ | +++ | ++ |
| 31 | + | 0 | * | * | +++ | ++ | +++ | ++ | ++ | ++ | +++ | +++ | +++ | ++ |
| 32 | * | * | * | * | * | * | * | * | * | * | * | * | ++ | +++ |
| yieldc | 59% | 30% | 59% | 48% | 74% | 52% | 74% | 56% | 70% | 67% | 81% | 70% | 89% | 74% |
a Expression levels given as: +++ = strongest band on SDS-PAGE gel, ++ = among the stronger bands, + = visible band, − = no visible band, * = incorrect size, blank = no data available.
b Solubility given as: +++ = majority in soluble fraction, ++ = roughly 50% in soluble fraction, + = minority in soluble fraction, 0 = nothing in soluble fraction, − = no expression, blank = no data available.
c Percentage of genes that are expressed (of 27) and percentage of gene products that are soluble (of 27). The actual levels are not taken into account.
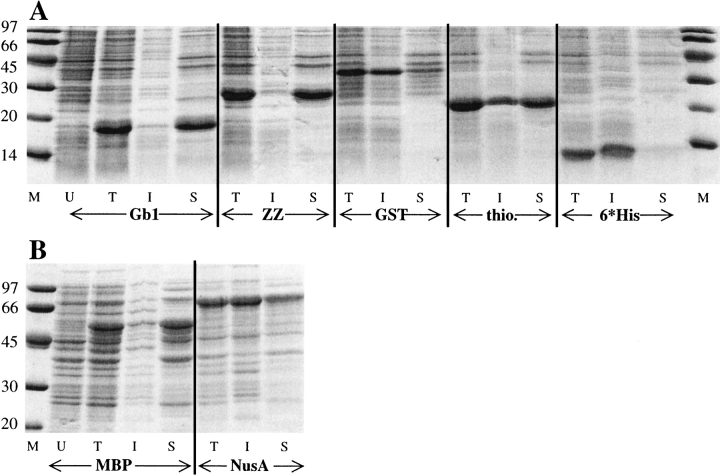
Fig. 1.
SDS-Polyacrylamide gels showing soluble and insoluble fractions after expressing gene number 2 at 37°C using seven different expression vectors. U, T, I, S, and M denote uninduced total fraction, induced total fraction, insoluble fraction, soluble fraction, and size marker, respectively. The insoluble fraction was desolved in a volume equal to that of the soluble fraction. Marker sizes are given in the left column. (A) A 16% gel with Gb1, ZZ, GST, thioredoxin, and 6*His tag fusions, the expected sizes are 17, 26, 36, 22, and 11 kD, respectively. (B) A 10% gel with MBP and NusA fusions, the expected sizes are 52 and 64 kD.
Out of 27 cloned genes, 26 gave expression levels detectable on polyacrylamide gels. Twenty-three of these 26 were expressed in at least two different vectors, and 11 were expressed in all seven different vectors. No single expression vector gave expression of all 26 expressing genes. This high number could only be achieved by a combination of at least three different vectors, for instance thioredoxin, NusA, and 6*His tag. With this combination, thioredoxin accounts for the largest success rate with expression of 24 genes, and the His tag and NusA complement this list by one additional expressed gene each.
We have solubility data for 26 proteins, of which 23 show at least some solubility and 20 at least 50% solubility when comparing soluble and insoluble fractions (Table 2). However, six of these 20 have a higher solubility in only one vector, namely genes 4, 10, and 12 with Gb1, genes 1 and 32 with thioredoxin, and gene 6 with NusA. The number of expression products that show at least some solubility for the different fusions are: 8 for 6*His tag, 13 for GST, 14 for NusA, 15 for ZZ, 18 for Gb1, 19 for MBP, and 20 for thioredoxin (see also comment below on gene 32).
It is interesting to compare the profound differences between 6*His tag fusions and thioredoxin fusions. The 6*His tag fusions are expressed for 16 of the 27 gene fusions, and 8 of these products have at least some solubility. With thioredoxin fusions, 24 of 27 gene fusions are expressed, and 20 of these products have at least some solubility. The differences are even larger when comparing the number of genes that have at least 50% of their product in the soluble fraction compared to the insoluble fraction: 3 for the 6*His tag compared to 16 for thioredoxin. It is conceivable that the large solubility of thioredoxin fusions might be linked to the correct formation of disulfide bonds, but such a correlation is not immediately apparent with the present set of proteins. It is clear that thioredoxin, or possibly the maltose binding protein, is the fusion protein of choice if one would aim to produce as many as possible of the 27 proteins included in the present screen. However, as with expression, these two best fusion partners would have to be complemented with other fusions to obtain all 23 soluble products. The statistical significance of the behavior of different fusions and the implications for setting up optimal screening protocols are discussed further below.
Other comments
Gene number 32 yields expression products with incorrect size in all but one case. The sizes of the incorrect products are much smaller than expected, and only somewhat larger than those of the fusion proteins with linkers. All expression constructs with gene number 32 show the correct size when they are PCR screened with a gene-specific and a vector-specific primer, so the architecture of all the gene fusions are correct. This and the consistent product sizes indicate that there is a nonsense mutation in the entry clone in this case, and that the single positive result for the thioredoxin fusion could be an artifact.
Expression of gene number 6 has only been detected when fused to NusA, in which case the cell density dropped after induction. Cells containing gene number 6 fused to Gb1 failed to grow; hence, the lack of data for this clone. Both these observations indicate that the product of gene number 6 is toxic to the cells. This could also account for the incorrect product sizes when fused to thioredoxin and MBP, as only cells that degrade the products will survive. For reasons unknown, gene products of five different expression clones not involving genes number 6 and 32 appear to be of wrong size on polyacrylamide gels, although they are correct on the DNA level (Table 2). All these are smaller than expected, so they could be stable degradation products. One of the NusA gene fusions, number 29, has not yet been isolated, and hence, we lack expression and solubility data for this clone. Otherwise, we have expression data for 187 of the 189 expression clones (98.9%).
Discussion
The different fusion proteins and their effect
It seems reasonable that high throughput protein production screening for structural genomics should rely on fusion proteins to allow for common first-step purification procedures based on affinity chromatography and similar methodologies. An important objective of the present work has been to conduct a comparison of many commonly used fusion proteins with regard to their relative effect on solubility of the products of human target genes. We chose to include a 6*His tag, GST, MBP, thioredoxin, NusA, the Gb1-domain, and a double Z-domain (ZZ) as N-terminal fusions in our study. The 43-kD MBP and 13-kD thioredoxin from E. coli are commonly used fusions with known solubility enhancing properties, and are therefore included. The 26-kD GST protein from Schistosoma japonicum is also very frequently used as a fusion in molecular biology research. The 55-kD NusA protein from E. coli was predicted and shown to be a very good fusion for increasing solubility (Davis et al. 1999). It is included because it represents an interesting but not yet commonly used fusion. The 7.5-kD Gb1 domain of protein G from group G Streptococcus and the 17-kD double-Z domain derived from Staphylococcus aureus protein A were included because they are solubilizing, can be used for affinity purification, and their sizes allow for characterization by NMR without proteolytic cleavage. Although the His tag is normally not a solubilizing tag we chose to include it as it is a commonly used tag, and it thus serves as a reference point for comparisons. Most bacterial structural genomics projects are using only the His tag.
The present results on a set of 27 small- and medium-sized human proteins indicate that several of the fusions perform very well, but that none is outstanding. Two-sample Student's t-tests for pair-wise comparisons of solubility allows for a few conclusions with regard to the characteristics of the different fusion proteins and the significance of our observations at a 95% confidence level. First, one can conclude that having only a His tag as an N-terminal fusion gives a lower chance of obtaining soluble product than with any other fusion. Second, a GST fusion increases the probability of obtaining soluble product compared to a 6*His tag, but the chance is lower than with any of the other fusion proteins. Third, NusA and ZZ fusions give higher probabilities than His tag and GST but lower than thioredoxin and, in the case of NusA, also lower than MBP and Gb1. Last, the differences between thioredoxin, MBP, and Gb1 are not statistically significant. Based on these differences a ranking of the fusion proteins according to their effect on solubility would be thioredoxin, Gb1/MBP, ZZ, NusA, GST, and the 6*His tag.
A somewhat different pattern is observed when the same comparisons are done for expression. His tag gives lower expression than ZZ, MBP, and thioredoxin. Gb1, GST, and MBP give lower expression than thioredoxin. Based on these differences a ranking of the fusion proteins according to their effect on expression would be thioredoxin, ZZ, NusA/MBP, GST/Gb1, and the 6*His tag. From our data on expression and solubility we suggest that thioredoxin, MBP, Gb1, and ZZ are the best fusion proteins with regard to solubility. These fusions can also be utilized for purification based on affinity chromatography.
It should be stressed, however, that there is a variation in what proteins are expressed and at what yield in the different fusion constructs. Thus, it could still be worthwhile to screen as many fusions as possible to optimize the product yield for a limited set of genes. In particular, it appears that there may be genes that can only be expressed with "odd" fusion tags. Examples of such genes are number 8 and 6 in the present set, which could only be expressed with a His tag and a NusA fusion, respectively.
Temperature effect
There are numerous single case studies showing increased solubility of recombinant proteins at lower cultivation temperatures (Baneyx 1999). We have not seen that effect on our set of proteins in the range of 20 to 37°C, and prefer to carry out solubility screening at the higher temperature. It is possible that this lack of temperature dependence is an effect of the bias towards small proteins in our set. It is reasonable to expect that larger proteins are more likely to have complex folding mechanisms, which render them more vulnerable to the aggregation associated with very high transcription rates at optimal growth temperatures. Impaired solubility of smaller proteins could, on the other hand, be an effect of less temperature-dependent factors such as missing cofactors, post-translational modifications, or folding partners.
Fusion proteins in structural genomics
Although we show very promising results for the solubility of eukaryotic proteins expressed in E. coli, it should be remembered that this does not per se mean that the proteins are correctly folded or soluble when the fusions have been cleaved off. There are examples of soluble fusion proteins lacking activity both before and after cleavage of the fusion, or gaining activity only after refolding (Saavedra-Alanis et al. 1994; Sachdev and Chirgwin 1998a). This effect of "false solubility" should not be considered as a problem in a high throughput project. The only effect is that these proteins will be discarded at a later step in the process. The extra work of taking these proteins a further step in the process should be counterbalanced by the gain of finding more proteins that are truly soluble, but difficult to produce in E. coli. The method of choice so far in structural genomics has been expression in E. coli with only a His tag fusion for purification, referred to as picking the "low hanging fruits" (Edwards et al. 2000). Proteins that are not soluble with only a His tag are thus discarded. This has still yielded sufficiently high success rates, possibly due to the fact that targets so far mainly have been chosen from prokaryotes and thermophiles (Christendat et al. 2000). However, as we have shown in this study it should not be concluded that previous success rates with His tags can be maintained when selecting targets from eukaryotes. Including a highly soluble fusion protein in the construct allows a larger number of targets to be kept in the project. The only cost for this inclusion is the need for a proteolytic cleavage step, a step that is already included in many cases to remove the His tag before further structural studies.
Finally, we note that the large success rate for solubility (85% given a PCR fragment) that we obtain in the present study is likely to decrease when also proteins larger than 20 kD are included in the target set. Most of the larger human proteins are multidomain proteins, and it is unlikely that optimal fragments for structure determination are going to be identified by the type of screening described here, without including additional procedures to identify domain limits.
Time aspects and parallelism
With our protocol for rapid subcloning and solubility screening we are able to considerably increase the throughput in the molecular biology parts of any structural biology project. The work described in this article has been performed by three persons with basic equipment and without any automation; thus, it is amendable for small labs. One bottleneck in the present work has been the isolation of expression clone plasmids from DH5α cells prior to transformation into BL-21 expression cells. This step was included to provide an extra control for the correct expression clones. It is likely that it may be omitted in future applications, and that the expression cells are instead directly transformed with the recombination reaction mixture, whereupon considerable time will be saved. The present protocol, if repeated, can be expected to require 1 to 2 wk per person for 96 subcloning reactions. The alternative, direct transformation into BL-21, should require about 2 d. The actual time for running 96 expression and solubility screens on a microtiter platform is about 1 wk for one person, given that the expression clones are available. The major bottlenecks here are sample preparation for gel analysis, which is labor intensive, and also the time it takes to carry out the gel electrophoresis, if it cannot be done with a high degree of parallelism. Hence, the total time it should take for one person to complete 96 expression and solubility screens, given PCR fragments, is either 3 wk with the protocol described here, or about 7 work days if direct transformation of BL-21 cells is instead performed. Automation can be expected to increase throughput, but not decrease the required time. The phase where major improvements in speed can be made is the detection of soluble product, by introducing an alternative to the present gel electrophoresis procedures.
Conclusions
We have demonstrated a methodology to rapidly subclone a large number of human genes and screen these for expression and protein solubility in E. coli. The procedures can be adopted to microtiter plate format and partly automated. We selected 32 human proteins to represent a realistic set of targets for structural studies by NMR. This set was used to estimate the yields for obtaining expression plasmids from cDNA, expressed, and soluble protein, and the relative effects of six different N-terminal fusion proteins and an N-terminal 6*His tag on expression and solubility. The subcloning yield was 100% for 27 genes for which a PCR fragment with the correct size could be obtained, and the overall yield for soluble protein product, given a correct PCR fragment, was high: 85%. We find large differences in the effect of fusion protein or tag on expression and solubility. Four of seven fusions—thioredoxin, MBP, the Gβ1 domain, and the ZZ domains—perform very well, and much better than a 6*His tag, but the combination of several fusion possibilities is still the key to the high overall success rate. We conclude that our methodology and expression vectors can be used for screening of genes for structural studies, and that it should be possible to obtain a large fraction of the NMR-sized and nonmembrane human proteins as soluble fusion proteins in E. coli.
Materials and methods
If not stated otherwise, all reagents and reactions are prepared as recommended by the suppliers or as described in Sambrook et al. (1989).
Target selection
Our target proteins were selected from the SWALL database covering SwissProt, SP-TrEMBL, and TrEMBL-New (Bairoch and Apweiler 2000). The selection criteria were human proteins without transmembrane regions, without links to the PDB or HSSP databases, and with links to the OMIM database (Hamosh et al. 2000). The selection was done using the Sequence Retrieval System SRS 6.0 (Etzold and Argos 1993). However, care has to be taken with entries from the TrEMBL database, as these entries are not completely annotated. Thus, there is one protein in our set whose structure was determined before the onset of our project. To ensure that homologs to proteins with known structures are excluded an extra BLAST search with a cutoff of E > 1 × 10−3 was done against the PDB database rather than relying on that links to PDB and HSSP are given for all entries in the SWALL database.
To assess the availability of the selected targets a BLAST search with the coding DNA sequences was done against the EST database (Boguski et al. 1993) using the network BLAST client Blastcl3 (Madden et al. 1996). Clones that were available from the IMAGE consortium (Lennon et al. 1996) and that were of full length were ordered from the German IMAGE clone distributor, RZPD. The gene specific parts of the PCR primers were designed using the program Web Primer (http://genome-www2.stanford.edu/cgi-bin/SGD/web-primer) to have a melting temperature as close as possible to 53°C with respect given to total GC content, self-end annealing, and pair-wise annealing. Full-length primers including the attB1 and attB2 overhangs required for recombinant cloning were ordered from Invitrogen/Life Technologies.
Cloning
Individual cDNA clones were delivered from RZPD as bacterial stab cultures. These were restreaked on fresh LA plates containing ampicillin, and single colonies were picked for plasmid preparation using the QIAprep Spin Miniprep Kit (Qiagen). The genes of interest were PCR amplified with gene-specific primers using the proofreading Vent DNA polymerase (New England Biolabs), analyzed on agarose gel, and PCR products of correct size were purified using the QIAquick PCR Purification Kit (Qiagen). Cloning into the different donor and destination vectors was performed as described in the Gateway manual (Invitrogen/Life Technologies) or with a downscaled protocol using 0.5 μL Clonase Enzyme mix, 0.5 μL Clonase Reaction Buffer, 0.5 μL Destination/Donor vector at 100–300 ng/μL, 1.0 μL PCR product/Entry clone at 20–200 ng/μL, and no Proteinase K after stopping the reaction, but otherwise as in the original protocol. The resulting entry and expression clones were transformed into DH5α cells, confirmed by PCR screening with the gene-specific primers or a combination of vector-specific and 3` gene-specific primers in the case of expression clones. Plasmids were prepared using QIAprep Spin Miniprep Kit (Qiagen). All steps are compatible with the 96-well format of microtiter plates, although for this limited set of genes we used single tube protocols in most cases.
Expression and solubility tests
Expression test were carried out in the strain BL-21 (DE3) Codon Plus RP (Stratagene), which has a plasmid that supplies extra tRNAs corresponding to codons that are rare in E. coli but common in humans. The culture volume was either 5 mL in a 50-mL Polypropylene tube (Falcon) or 1 mL in a 96 × 1.2-mL polypropylene microtiter plate (Labora). There were only minor differences in results between growing in microtiter plates or single tubes. The 5-mL cultures were inoculated with 10–20 fresh-picked colonies from LA plates with appropriate antibiotics. Transformation in microtiter plates was done as follows: 1 μL of LR reaction or prepared plasmid was transformed into 5 μL Ca2+-competent BL-21 (DE3) Codon Plus RP using standard heat-shock; 200-μL LB was added, and the cells were grown at 37°C for 1 h, after which appropriate antibiotics were added. The cultures were grown at 37°C overnight and 50 μL were used to inoculate the expression cultures. Transformation in microtiter plates instead of plating on LA plates did not interfere with antibiotic selection. Expression cultures were grown at 37°C to mid-log phase and induced by adding isopropyl-β-d-thiogalactoside (Boehringer Mannheim) to a final concentration of 1 mM. Cells were harvested by centrifugation after 1 to 4 h of induction. Cell pellets were lysed using Bacterial-Protein Extraction Reagent (Pierce), the soluble and insoluble fractions were separated by centrifugation, and all samples were analyzed using SDS-PAGE gels stained with Coomassie stain. The levels of expression were quantified by comparing them to endogenous E. coli protein levels. The solubility was quantified by comparing soluble and insoluble fractions as described in Table 2.
Vector conversion
The original vectors were cut with a restriction enzyme as close to the 3` end of the fusion gene as possible (Table 3). They were then treated with either mung bean nuclease (New England Biolabs) or with Klenow polymerase (Promega) to create blunt ends. The 5` phosphate groups of the ends were removed with calf intestinal alkaline phosphatase (Promega). The appropriate Gateway cloning cassette was chosen as to retain the reading frame of the fusion gene over the recombination site. Ligation reactions were set up using T4 DNA ligase (New England Biolabs). The ligation mixes were transformed into DB3.1 cells, and positives were confirmed by colony PCR, restriction analysis, and sequencing.
Table 3.
Description of the vectors, enzymes, and reading frame cassettes that were used to create new destination vectors for the Gateway system
| Original vector | New vector | Fusion protein | Promoter | Restriction enzyme | Blunt ends | Reading frame |
| pMAL-c2a | pDEST-TH1 | MBP | tac | NcoI | Nuclease | C |
| pET-43ab | pDEST-TH2 | NusA | T7Iac | SpeI | Nuclease | C |
| pGβ1c | pDEST-TH3 | Gb1 | T7Iac | NcoI | Nuclease | C |
| pT7-ZZAd | pDEST-TH5 | ZZ | T7Iac | BamHI | Klenow | B |
a New England Biolabs.
b Novagen.
c Dr. K. Gardner, Dept. of Biochemistry, UT Southwestern Medical Center.
d Larsson et al. 1996.
Acknowledgments
We thank Dr. Kevin Gardner for contributing the Gb1 fusion vector pGβ1, Dr. Per-Åke Nygren for contributing the ZZ fusion vector pT7-Zza, and Invitrogen/Life Technologies for contributing the thioredoxin fusion vector pDEST-16. This work was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, and the Swedish Foundation for Strategic Research (SSF).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.22102.
References
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10 411–421. [DOI] [PubMed] [Google Scholar]
- Bedouelle, H. and Duplay, P. 1988. Production in Escherichia coli and one-step purification of bifunctional hybrid proteins which bind maltose. Export of the Klenow polymerase into the periplasmic space. Eur. J. Biochem. 171 541–549. [DOI] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. 1993. dbEST—Database for "expressed sequence tags." Nat. Genet. 4 332–333. [DOI] [PubMed] [Google Scholar]
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Savchenko, A., Cort, J.R., Booth, V., Mackereth, C.D., Saridakis, V., Ekiel, I., Kozlov, G., Maxwell, K.L., Wu, N., McIntosh, L.P., Gehring, K., Kennedy, M.A., Davidson, A.R., Pai, E.F., Gerstein, M., Edwards, A.M., and Arrowsmith, C.H. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7 903–909. [DOI] [PubMed] [Google Scholar]
- Davis, G.D., Elisee, C., Newham, D.M., and Harrison, R.G. 1999. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 65 382–388. [PubMed] [Google Scholar]
- di Guan, C., Li, P., Riggs, P.D., and Inouye, H. 1988. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 67 21–30. [DOI] [PubMed] [Google Scholar]
- Edwards, A.M., Arrowsmith, C.H., Christendat, D., Dharamsi, A., Friesen, J.D., Greenblatt, J.F., and Vedadi, M. 2000. Protein production: Feeding the crystallographers and NMR spectroscopists. Nat. Struct. Biol. (Suppl.) 7 970–972. [DOI] [PubMed] [Google Scholar]
- Etzold, T. and Argos, P. 1993. SRS—An indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 9 49–57. [DOI] [PubMed] [Google Scholar]
- Gentz, R., Certa, U., Takacs, B., Matile, H., Döbeli, H., Pink, R., Mackay, M., Bone, N., and Scaife, J.G. 1988. Major surface antigen p190 of Plasmodium falciparum: Detection of common epitopes present in a variety of plasmodia isolates. EMBO J. 7 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh, A., Scott, A.F., Amberger, J., Valle, D., and McKusick, V.A. 2000. Online Mendelian inheritance in man (OMIM). Hum. Mutat. 15 57–61. [DOI] [PubMed] [Google Scholar]
- Hochuli, E., Bannwarth, W., Döbeli, H., Gentz, R., and Stüber, D. 1988. Genetic approach to facilitate purifaction of recombinant proteins with a novel metal chelate adsorbent. Biotechnology 6 1321–1325. [Google Scholar]
- Huth, J.R., Bewley, C.A., Jackson, B.M., Hinnebusch, A.G., Clore, G.M., and Gronenborn, A.M. 1997. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust, R.B. and Waugh, D.S. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, M., Brundell, E., Nordfors, L., Höög, C., Uhlén, M. ,and Ståhl, S. 1996. A general bacterial expression system for functional analysis of cDNA-encoded proteins. Protein Expr Purif. 7 447–457. [DOI] [PubMed] [Google Scholar]
- LaVallie, E.R., DiBlasio, E.A., Kovacic, S., Grant, K.L., Schendel, P.F., and McCoy, J.M. 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology 11 187–193. [DOI] [PubMed] [Google Scholar]
- Lennon, G., Auffray, C., Polymeropoulos, M., and Soares, M.B. 1996. The I.M.A.G.E. Consortium: An integrated molecular analysis of genomes and their expression. Genomics 33 151–152. [DOI] [PubMed] [Google Scholar]
- Madden, T.L., Tatusov, R.L., and Zhang, J. 1996. Applications of network BLAST server. Methods Enzymol. 266 131–141. [DOI] [PubMed] [Google Scholar]
- Nilsson, B., Moks, T., Jansson, B., Abrahmsén, L., Elmblad, A., Holmgren, E., Henrichson, C., Jones, T.A., and Uhlén, M. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1 107–113. [DOI] [PubMed] [Google Scholar]
- Saavedra-Alanis, V.M., Rysavy, P., Rosenberg, L.E., and Kalousek, F. 1994. Rat liver mitochondrial processing peptidase. Both alpha- and beta-subunits are required for activity. J. Biol. Chem. 269 9284–9298. [PubMed] [Google Scholar]
- Sachdev, D. and Chirgwin, J.M. 1998a. Order of fusions between bacterial and mammalian proteins can determine solubility in Escherichia coli. Biochem. Biophys. Res. Commun. 244 933–937. [DOI] [PubMed] [Google Scholar]
- ———. 1998b. Solubility of proteins isolated from inclusion bodies is enhanced by fusion to maltose-binding protein or thioredoxin. Protein Expr. Purif. 12 122–132. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maneatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shapiro, L. and Lima, C.D. 1998. The Argonne Structural Genomics Workshop: Lamaze class for the birth of a new science. Structure 6 265–267. [DOI] [PubMed] [Google Scholar]
- Smith, D.B. and Johnson, K.S. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67 31–40. [DOI] [PubMed] [Google Scholar]
- Smith, M.C., Furman, T.C., Ingolia, T.D., and Pidgeon, C. 1988. Chelating peptide-immobilized metal ion affinity chromatography. A new concept in affinity chromatography for recombinant proteins. J. Biol. Chem. 263 7211–7215. [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328 575–592. [DOI] [PubMed] [Google Scholar]
- Wang, C., Castro, A.F., Wilkes, D.M., and Altenberg, G.A. 1999. Expression and purification of the first nucleotide-binding domain and linker region of human multidrug resistance gene product: Comparison of fusions to glutathione S-transferase, thioredoxin and maltose-binding protein. Biochem J. 338 77–81. [PMC free article] [PubMed] [Google Scholar]