Abstract
The polypeptide hormone amylin forms amyloid deposits in Type 2 diabetes mellitus and a 10-residue fragment of amylin (amylin20–29) is commonly used as a model system to study this process. Studies of amylin20–29 and several variant peptides revealed that low levels of deamidation can have a significant effect on the secondary structure and aggregation behavior of these molecules. Results obtained with a variant of amylin20–29, which has the primary sequence SNNFPAILSS, are highlighted. This peptide is particularly interesting from a technical standpoint. In the absence of impurities the peptide does not spontaneously aggregate and is not amyloidogenic. This peptide can spontaneously deamidate, and the presence of less than 5% of deamidation impurities leads to the formation of aggregates that have the hallmarks of amyloid. In addition, small amounts of deamidated material can induce amyloid formation by the purified peptide. These results have fundamental implications for the definition of an amyloidogenic sequence and for the standards of purity of peptides and proteins used for studies of amyloid formation.
Keywords: Amylin, islet amyloid polypeptide, IAPP, amyloid, deamidation, protein aggregation, diabetes mellitus, chemical modification
Amyloid formation has been implicated in the pathology of many different diseases (Sipe 1994). The protein or peptide component of the insoluble amyloid fibrils varies from disease to disease, but in each case the fibrillar deposits cause adverse physiological consequences. The mechanism by which a polypeptide self-assembles into amyloid fibrils is still unknown, but the characterization of this process is important in the development of effective treatments for amyloid associated diseases.
Amyloidogenic proteins and polypeptides are notoriously difficult to prepare, purify, and analyze. Consequently, many groups have used smaller peptide fragments derived from the protein of interest as convenient model systems (Halverson et al. 1991; Shen et al. 1993; Baumann et al. 1996; Inouye and Kirschner 1997; Guijarro et al. 1998; Sherzinger et al. 1999). Our own work has focused on the polypeptide hormone, amylin, which is responsible for the amyloid deposits in the pancreas of patients with Type 2 diabetes mellitus. A peptide corresponding to residues 20 to 29 of human amylin (amylin20–29, sequence SNNFGAILSS) is widely used as a model system to study amyloid formation by amylin (Glenner et al. 1988; Betsholtz et al. 1990; Westermark et al. 1990; Ashburn et al. 1992; Ashburn and Lansbury 1993; Griffiths et al. 1995). In this article we report that very low levels of asparagine deamidation can lead to significant changes in the propensity of amylin derived peptides to aggregate.
During the course of a study involving the replacement of the glycine residue in SNNFGAILSS with other amino acids we detected small amounts of deamidated material in several samples. Particularly dramatic effects were observed for a peptide with the sequence SNNFPAILSS. Strikingly, the peptide did not aggregate in the absence of the impurities, but spontaneous deamidation of trace amounts of material lead to aggregation. The amount of deamidated material that was required to cause this change in behavior was very small (less than 5% of the total sample), and would typically not be detected by standard analytical methods. Deamidation lead to an unexpected pH dependence of the aggregation behavior, and furthermore, trace amounts of deamidated material can induce aggregation when added to freshly purified samples. Asparagine deamidation is one of the most common nonenzymatic modifications, and the results presented here indicate that it can, at least in some cases, have striking effects on the ability of peptides to aggregate.
Results
Low levels of deamidation were detected by analytical HPLC and MALDI-MS
Samples of the purified peptides slowly deamidated in solution to produce small amounts of impurities. Deamidation also occurred when the peptides were stored as lyophilized powders. Analytical reversed phase-high performance liquid chromatography (RP-HPLC) in phosphate buffers resolved a minimum of two impurity peaks in each sample, suggesting a mixture of products were produced. Deamidation of Asn residues can result in fragmentation, formation of l- or d-aspartic acid (Asp), formation of l- or d-isoaspartic acid (iso-Asp), or a mixture of these products (Wright 1991). Thus, deamidation of SNNFGAILSS or SNNFPAILSS could result in the formation of l- or d-Asp or l- or d-iso-Asp at one or both of the two Asn residues leading to 16 possible products (excluding fragmentation products).
The impurities in these samples were identified as deamidation products using two methods, analytical HPLC, and a methyl esterification assay coupled with matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS). These experiments were performed on all the peptides in this study. The results reported here describe in detail the analysis of the SNNFPAILSS peptide.
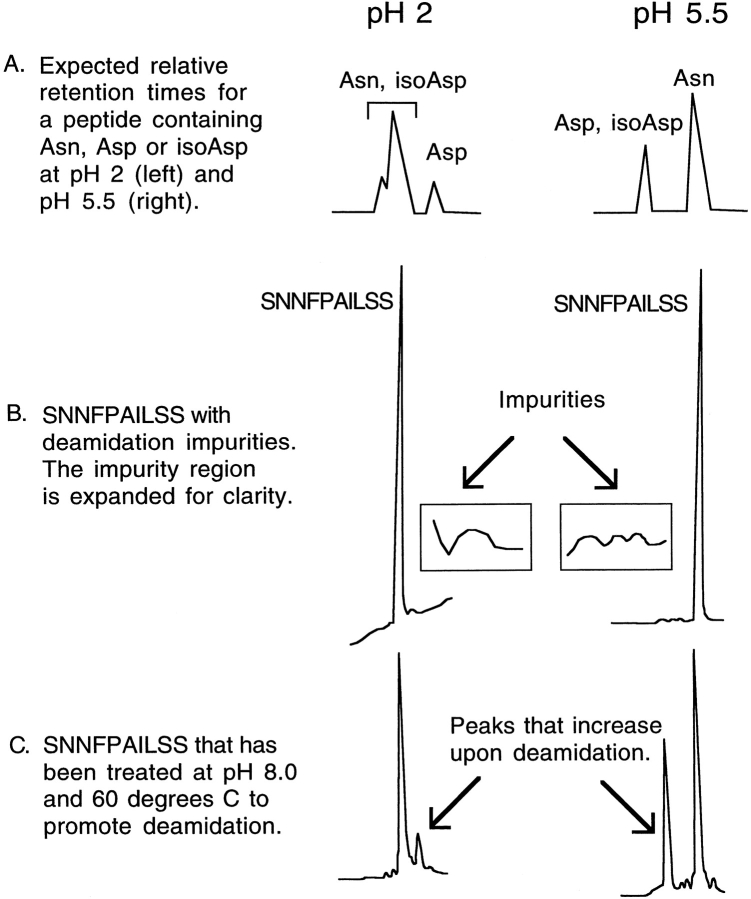
HPLC has been used to characterize deamidation in other peptide systems, and the relative retention times for deamidated peptides and unmodified peptides have been reported (Aswad and Guzetta 1995). At pH 2, the Asn and iso-Asp derivatives normally elute first followed by the Asp derivatives. At pH 6, the Asp and iso-Asp derivatives will be deprotonated and typically elute first, followed by the Asn derivative (Fig. 1A ▶). The small impurities for SNNFPAILSS observed by HPLC eluted after the Asn containing peptide peak at pH 2 but before the Asn containing peptide peak at pH 5.5 (Fig. 1B ▶). This corresponds to the expected pH-dependent relative retention times for Asp/isoAsp derivatives. If the sample is subjected to conditions that promote deamidation (pH 8 and 60°C), the relative intensity of the peaks due to the impurities increases, providing additional indirect evidence that the minor peaks represent deamidation products (Fig. 1C ▶). The prominent deamidation peak observed in the HPLC trace of this sample run with the low pH mobile phase is smaller than the peak observed using the phosphate-based buffer system. This reflects the fact that the peak in the low pH buffers represents only the Asp peptide, but in the phosphate buffer system the peak represents Asp plus iso-Asp (see Fig. 1A ▶). When deamidation is promoted by this method, the ratio of iso-Asp to Asp is typically about 3:1, which is consistent with the observed relative peak heights. It is important to note that this experiment was conducted to confirm the relative retention times of deamidated and nondeamidated material. It should not be assumed that the same products or ratios of products would necessarily be formed as a result of spontaneous deamidation of a sample of the peptide in solution at lower temperature and neutral pH. Finally, a synthetic peptide with the sequence SNDFPAILSS coeluted with the impurities providing additional evidence that they arose from Asn deamidation.
Fig. 1.
HPLC traces showing the presence of deamidation impurities. (A) A schematic diagram depicting the relative retention time of Asn, Asp, and iso-Asp containing peptides at pH 2 or pH 5.5. Adapted from Aswad and Guzetta (1995). (B) HPLC traces of the SNNFPAILSS peptide that show the small impurities. The region corresponding to the impurities is expanded in the boxes for clarity. Note that these impurities constitute less than 5% of the total sample. (C) HPLC trace that confirms that the impurities are due to deamidation by demonstrating that the impurity peaks increase upon further deamidation. Additional deamidation was promoted by heating the sample to 60°C at pH 8.0. The prominent deamidation peak at pH 2 is smaller than the peak at pH 5.5. This is because the peak at pH 2 represents only the Asp peptide, while at pH 5.5 in phosphate the peak represents Asp plus iso-Asp [see (A)].
The SNNFPAILSS peptide was further characterized using mass spectrometry in conjunction with a methyl esterification assay. The carboxylate of an Asp or iso-Asp side chain is susceptible to esterification while the Asn side chain is not, and this difference in chemical reactivity can be exploited to provide a simple method for detecting deamidation. Methyl esterification using methanolic HCl has been previously used to detect deamidated material (Tuong et al. 1992). Esterification of a peptide containing Asp or iso-Asp will result in an increase in mass of +14 or +15, depending upon the pH. Asparagine, in contrast, will not react, and will not show a change in molecular weight. The calculated molecular weight of SNNFPAILSS is 1048.1 g/mol, but this mass was rarely detected; instead, the observed mass for untreated SNNFPAILSS corresponded to the peptide plus sodium (MW ≃ 1048 + 23 = 1071 g/mol). The formation of a complex with sodium is commonly observed in MALDI-MS. Methyl esterification of SNNFPAILSS resulted in the detection of two peaks corresponding to molecular weights of 1071 ± 2 g/mol and 1086 ± 2 g/mol. These peaks are due to SNNFPAILSS plus sodium and SNNFPAILSS plus sodium plus 14 from the methyl esterification (Fig. 2 ▶). There is a formal possibility that the 1086 ± 2 g/mol peak could correspond to SNNFPAILSS plus potassium (another ion commonly detected in complexes with peptides in MALDI-MS), although no comparable peak was detected in the untreated sample. To ensure that the assay was detecting esterification, a set of experiments were performed in which the ratio of the 1086 peak to the 1071 peak was monitored as a function of the esterification reaction time. Although MALDI-MS is not quantitative, the ratio will increase during the course of the reaction if the 1086 peak is due to esterification but will remain unchanged if the peak is due to the addition of potassium. The ratio does increase with reaction time, confirming that the 1086 peak corresponds to an aspartic acid methyl ester or an iso-aspartic acid methyl ester (Fig. 2 ▶). As a further control, MALDI-MS was used to monitor the esterification reaction of a sample of the SNDFPAILSS peptide, and similar changes in peak ratios were observed.
Fig. 2.
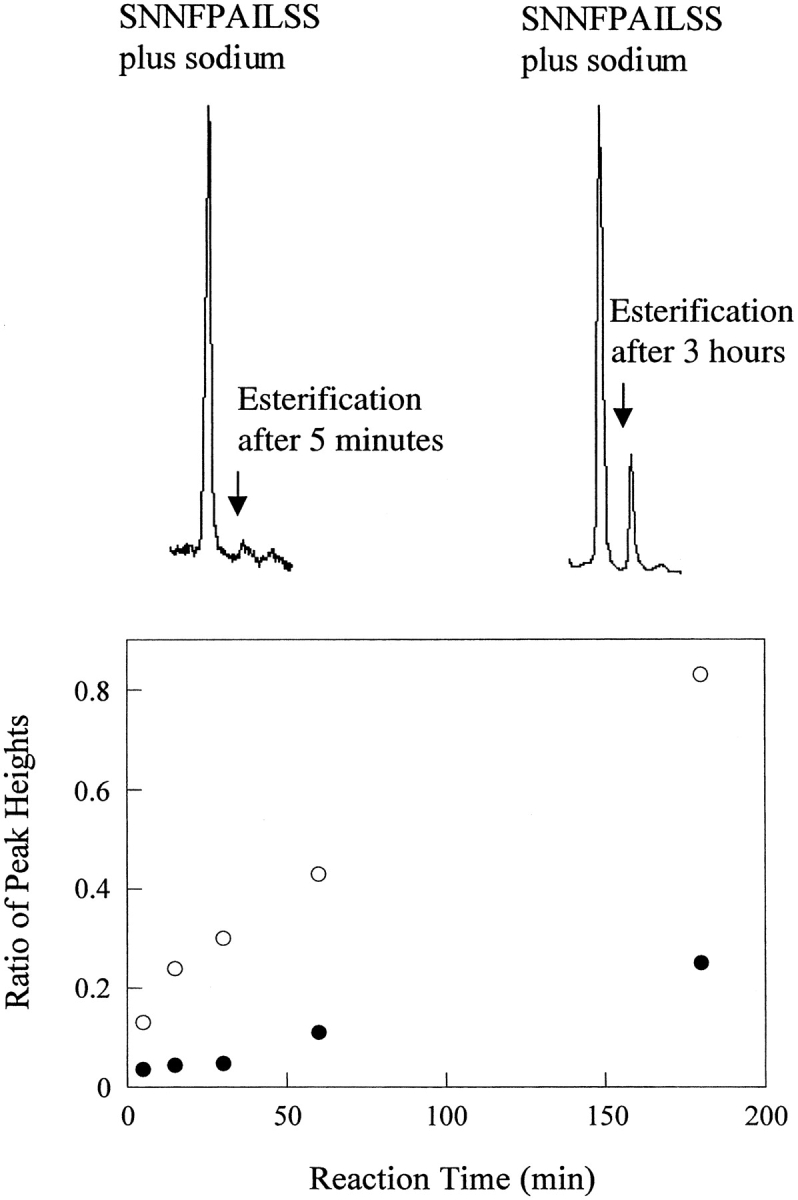
MALDI-MS detected methyl esterification. The top figures show representative MALDI-MS traces. The major peaks correspond to SNNFPAILSS plus sodium. The minor peaks that result from esterification (after either 5 min or 3 h of reaction with MeOH: HCl) are labeled. The bottom plot shows a time course of methyl esterification of SNNFPAILSS (filled circles) and SNDFPAILSS (open circles) followed by MALDI-MS. The ratio of the 1071 peak to the 1086 peak is plotted as a function of reaction time. The larger change in the ratio for the control SNDFPAILSS sample simply reflects the fact that the entire sample is capable of undergoing esterification. In contrast, only the deamidated material is susceptible to esterification in the SNNFPAILSS peptide.
Samples of the SNNFPAILSS peptide without deamidation impurities do not form fibrils, but the presence of less than 5% deamidation impurities leads to amyloid-like deposits
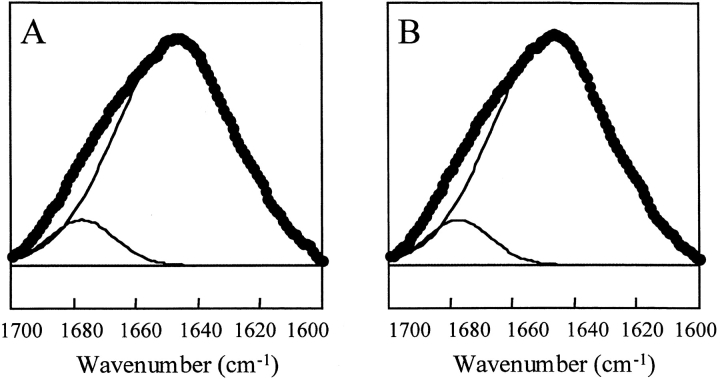
Purification of SNNFPAILSS to remove the deamidation impurities resulted in a sample that did not aggregate, and remained unstructured in aqueous solution for at least 8 d. Pure samples of SNNFPAILSS were examined at pD 2.29 and at pD 5.47 by Fourier transform infrared spectroscopy (FTIR), transmission electron microscopy (TEM), and Congo Red staining. The FTIR spectra recorded at the two pD values are very similar. Curve fitting revealed bands at 1647 cm−1 and 1677 cm−1, which are characteristic of an unstructured peptide in D2O (Fig. 3 ▶). TEM revealed no fibrillar aggregates and no significant difference between the pD 2.29 and pD 5.47 samples. Congo Red staining revealed no birefringence.
Fig. 3.
FTIR spectra of a sample of SNNFPAILSS that contains no deamidation impurities. (A) FTIR amide I band of SNNFPAILSS recorded at pD 2.29. Experimental data is shown in black circles. Component bands identified by curve fitting are shown as solid lines (1647 cm−1, 1677 cm−1). (B) FTIR amide I band of SNNFPAILSS recorded at pD 5.47. Experimental data is shown in black circles. Component bands identified by curve fitting are shown as solid lines (1647 cm−1, 1677 cm−1). TEM micrographs of these samples contained few ordered deposits and are not shown.
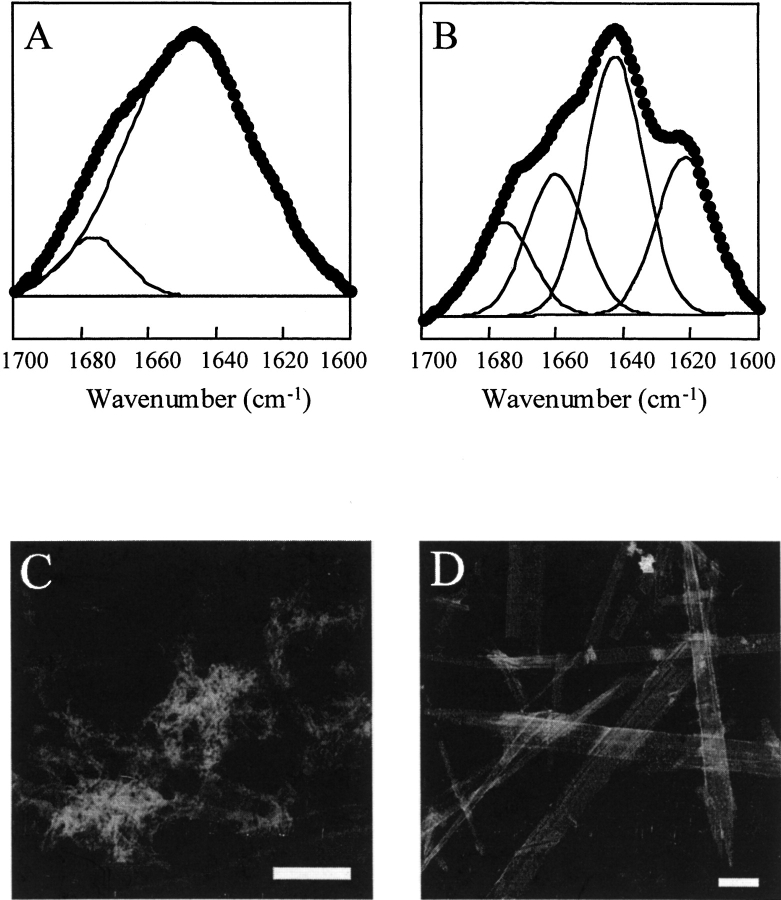
In contrast, the presence of small amounts of deamidated material lead to large changes in the properties of the peptide. Samples that contained less than 5% deamidation impurities (determined by analytical HPLC) formed amyloid-like deposits and the aggregation was pH dependent (Fig. 4 ▶). At pD 2.25, the sample with the deamidation impurities behaved in a similar fashion to the pure sample. The FTIR spectrum recorded at pD 2.25 showed two bands (1647 cm−1, 1676 cm−1), which are characteristic of a random coil peptide in D2O (Fig. 4A ▶). Completely different behavior was observed at pD 5.71. At pD 5.71, the sample containing deamidation impurities showed evidence of β-sheet secondary structure and extensive formation of ordered aggregates. The spectrum recorded at pD 5.71 was very different from the low pD spectrum and showed two additional peaks, one near 1620 cm−1 and another at 1660 cm−1 (Fig. 4B ▶). The 1620 cm−1 peak is normally assigned to β-sheet secondary structure. The 1660 cm−1 peak may be due to the Asn side chains, which might not have been resolved in the pD 2.25 spectrum or from transition dipole coupling associated with an antiparallel β-sheet (Chirgadze et al. 1973, 1975; Jackson and Mantsch 1995; Middaugh et al. 1995). An increase in β-sheet content was also observed with time and upon drying. It was, in fact, this pH dependence that was the first clue that the SNNFPAILSS sample had spontaneously deamidated. There are no groups in this peptide that titrate over this pH range (the C-terminus is amidated) and deamidation seemed to be a likely explanation for the pH dependent behavior.
Fig. 4.
FTIR spectra and TEM images of a sample of SNNFPAILSS that contains deamidation impurities. (A) FTIR amide I band of SNNFPAILSS recorded in D2O at pD 2.25. Experimental data is shown in black circles. Component bands identified by curve fitting are shown as solid lines (1647 cm−1, 1676 cm−1). TEM micrographs of the pD 2.25 sample showed few ordered deposits and are not shown. (B) FTIR amide I band of SNNFPAILSS recorded at pD 5.71. Experimental data is shown in black circles. Component bands identified by curve fitting are shown as solid lines (1620 cm−1, 1643 cm−1, 1660 cm−1, and 1675 cm−1). (C) TEM micrograph of SNNFPAILSS at pD 5.71 showing amyloid-like fibrils. The scale bar represents 100 nm. (D) TEM micrograph of SNNFPAILSS at pD 5.71 showing the sheet-like assemblies. The scale bar represents 1 μm.
TEM analysis of the pD 2.25 sample revealed some small, relatively unstructured aggregates. In contrast, the pD 5.71 sample aggregated into well-ordered structures (Fig. 4C,D ▶). Some of the aggregates resemble amyloid fibrils, but others are lateral assemblies of fibrils that form very large, flat, sheet-like structures. The morphology of the larger aggregates does not conform to the classic amyloid fibril morphology, but it is worth noting that amyloid fibrils formed by wild-type amylin20–29 do not always exhibit the classic morphology observed with other amyloid-forming peptides. The wild-type peptide has been reported to have a strong tendency to self-assemble in a lateral fashion to generate broad ribbons (Tenidis et al. 2000). The samples did demonstrate birefringence after staining with Congo Red, although the birefringence was not as distinct as we have observed for some other amyloid forming peptides.
It is important to note that independent aggregation of the deamidated impurity cannot account for the observed changes in the FTIR spectra or the TEM images. Only 5% of the material is deamidated, yet the β-sheet peak in the FTIR spectrum corresponds to roughly 25% of the total spectral intensity, suggesting that on the order of 25% of the material has aggregated. The analysis assumes that the extinction coefficients for the β-sheet and random coil bands are approximately equal. This is a common assumption in the interpretation of FTIR spectra. Qualitative examination of the TEM grids revealed that virtually the entire sample grid was covered with deposits, again arguing that more than just 5% of the material had aggregated. These observations are corroborated by the fact that the deamidated material can induce the nondeamidated sample to aggregate (Fig. 5 ▶).
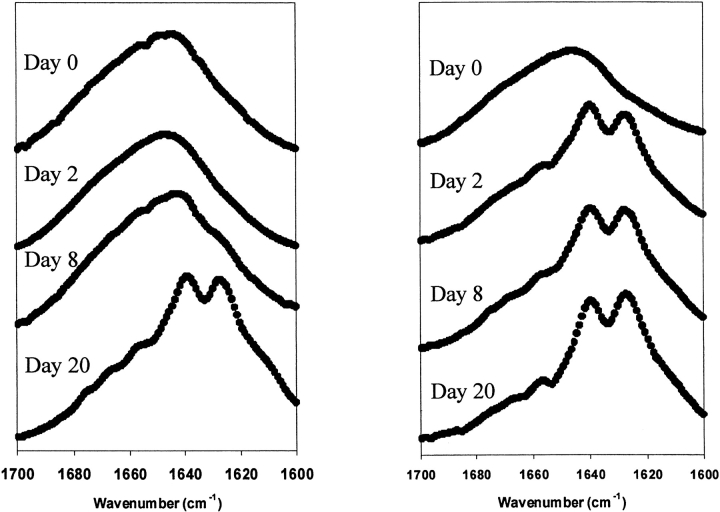
Fig. 5.
Time course of seeding experiments followed by FTIR. FTIR spectra of pure SNNFPAILSS (left) and SNNPAILSS seeded with deamidation impurities (right). FTIR spectra were recorded on the day of sample preparation (designated day 0) and on days 1, 2, 3, 4, 8, and 20.
Deamidation impurities seed the aggregation of a pure sample of SNNFPAILSS
The ability of the deamidation material to induce the aggregation of a pure sample of the SNNFPAILSS peptide was investigated at pD 5.65. A freshly purified sample of the peptide was prepared and divided into two aliquots of equal concentration. Deamidated material that had been isolated by HPLC was added to one of the aliquots and both samples were monitored over time by FTIR (Fig. 5 ▶). This protocol, while clearly a seeding experiment, differs somewhat from traditional seeding experiments in which preformed fibrils are added to a sample. Our goal was to test if trace amounts of deamidated material could induce aggregation; hence, the choice of experimental design. The initial spectrum recorded for each sample (day 0) was dominated by an intense band at the random coil position. No significant changes were observed after 24 h. After 48 h the spectrum of the unseeded sample had still not changed, while the spectrum of the seeded sample exhibited intense bands indicative of β-sheet structure. The results demonstrate that small amounts of deamidation impurities can induce the unmodified peptide to aggregate. No further significant changes were observed over the course of 8 d, and the unseeded sample showed no evidence of the development of β-structure. After 20 d, the FTIR spectrum of the unseeded sample revealed the presence of β-sheet secondary structure, implying either that over a long enough period of time the peptide can aggregate or that sufficient amounts of the peptide had spontaneously deamidated and induced aggregation.
Deamidation is not restricted to the SNNFPAILSS peptide
We also examined several other peptides for the presence of deamidated material. Samples tested included variants in which the Gly residue of the wild-type sequence was replaced by l- or d-Ala and by d-Pro. All of the peptides with the exception of the d-Ala sample underwent spontaneous deamidation that lead to detectable amounts of material. The presence of trace amounts of deamidated material lead to clear changes in the aggregation of the d-Pro sample. We also observe effects, albeit more subtle, with the wild-type sequence. Deamidation of amylin20–29 did result in modest changes in the FTIR spectra, but did not affect the ability of the peptide to form amyloid at high or low pH. It is likely that the intrinsic tendency of the SNNFGAILSS sequence to aggregate is so high that the presence or absence of deamidated material has little detectable effect on the observed aggregation behavior. The presence of low levels of impurities might, however, still affect the kinetics of aggregation of the wild-type sequence.
Discussion
The results presented here are directly relevant to studies of amyloid formation; in particular to the way in which in vitro experiments are performed. Studies of amyloid formation in vitro often make use of proteins or peptides that are either "aged" in phosphate buffer or exposed to extremes of pH and/or temperature to promote aggregation (Harper et al. 1997; Guijarro et al. 1998; Litvinovich et al. 1998; Kowalewski and Holtzman 1999; Cribbs et al. 2000; Higham et al. 2000; Morozova-Roche et al. 2000; Tenidis et al. 2000; Fandrich et al. 2001). In many cases the "aging" effect is likely due to the reaction kinetics of the system but the conditions used are known to promote deamidation, giving rise to the possibility that deamidation of Asn or Gln residues could play a role (Robinson and Rudd 1974; Tyler-Cross and Schirch 1991; Jost et al. 2001). In addition, it is worth noting that phosphate is known to be particularly effective at promoting deamidation (Tyler-Cross and Schirch 1991; Johnson and Aswad 1995). The results presented in this study clearly show that low levels of deamidation impurities can induce the aggregation of certain amylin derivatives, and it is possible that unanticipated deamidation might also induce aggregation and amyloid formation in other systems. It is also known that isomerization of Asp residues to iso-Asp or D-Asp can influence the extent and rate of amyloid formation in some systems (Fabin et al. 1994; Shimizu et al. 2000). Hence, if amyloid formation is observed, it could be the case that the wild-type sequence is not amyloidogenic, but rather that nonenzymatic modifications create a chemically modified "mutant" sequence that is amyloidogenic. The SNNFPAILSS peptide provides a particularly striking example of this sort of behavior. Initial studies revealed that samples that were 95% pure (a level of purity that often exceeds that used in other studies) aggregated extensively at pH 5.5. These initial results might have lead to the erroneous conclusion that SNNFPAILSS was an amyloidogenic sequence, and that the substitution of the central glycine by a proline had no effect on the aggregation. Further investigation ultimately revealed that trace impurities derived from spontaneous deamidation drove the aggregation, and that removal of this material resulted in a peptide that did not aggregate. The results presented here argue that the purity of peptides and proteins being used to study amyloid formation in vitro needs to be held to a higher standard than is necessary for other studies.
In principle, the deamidation of a peptide could promote, inhibit, or have no effect on the aggregation behavior. In the case of the SNNFPAILSS peptide, it is clear that deamidation promotes aggregation, and it is interesting to speculate on the possible mechanism by which this peptide aggregates. Our pH-dependent studies indicate that the protonation state of the deamidation impurity plays a key role. This, in turn, suggests that any potential alterations in the backbone induced by the possible conversion of an Asn residue to an iso-Asp residue are likely to be of lesser importance than the ionization state of the newly introduced acidic side chain. At pD 2.25, the affected side chain of the deamidated impurities will be largely protonated and neutral. At pD 5.71, the affected side chain will be deprotonated and have a negative charge. This negative charge may initiate the aggregation via an electrostatic interaction with the positively charged N-terminus on another peptide molecule. Alternatively, a singly deamidated peptide would have no net charge at pD 5.71 but be positively charged at pD 2.25, which may explain the preferential aggregation at pD 5.71.
Materials and methods
Peptide synthesis and purification
The peptides were synthesized by solid-phase peptide synthesis using standard Fmoc chemistry. Synthesis procedures are described elsewhere (Moriarty and Raleigh 1999). The peptides were purified by RP-HPLC using a C18 column (Vydac) and a two-buffer solvent system. The initial purification of the crude peptides was performed using Buffer A, which contained 5 mM HCl in H2O and Buffer B, which contained 5 mM HCl, 20% H2O and 80% acetonitrile (ACN). This buffer system, however, was not sufficient to remove deamidation impurities. To remove deamidation impurities, a phosphate buffer system was used followed by desalting with the HCl buffers. Buffer A contained 10 mM phosphate in H2O, pH 5.5. Buffer B contained the same amount of phosphate in 20% H2O and 80% ACN.
Identification of deamidated peptides
Deamidation was characterized by analytical RP-HPLC and by a methyl esterification assay. The HPLC experiments were performed using a C18 analytical column (Vydac) and the phosphate and HCl buffers described in the peptide purification section. Methyl esterification of the peptides was used to detect the presence of deamidated material. The esterification was followed by MALDI-MS using a modification of the procedure outlined by Tuong et al. (1992). Methanolic HCl (2 M) was prepared by the addition of 5 mL acetyl chloride to 30 mL MeOH. The addition was performed under N2 at 0°C. The SNNFPAILSS peptide was placed in an eppendorf tube and 100 μL of methanolic HCl was added. The solution was briefly vortexed and the reaction performed at room temperature. Aliquots of the reaction solution were removed, quenched with a fourfold volume of water, and stored at 0°C until analyzed. Sample aliquots were analyzed by MALDI-MS using α-cyano-4-hydroxycinnamic acid (CCA) as the matrix. The CCA matrix solution was prepared by dissolving the CCA in a solution of methanolic HCl/H2O (1:4). Measured peak heights were used to estimate the relative amounts of deamidated and nondeamidated material. MALDI-MS was performed on a Bruker TOF instrument. Calibration was performed using at least three standards. All spectra were obtained in positive ion/reflectron mode.
Characterization of peptides by FTIR, TEM, and Congo Red birefringence
Most experiments were performed twice to ensure reproducibility. Peptide concentrations ranged from 3.5 to 5.1 mM. The pD value of the samples ranged from pD 1.13 to 2.29 and pD 5.47 to 5.80 and were corrected for isotope effects. Samples were dissolved in D2O, incubated at room temperature for 30 min, and lyophilized overnight to remove residual water. The samples were then redissolved in D2O, and the FTIR spectra were recorded and corrected for background. TEM was performed after 1 or 7 d of incubation at room temperature. Experimental details of the FTIR, TEM, and amino acid analysis procedures can be found elsewhere (Nilsson and Raleigh 1999). In brief, FTIR was performed on a Biorad FTS-40A spectrometer using a DTGS detector with 2 cm−1 resolution at 23°C. A dismountable sample cell (CaF2 plates) was used with a 0.05 mm Teflon spacer. The amide I band (1600–1700 cm−1) was fit using the Biorad software with a linear baseline correction. All parameters were allowed to vary and successive iterations resulted in a reasonable fit. The spectrum of the pD 5.71 sample of SNNFPAILSS that contains deamidation impurities, however, required the values for the full width at half height to be fixed to achieve a reasonable fit. TEM was performed at the University Microscopy Imaging Center at the State University of New York at Stony Brook. Samples were placed on a carbon-coated Formvar 200 mesh copper grid and negatively stained with uranyl acetate and imaged using a JOEL 1200EX TEM operating at 80 kEV. Amino acid analysis was performed at Commonwealth Biotechnologies, Inc. Congo Red staining was performed on a peptide sample that was air dried on a superfrost microscope slide. The peptide was stained using an 80% ethanol solution that was saturated with sodium chloride and Congo Red. Excess staining solution was removed and the slides were analyzed using either a Nikon SMZ-2T polarizing microscope (10 to 60×) or a Nikon Labophot-pol laboratory polarizing microscope (50 to 400×). The birefringence was very weak using the Nikon SMZ-2T microscope, possibly due to imperfections in the lens and the low magnification, but the Labophot-pol microscope revealed birefringence that was significantly more pronounced.
Seeding experiments
Deamidation impurities were isolated by HPLC. Phosphate buffers were used for separation because the peak resolution is much better under these conditions (Fig. 1 ▶). The samples were desalted by HPLC using HCl buffers. Both samples were then dissolved in D2O, lyophilized to remove residual water, resuspended in D2O, and the pD adjusted to pD 5.65 using DCl/NaOD. The sample of pure SNNFPAILSS peptide was divided in half to generate two samples of equal concentration at the same pD. The deamidation impurities were suspended in a minimal volume of D2O and added to one sample of the SNNFPAILSS. Both samples were incubated at 25°C and monitored by FTIR. FTIR spectra were recorded on the day of sample preparation (designated day 0) and on days 1, 2, 3, 4, 8, and 20.
Acknowledgments
We thank Professor Peter Tonge for use of his FTIR, and Professors Joseph Lauher, Bill Fowler, and Troy Rasbury for use of their polarizing light microscopes. TEM was performed at the University Microscopy Imaging Center at SUNY Stony Brook with the assistance of Greg Rudomen. MALDI-MS data was collected at the CASM facility at SUNY Stony Brook. Dan Moriarty synthesized the SNNFPAILSS peptide for initial characterization. Mona Becker provided technical assistance with the polarized light microscopy. This work was supported in part by NIH grant GM54233 to D.P.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ACN, acetonitrile
Asn, asparagine
Asp, aspartic acid
CCA, α-cyano-4-hydroxycinnamic acid
FTIR, Fourier transform infrared spectroscopy
isoAsp, isoaspartic acid
MALDI-MS, matrix-assisted laser desorption ionization mass spectrometry
RP-HPLC, reversed-phase high-performance liquid chromatography
TEM, transmission electron microscopy
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.48702.
References
- Ashburn, T.T. and Lansbury, P.T., Jr. 1993. Interspecies sequence variations affect the kinetics and thermodynamics of amyloid formation: Peptide models of pancreatic amyloid. J. Am. Chem. Soc. 11511012–11013. [Google Scholar]
- Ashburn, T.T., Auger, M., 1st., and Lansbury, P.T., Jr. 1992. The structural basis of pancreatic amyloid formation: Isotope-edited spectroscopy in the solid state. J. Am. Chem. Soc. BO114: 790–791. [Google Scholar]
- Aswad, D.W. and Guzetta, A.W. 1995. Methods for analysis of deamidation and isoaspartate formation in peptides and proteins. In Deamidation and isoaspartate formation in peptides and proteins (ed. D.W. Aswad), pp. 8–29. CRC Press, Ann Arbor, MI.
- Baumann, M.H., Wisniewski, T., Levy, E., Plant, G.T., and Ghiso, J. 1996. C-terminal fragments of alpha- and beta-tubulin form amyloid fibrils in vitro and associate with amyloid deposits of familial cerebral amyloid angiopathy, British type. Biochem. Biophys. Res. Commun. 219238–242. [DOI] [PubMed] [Google Scholar]
- Betsholtz, C., Christmansson, L., Engstrom, U., Rorsman, F., Jordan, K., O'Brien, T.D., Murtaugh, M., Johnson, K.H., and Westermark, P. 1990. Structure of cat islet polypeptide and identification of amino acid residues of potential significance for islet amyloid formation. Diabetes 39 118–122. [DOI] [PubMed] [Google Scholar]
- Chirgadze, Y.N., Fedorov, O.V., and Trushina, N.P. 1975. Estimation of amino acid residue side-chain absorption in the infrared spectra of protein solutions in heavy water. Biopolymers 14679–694. [DOI] [PubMed] [Google Scholar]
- Chirgadze, Y.N., Shestopalov, B.V., and Venyaminov, S.Y. 1973. Intensities and other spectral parameters of infrared amide bands of polypeptides in the β- and random forms. Biopolymers 121337–1351. [DOI] [PubMed] [Google Scholar]
- Cribbs, D.H., Azizeh, B.Y., Cotman, C.W., and LaFerla, F.M. 2000. Fibril formation and neurotoxicity by a herpes simplex virus glycoprotein B fragment with homology to the Alzheimer's Aβ peptide. Biochemistry 395988–5994. [DOI] [PubMed] [Google Scholar]
- Fabin, H., Szendrei, G.I., Mantsch, H.H., Greenberg, B.D., and Otovos, L., Jr. 1994. Synthetic post-translationally modified human Aβ peptide exhibits a markedly increased tendency to form β-pleated sheets in vitro. Eur. J. Biochem. 221959–964. [DOI] [PubMed] [Google Scholar]
- Fandrich, M., Fletcher, M.A., and Dobson, C.M. 2001. Amyloid fibrils from muscle myoglobin. Nature 410165–166. [DOI] [PubMed] [Google Scholar]
- Glenner, G.G., Eanes, E.D., and Wiley, C.A. 1988. Amyloid fibrils formed from a segment of the pancreatic-islet amyloid protein. Biochem. Biophys. Res. Commun. 155608–614. [DOI] [PubMed] [Google Scholar]
- Griffiths, J.M., Ashburn, T.T., Auger, M., 1st, Costa, P.R., Griffin, R.G., and Lansbury, P.T., Jr. 1995. Rotational resonance solid-state NMR elucidates a structural model of pancreatic amyloid. J. Am. Chem. Soc. 1173539–3546. [Google Scholar]
- Guijarro, J.I., Sunde, M., Jones, J.A., Campbell, I.D., and Dobson, C.M. 1998. Amyloid fibril formation by an SH3 domain. Proc. Natl. Acad. Sci. 954224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson, K.J., Sucholeiki, I., Ashburn, T.T., and Lansbury, P.T., Jr. 1991. Location of β -sheet-forming sequences in amyloid proteins by FTIR. J. Am. Chem. Soc. 1136701–6703. [Google Scholar]
- Harper, J.D., Wong, S.S., Lieber, C.M., and Lansbury, P.T., Jr. 1997. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem. Biol. 4119–125. [DOI] [PubMed] [Google Scholar]
- Higham, C.E., Jaikaran, E.T.A.S., Fraser, P.E., Gross, M., and Clark, A. 2000. Preparation of synthetic human islet amyloid polypeptide (IAPP) in a stable conformation to enable study of conversion to amyloid-like fibrils. FEBS Lett. 47055–60. [DOI] [PubMed] [Google Scholar]
- Inouye, H. and Kirschner, D.A. 1997. X-ray diffraction analysis of scrapie prion: Intermediate and folded structures in a peptide containing two putative alpha-helices. J. Mol. Biol. 268375–389. [DOI] [PubMed] [Google Scholar]
- Jackson, M. and Mantsch, H.H. 1995. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 3095–120. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A. and Asward, D.W. 1995 Deamidation and isoaspartate formation during in vitro aging of purified proteins In Deamidation and isoaspartate formation in peptides and proteins (ed. D.W. Aswad), pp. 92–113. CRC Press, Ann Arbor, MI.
- Jost, K., Varga, J., Penke, B., and Zarandi, M. 2001. In vitro degradation of β-amyloid [25–35] peptide. Protein Pept. Lett. 8423–429. [Google Scholar]
- Kowalewski, T. and Holtzman, D.M. 1999. In situ atomic force microscopy study of Alzheimer's β-amyloid peptide on different substrates: new insights into mechanism of β-sheet formation. Proc. Natl. Acad. Sci. 963688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, Y.-M., Emmerling, M.R., Woods, A.S., Cotter, R.J., and Roher, A.E. 1997. Isolation, chemical characterization, and quantitation of Aβ 3-pyroglutamyl peptide from neuritic plaques and vascular amyloid deposits. Biochem. Biophys. Res. Commun. 237188–191. [DOI] [PubMed] [Google Scholar]
- Litvinovich, S.V., Brew, S.A., Aota, S., Akiyama, S.K., Haudenschild, C., and Ingham, K.C. 1998. Formation of amyloid-like fibrils by self-association of a partially folded fibronectin type III module. J. Mol. Biol. 280245–258. [DOI] [PubMed] [Google Scholar]
- Middaugh, C.R., Mach, H., Ryan, J.A., Sanyal, G., and Volkin, D.B. 1995. Methods in molecular biology, vol. 40: Protein stability and folding: Theory and practice (ed. B.A. Shirley). Humana Press Inc., Totowa, NJ.
- Miyata, T., Iida, Y., Ueda, Y., Shinzato, T., Seo, H., Monnier, V.M., Maeda, K., and Wada, Y. 1996. Monocyte/macrophage response to beta 2-microglobulin modified with advanced glycation end products. Kidney Int. 49538–550. [DOI] [PubMed] [Google Scholar]
- Moriarty, D.F. and Raleigh, D.P. 1999. Effects of sequential proline substitutions on amyloid formation by human amylin 20–29. Biochemistry 381811–1818. [DOI] [PubMed] [Google Scholar]
- Morozova-Roche, L.A., Zurdo, J., Spencer, A., Noppe, W., Receveur, V., Archer, D.B., Joniau, M., and Dobson, C.M. 2000. Amyloid fibril formation and seeding by wild-type human lysozyme and its disease-related mutational variants. J. Struct. Biol. 130339–351. [DOI] [PubMed] [Google Scholar]
- Nilsson, M.R. and Raleigh, D.P. 1999. Analysis of amylin cleavage products provides new insights into the amyloidogenic region of human amylin. J. Mol. Biol. 294 1375–1385. [DOI] [PubMed] [Google Scholar]
- Robinson, A.B. and Rudd, C.J. 1974. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. In Current topics in cellular regulation (eds. B.L. Horecker and E.R. Stadtman), p. 8. Academic Press, New York. [DOI] [PubMed]
- Shen, C.-L., Scott, G.L., Merchant, F., and Murphy, R.M. 1993. Light scattering analysis of fibril growth from the amino-terminal fragment beta(1–28) of beta-amyloid peptide. Biophys. J. 65 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T., Wantanabe, A., Ogawara, M., Mori, H., and Shirasawa, T. 2000 Isoasparate formation and neurodegeneration in Alzheimer's disease. Arch. Biochem. Biophys. 381 225–234. [DOI] [PubMed]
- Sherzinger, E., Sittler, A., Schweiger, K., Heiser, V., Lurz, R., Hasenbank, R., Bates, G.P., Lehrach, H., and Wanker, E.E. 1999. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. 96 4604–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe, J.D. 1994. Amyloidosis. Crit. Rev. Clin. Lab. Sci. 31 325–354. [DOI] [PubMed] [Google Scholar]
- Soto, C., Castaño, E.M., Kumar, R.A., Beavis, R.C., and Frangione, B. 1995. Fibrillogenesis of synthetic amyloid-β peptides is dependent on their initial secondary structure. Neurosci. Lett. 200 105–108. [DOI] [PubMed] [Google Scholar]
- Tenidis, K., Waldner, M., Bernhagen, J., Fischle, W., Bergmann, M., Weber, M., Merkle, M.-L., Voelter, W., Brunner, H., and Kapurniotu, A. 2000. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol. 295 1055–1071. [DOI] [PubMed] [Google Scholar]
- Tuong, A., Maftouh, M., Ponthus, C., Whitechurch, O., Roitsch, C., and Picard, C. 1992. Characterization of the deamidated forms of recombinant hirudin. Biochemistry 31 8291–8299. [DOI] [PubMed] [Google Scholar]
- Tyler-Cross, R. and Schirch, V. 1991. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J. Biol. Chem. 266 22549–22556. [PubMed] [Google Scholar]
- Westermark, P., Engström, U., Johnson, K.H., Westermark, G.T., and Betsholtz, C. 1990. Islet amyloid polypeptide: Pinpointing amino acid residues linked to amyloid fibril formation. Proc. Natl. Acad. Sci. USA 87 5036–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, H.T. 1991. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 26 1–52. [DOI] [PubMed] [Google Scholar]