Abstract
Malic enzymes are widely distributed in nature, and have important biological functions. They catalyze the oxidative decarboxylation of malate to produce pyruvate and CO2 in the presence of divalent cations (Mg2+, Mn2+). Most malic enzymes have a clear selectivity for the dinucleotide cofactor, being able to use either NAD+ or NADP+, but not both. Structural studies of the human mitochondrial NAD+-dependent malic enzyme established that malic enzymes belong to a new class of oxidative decarboxylases. Here we report the crystal structure of the pigeon cytosolic NADP+-dependent malic enzyme, in a closed form, in a quaternary complex with NADP+, Mn2+, and oxalate. This represents the first structural information on an NADP+-dependent malic enzyme. Despite the sequence conservation, there are large differences in several regions of the pigeon enzyme structure compared to the human enzyme. One region of such differences is at the binding site for the 2`-phosphate group of the NADP+ cofactor, which helps define the cofactor selectivity of the enzymes. Specifically, the structural information suggests Lys362 may have an important role in the NADP+ selectivity of the pigeon enzyme, confirming our earlier kinetic observations on the K362A mutant. Our structural studies also revealed differences in the organization of the tetramer between the pigeon and the human enzymes, although the pigeon enzyme still obeys 222 symmetry.
Keywords: Malic enzyme, oxidative decarboxylase, cofactor selectivity, protein structure
Malic enzymes (ME) catalyze the reversible oxidative decarboxylation of l-malate to produce pyruvate and CO2, coupled with the reduction of the dinucleotide cofactor NAD+ or NADP+:
l-Malate + NAD(P)+ ⇔ pyruvate + CO2 + NAD(P)H
The enzymes also require the presence of divalent cations (most commonly Mg2+ or Mn2+) for their catalytic activity. Most malic enzymes can use only NAD+ or NADP+ as the cofactor, and they are thereby classified as NAD+- or NADP+-dependent malic enzymes. Malic enzymes are widely distributed in nature, having been identified in bacteria, yeast, fungi, plants, animals, and humans. Their amino acid sequences are highly conserved among the various living organisms, suggesting that malic enzymes may have important biological functions.
Malic enzymes are generally homo-tetramers of 60 kD monomers. The conversion of malate to pyruvate by these enzymes generally proceeds in two steps: oxidation (dehydrogenation) of malate to produce oxaloacetate, and then decarboxylation of oxaloacetate to produce pyruvate and CO2. Many substrate- and transition-state analog inhibitors have been identified for these enzymes, including d-malate, tartronate (−OOCCH(OH)COO−), ketomalonate (−OOCCOCOO−) and oxalate (−OOCCOO−).
We recently reported crystal structures of the human mitochondrial NAD(P)+-dependent malic enzyme (m-NAD-ME) (Xu et al. 1999; Yang et al. 2000; Yang and Tong 2000). This isoform of the enzyme has "dual specificity" and can use both cofactors, although it prefers NAD+ at physiological conditions (Loeber et al. 1991). The structures establish malic enzymes as a new class of oxidative decarboxylases (Xu et al. 1999), consistent with the fact that their amino acid sequences show no detectable homology to other proteins in the database. The structure of the monomer can be divided into four domains (A, B, C, and D), with the active site of the enzyme located at the interface of domains B and C. Residues in domains A and D mostly participate in the formation of the dimers and tetramers of the enzyme. The tetramer of the human ME can be described as a dimer of dimers.
There is a large conformational change in the ME monomer when the divalent cation and transition-state analogs (oxalate, tartronate, and ketomalonate) are bound to the enzyme, creating the so-called closed conformation (Yang et al. 2000). The side chains of Glu255, Asp256, and Asp279 provide three ligands to the cation, and the transition-state analogs provide two ligands. A solvent water molecule then completes the octahedral coordination of the cation (Yang et al. 2000). The structures reveal that conserved residues Tyr112 and Lys183 may have important roles in the catalytic mechanism of the enzyme. Consistent with this, the Y112F and K183M mutants have dramatically reduced catalytic activity (Yang and Tong 2000).
Malic enzyme activity was first characterized in the pigeon liver (Ochoa et al. 1947). This cytosolic NADP+-dependent malic enzyme (c-NADP-ME, EC 1.1.1.40) is one of the most abundant proteins in that tissue, and accounts for about 0.6% of the total cytosolic protein (Hsu 1982). This allowed the purification of 1–2 mg of the enzyme per pigeon liver and enabled extensive biochemical and biophysical studies on this enzyme over the years (Hsu 1982). The kinetic analysis demonstrated a half-of-the-sites reactivity for the tetramer of the pigeon enzyme, suggesting possible anticooperativity among the four active sites (Pry and Hsu 1980). In the current study, we have determined the crystal structure of the pigeon cytosolic NADP+-dependent malic enzyme (c-NADP-ME) in a quaternary complex with NADP+, Mn2+, and the transition-state analog oxalate. This represents the first structural information on an NADP+-dependent malic enzyme, and it reveals the possible molecular determinants for cofactor selectivity. In addition, the structural information provides a foundation for understanding the large body of kinetic, biochemical and biophysical information on this enzyme.
Results and Discussion
Crystallization of pigeon malic enzyme
Crystals of pigeon c-NADP-ME were first reported more than 30 years ago (Hsu and Lardy 1967), although those crystals were not characterized by X-ray diffraction studies. Recently, we reported two new crystal forms of this enzyme, grown in the presence of d-malate as a substrate-analog inhibitor (Tsai et al. 1999). Although one of these crystal forms only diffracted X-rays to 4-Å resolution, a data set to 2.9-Å resolution was collected for the other crystal form (Tsai et al. 1999). The structure solution by the molecular replacement method was able to locate only one monomer of the enzyme in the asymmetric unit. Even though this corresponded to a Vm of 5.8 Å3/Dalton, or about 80% solvent content, examination of the crystal packing showed that the solution could actually be correct. The tetramer of the enzyme can be generated by the crystallographic symmetry from the monomer, and there are packing contacts among the tetramers in the crystal. Crystal structure refinement based on this molecular replacement solution was not successful, however, due to the poor quality of the diffraction data (Tsai et al. 1999).
For the current studies, we have produced two additional crystal forms of the pigeon ME, this time grown with oxalate as a transition-state analog inhibitor. The presence of a large number of crystal forms for the pigeon ME is reminiscent of the situation with the human ME, where close to 10 different crystal forms have been observed so far (Bhargava et al. 1999; Z. Yang, G. Bhargava, and L. Tong, unpubl.). X-ray diffraction studies, however, showed that these new pigeon ME crystals are often heavily twinned microscopically. After screening through many crystals, a good-quality X-ray diffraction data set to 2.5-Å resolution was collected on a triclinic crystal that displayed no twinning. This triclinic unit cell is extremely large, containing four unique tetramers. This corresponds to about 9000 amino acid residues, or molecular weight of about 1 million Daltons, in this triclinic unit cell.
The overall structure
The crystal structure of the pigeon cytosolic NADP+-dependent malic enzyme (c-NADP-ME), in complex with NADP+, oxalate and Mn2+, has been determined at 2.5-Å resolution (Table 1). The atomic model is of good quality, with a crystallographic R factor of 21.1% and low deviations from ideal bond length and bond angle parameters (Table 1); 90.8% of the residues are in the most favored regions of the Ramachandran plot, and 12.0% are in the additionally allowed regions. The atomic coordinates have been deposited at the Protein Data Bank (accession code 1GQ2).
Table 1.
Summary of crystallographic information
| Maximum resolution (Å) | 2.5 |
| Number of observations | 570,674 |
| Rmerge (%)a | 8.2 (21.4) |
| Resolution range used for refinement | 20–2.5 |
| Number of reflections | 314,771 |
| Completeness (%) (2σ cutoff) | 83 (76) |
| R factorb (%) | 21.1 (30.2) |
| Free R factor (%) | 25.8 (34.2) |
| rms deviation in bond lengths (Å) | 0.008 |
| rms deviation in bond angles (°) | 1.3 |
a Rmerge= ∑h∑i|Ihi − 〈Ih〉|∑h∑iIhi.The numbers in the parentheses are for the highest resolution shell (2.6–2.5 Å).
b R = ∑h|Fho − Fho|∑hFho.
There are four tetramers of pigeon c-NADP-ME in the asymmetric unit of the crystal. The conformations of the 16 monomers are similar to each other. The root mean square (RMS) distance between equivalent Cα atoms is about 0.15 Å when any pair of the monomers are superimposed. The four independent tetramers also have similar organizations. The RMS distance between equivalent Cα atoms is about 0.3 Å when any pair of the tetramers is superimposed. Consequently, we will focus on only one tetramer in our subsequent discussions.
To facilitate the structural comparison with the human malic enzyme, we have numbered the residues in the pigeon ME according to their structural equivalents in the human ME. We propose that this numbering scheme be adopted for the other malic enzymes as well, using structural or sequence alignment to the human ME. In this system, the pigeon ME residues are numbered from 22 to 581. There are two breaks in the numbers, due to deletions of 1 and 2 residues after residue 354 and 370 in the pigeon ME, respectively. There are no insertions in the pigeon ME sequence relative to the human ME.
The atomic models of the monomers contain residues 23 through 580 of the pigeon ME. At the C-terminus, only one residue (Leu581) is missing from the model due to disorder. At the N-terminus, the initiator Met22 residue is not observed in the electron density, although it is not known whether this residue is present in the protein as purified from E. coli. Two segments of the protein have weak electron density, corresponding to residues 352–357 and 455–460. Both segments are on the surface of the protein, and the 352–357 region is also where there is a one-residue deletion in the pigeon ME.
Conformational differences to the human malic enzyme
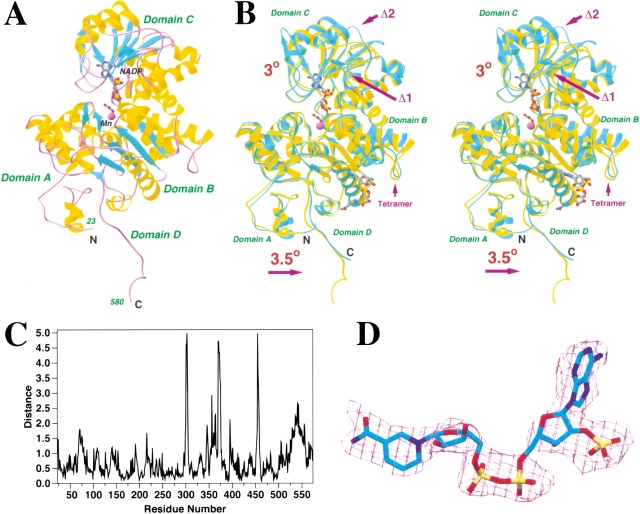
The overall backbone fold of the monomer of pigeon c-NADP-ME is identical to that of the human m-NAD-ME (Fig. 1A ▶). All the secondary structure elements that were observed in the human ME structure are also observed here in the pigeon enzyme. Analogous to the human ME, the pigeon ME monomer can be divided into four domains. Domain A (residues 23–130) is mostly helical in structure. Domain B (residues 131–277, and 467–538) contains a central five-stranded parallel β-sheet surrounded by several helices on both faces. The polypeptide backbone fold of this domain has so far only been observed in the malic enzymes (Yang and Tong 2000). Domain C (278–466) contains a dinucleotide-binding Rossmann fold, with the modification that strand β3 is replaced by a short antiparallel β-structure (Yang and Tong 2000). The two deletions in the pigeon ME sequence are both located in surface loops in this domain (Fig. 1B ▶). Domain D (539–580) encompasses the C-terminal segment of enzyme. It extends beyond the ordered portion of the human enzyme structure, which stops at residue 573.
Fig. 1.
Crystal structure of pigeon cytosolic NADP+-dependent malic enzyme. (A) Schematic drawing of the structure of pigeon c-NADP-ME in complex with NADP+, oxalate, and Mn2+. The β strands are shown in cyan, α helices in yellow, and the connecting loops in purple. NADP+ and oxalate are shown as stick models, and Mn2+ is shown as a purple sphere. The NAD+ molecule in the second site is present only in the human ME structure (Xu et al. 1999). (B) Structure comparison between human m-NAD-ME (shown in cyan) and pigeon c-NADP-ME (in yellow). The two regions where there are deletions in the pigeon enzyme are also indicated. (C) Plot of the distances between equivalent Cα atoms of the pigeon and human ME structures. (D) Electron density for the NADP+ molecule after noncrystallographic symmetry averaging over the four tetramers. The refined atomic model is shown for reference. (A) and (B) were created with Ribbons (Carson 1987), and (D) was created with SETOR (Evans 1993).
The current structure shows the pigeon ME in a quaternary complex with NADP+, oxalate, and Mn2+. Similar to the quaternary complex of the human enzyme (Yang et al. 2000), the pigeon enzyme in the quaternary complex also exhibits a closed conformation (Figs. 1A,B ▶). Structure comparison showed a rms distance of 0.9 Å between 526 out of 541 equivalent Cα atoms of the pigeon and the human enzymes (Fig. 1C ▶). In addition, several regions of the pigeon ME structure show large differences to the human ME structure, despite the strong sequence homology (57% identity) between the two proteins (Fig. 1C ▶, and see below).
Structural studies of the human ME showed that the changes between the open and closed conformations are mostly mediated by rigid-body movements of the four domains relative to each other (Yang et al. 2000; Yang and Tong 2000). For a detailed comparison between the pigeon and the human enzymes, residues in the different domains of the two structures were superimposed separately. This showed that residues 78–277 and 467–515 of pigeon ME (domain B and part of domain A) have a RMS distance of 0.6 Å to their equivalents in the human enzyme (Fig. 1B,C). In this superposition, residues 215–216 in the pigeon structure showed distances of greater than 1.2 Å to their equivalents in the human ME (Fig. 1C ▶). These residues are located near the dimer interface of the tetramer (see below).
With residues in domain B in superposition, a further rotation of about 3° is needed to superimpose residues in domain C (278–466) between the pigeon and human ME structures (Fig. 1B ▶). In contrast to the open–closed transition, this rotation does not affect the closure of the active site. It is rather a sliding motion between domains B and C (Fig. 1B ▶), and represents another aspect of the conformational differences between the pigeon and the human enzymes. The RMS distance for 161 out of 186 equivalent Cα atoms of this domain is 0.6 Å (Fig. 1C ▶). Several loops of pigeon ME showed large differences to the human ME, including 299–304, 345–348, 360–364, 370–375, and 454–458 (Fig. 1B,C ▶). The 370–375 region contains a deletion of two residues in the pigeon ME sequence, whereas the 345–348 and 360–364 regions surround the other deletion in pigeon ME. Most important among these differences is the segment 345–348, which is located near the 2`-phosphate of the NADP+ cofactor, and contributes to the different cofactor selectivity of the pigeon enzyme (see below).
With domain B in superposition, an independent rotation of 3.5° is needed to superimpose residues 23–67 (domain A) and 559–573 (domain D) between the pigeon and human ME structures (Fig. 1B ▶). The RMS distance for these 60 equivalent Cα atoms is 0.7 Å (Fig. 1C ▶). In the superposition described above, two segments of the enzyme, residues 68–77 and 516–558, are not included because they show large distances to their equivalent residues in the human ME structure (Fig. 1B,C ▶). Residues 68–77 are located in the dimer interface, whereas residues 516–558 are in the tetramer interface. The conformational differences for these residues are likely related to the differences in tetramer organization of the pigeon and human enzymes (see below).
Binding mode of NADP+ and molecular mechanism for differences in cofactor selectivity
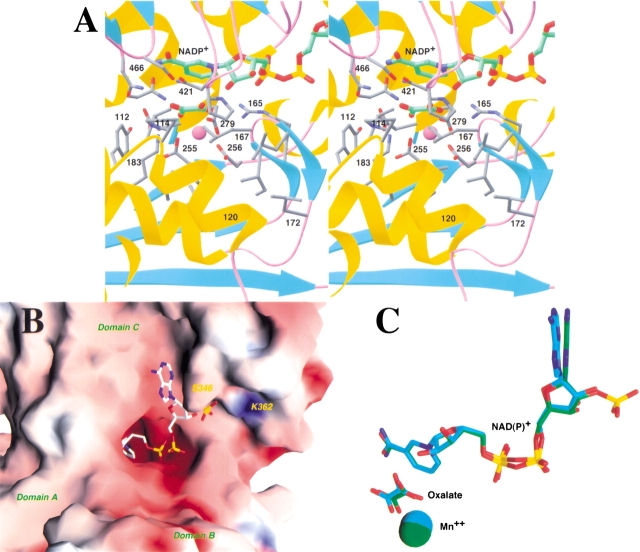
The dinucleotide cofactor NADP+ is associated with the Rossmann fold in domain C. The nicotinamide portion of the cofactor, in the active site of the enzyme, is shielded from the solvent by residues from domains B and C in this closed form of the enzyme (Fig. 2A,B ▶). The binding mode of this portion of the NADP+ cofactor is similar to that observed for NAD+ in the human m-NAD-ME (Fig. 2C ▶). Residues lining the nicotinamide binding pocket are generally conserved between the human and the pigeon enzymes.
Fig. 2.
The active site of the pigeon malic enzyme. (A) Stereo drawing showing the active site of pigeon c-NADP-ME. The oxalate molecule is shown in green for carbon atoms, NADP+ in green, and protein residues in gray. The Mn2+ cation is shown as a purple sphere. (B) Molecular surface of pigeon c-NADP-ME near the active site region, colored according to electrostatic potential. The NADP+ molecule is shown as a stick model. (C) Comparison of the binding modes of NADP+ to the pigeon ME (in green for carbon atoms) and of NAD+ to the human ME (in cyan). The oxalate and the Mn2+ ion are also shown in the comparison. (A) was created with Ribbons (Carson 1987), and (B) and (C) were created with Grasp (Nicholls et al. 1991).
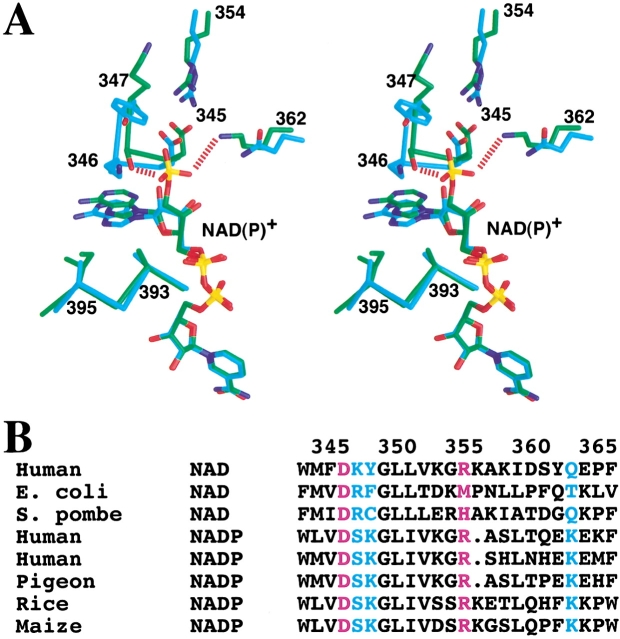
However, the binding mode of the adenine portion of the cofactor shows significant differences between the human and pigeon ME structures (Fig. 2C ▶). The adenine ring is placed between the loop segments that immediately follow strands β2 and β4 in the Rossmann fold (Fig. 3A ▶). Both of these segments show conformational and amino acid sequence differences between the pigeon and human enzymes (Fig. 3A,B ▶). Residues 346–347, immediately following strand β2, moved by about 2 Å in the pigeon ME structure compared to the human enzyme (Fig. 3A ▶). In addition, the sequence Lys–Tyr for these two residues in the human enzyme is replaced by Ser–Lys in the pigeon ME. The Ser346 side-chain hydroxyl is hydrogen bonded to the 2`-phosphate, but the side chain of Lys347 in pigeon ME does not interact with the NADP+ molecule. This is consistent with our previous mutagenesis experiments showing that the K347A mutant has a less than twofold change in the Km for NADP+ (Kuo et al. 2000). The conformational difference observed in this region is unlikely to be due solely to the addition of the 2`-phosphate of the cofactor. The structure of human m-NAD-ME in complex with NADP+ has essentially the same conformation as that of the NAD+ complex in this region (unpublished data).
Fig. 3.
A possible molecular mechanism for cofactor selectivity. (A) Stereo drawing showing the structure comparison between human m-NAD-ME (in cyan for carbon atoms) and pigeon c-NADP-ME (in green) near the 2`-phsophate of NADP+. (B) Alignment of ME sequences near the binding site for the 2`-phosphate group of NADP+. The cofactor dependence of the various malic enzymes is indicated. The Asp345:Arg354 ion-pair is shown in purple. (A) was created with Grasp (Nicholls et al. 1991).
The structure suggests a possible molecular mechanism for the NADP+ selectivity of the pigeon liver ME compared to the NAD+ preference for the human m-NAD-ME. The 2`-phosphate group is placed on the surface of the c-NADP-ME protein (Fig. 2B ▶), and interacts with residues Ser346 (as discussed above) and the side-chain ammonium group of Lys362 (Fig. 3A ▶). Residue 362 is Gln in the human enzyme (Fig. 3A ▶), which prefers NAD+, but is generally conserved as Lys in the NADP+-dependent malic enzymes (Fig. 3B ▶). Our earlier mutagenesis studies showed that the K362A mutant of the pigeon enzyme has a 70-fold increase in the Km for NADP+, whereas the Km for malate and Mn2+ and the kcat of the enzyme are not affected by the mutation (Kuo et al. 2000). This provides additional confirmation that residue Lys362 is an important factor in the NADP+ selectivity of pigeon ME. It would be interesting to determine whether the K362Q mutant can utilize NAD+ as a cofactor. Sequence comparisons showed that several other residues near the 2`-phosphate group have concerted variations between NADP+ and NAD+-dependent malic enzymes (Fig. 3B ▶). It may be possible that these residues, such as Ser346 described above, also play a role in the cofactor selectivity.
Residue 345 is at the end of strand β2 in the Rossmann fold of domain C. It is conserved as Asp in almost all the malic enzymes (Fig. 3A,B ▶). Earlier studies had suggested that this residue would indicate a NAD+ preference (Wierenga et al. 1986; Scrutton et al. 1990), due to potential charge repulsion with the 2`-phosphate of NADP+. However, the pigeon c-NADP-ME structure shows that this residue is pointed away from the 2`-phosphate group and forms an ion-pair with Arg354 (Fig. 3A ▶). This interaction is identical to that observed in the human ME structure (Xu et al. 1999; Yang et al. 2000). Therefore, our studies suggest that the presence of an Asp residue at the end of strand β2 does not necessarily indicate NAD+ preference.
Binding modes of Mn2+ and oxalate, and relevance for catalytic mechanism
The binding modes of the divalent cation and the transition-state analog inhibitor oxalate to the pigeon ME are essentially the same as those observed for human m-NAD-ME (Fig. 2C ▶) (Yang et al. 2000). These conserved binding modes suggest that the catalytic mechanism of the pigeon enzyme is likely to be the same as that for the human enzyme (Yang et al. 2000). The Mn2+ is liganded by the side-chain carboxylate groups of Glu255, Asp256, and Asp279, as well as the carboxylate oxygen atoms of the oxalate molecule (Fig. 2A ▶). The sixth ligand is a water molecule in the human enzyme structure (Yang et al. 2000). The presence of this water molecule in the pigeon enzyme cannot be confirmed based on the current structure due to the moderate resolution of the X-ray diffraction data.
Using the Fe–ascorbate affinity cleavage system, we had shown earlier that Asp279 is a ligand of the cation in the pigeon ME (Wei et al. 1994), which we later confirmed by mutagenesis experiments (Wei et al. 1995). Lys183 has been proposed to be a crucial residue in the catalysis by malic enzymes (Fig. 2A ▶) (Yang et al. 2000). Our mutagenesis studies showed that the K183A mutant of the pigeon enzyme has a 230-fold loss in the kcat, whereas the Km for malate, NADP+, and Mn2+ are not affected (Kuo et al. 2000), thereby confirming the functional importance of the Lys183 residue in pigeon ME. The equivalent Lys residue in Ascaris suum ME has been proposed to be the general acid in the catalysis (Liu et al. 2000).
Chemical labeling experiments with the pigeon and other malic enzymes have identified the presence of crucial residues in the active site. The presence of Arg (residue 165), Tyr (112), and Cys (120) residues in or near the active was confirmed from the structural analysis (Chang and Huang 1980, 1981; Hsu 1982; Rao et al. 1991; Satterlee and Hsu 1991; Yang and Tong 2000).
Differences in the organization of the tetramer compared to the human enzyme
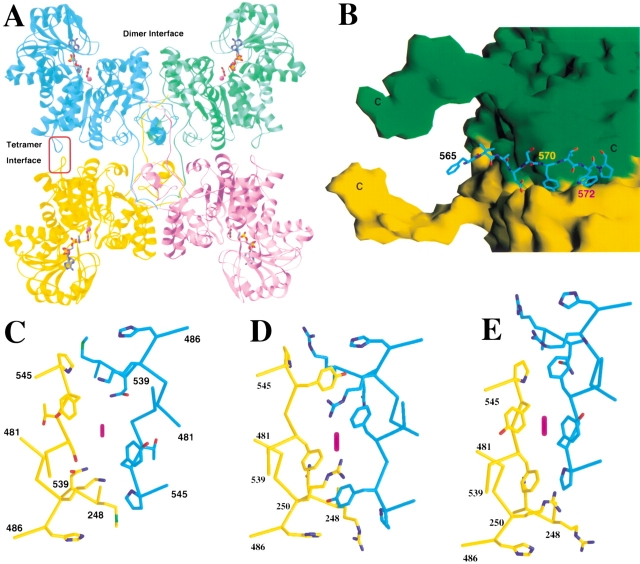
Earlier biochemical and biophysical studies showed that pigeon ME is a tetramer in solution (Hsu 1982). Electron microscopy experiments showed that the tetramer has square-planar arrangement of the four monomers, and the dimensions of the tetramer was estimated to be 100 × 110 × 70 Å3 (Nevaldine et al. 1974). These earlier observations are confirmed from the current structural analysis. The four monomers in the tetramer are arranged at the corners of a square, and the dimensions of the tetramer are about 110 × 110 × 55 Å3 (Fig. 4A ▶) (Xu et al. 1999). Structural studies also showed that the pigeon enzyme tetramer is a dimer of dimers (Fig. 4A,B ▶) (Xu et al. 1999).
Fig. 4.
The tetramer of the pigeon malic enzyme. (A) Schematic drawing showing the tetramer of the pigeon ME. The four monomers are given different colors. The dimer and the tetramer interfaces are labeled. (B) The interactions of the C-terminal tail in one molecule (shown as stick models in cyan) with the other dimer of the tetramer (shown as molecular surfaces colored green and yellow, with their C termini labeled with the letter "C"). (C–E). Detailed structure comparisons of residues 541–546, in the tetramer interface, between the pigeon (C) and the closed (D) and open (E) forms of the human malic enzymes. These residues are located in the tetramer interface, indicated by the red oval in (A). The twofold axis is indicated with the purple oval. (A) was created with Ribbons (Carson 1987), (B–E) were created with Grasp (Nicholls et al. 1991).
There are differences in the organization of the pigeon and human ME tetramers. This is consistent with the differences in the relative positions of domains A and D in the pigeon enzyme, as described above (Fig. 1B ▶). In addition, structural comparisons revealed that a portion of the tetramer interface, involving residues 539–545, has large differences between the human enzyme and the pigeon enzyme (Fig. 4C ▶–E). Some of these structural differences between the pigeon and human enzymes may possibly be related to the cooperative behavior of the human m-NAD-ME, although our structure of the human enzyme was obtained in the presence of fumarate, where it appears to lose the cooperative behavior.
There are intimate contacts within the dimers, and 1950 Å2 of surface area are buried for each monomer upon the formation of the dimer. The N-terminus of the enzyme, residues 23–31, has large contributions to the dimer interface, and smaller contributions to the tetramer interface. Our previous mutagenesis studies showed that these N-terminal residues are important for the stability of the enzyme (Chou et al. 1997,Chou et al. 1998; Huang et al. 1998). In addition, residue Phe40 is important for the stability of the pigeon enzyme (Chou et al. 1996). However, this residue is not directly located in the dimer or tetramer interface. Instead, it may be important for stabilizing the conformation of domain A, as it is a part of the hydrophobic core of this domain (Xu et al. 1999).
In contrast, interactions at the tetramer interface are mostly mediated by the C-terminal segment of one monomer packing against the other dimer of the tetramer (Fig. 4B ▶). An important residue in this interface is Trp572, which is located in a deep, hydrophobic pocket (Fig. 4B ▶). Our previous studies showed that the pigeon enzyme tetramer can be dissociated into dimers and monomers at lower pH (Chang et al. 1988; Lee and Chang 1990). In contrast, preliminary results showed that the human enzyme tetramer is more stable at lower pH (unpublished data). Examination of the pigeon enzyme structure shows the presence of a pair of His residues, His138 and His223, with His138 in the binding pocket for Trp572. It is likely that the protonation of these His residues at lower pH can cause a change in their conformation, which in turn, can affect the binding of the Trp572 residue in the tetramer interface. There is another pair of His residues (His33 and His196) that are located in close proximity in the pigeon ME structure, but not in the human ME structure. Protonation of this pair of residues at lower pH might also be detrimental to the stability of the pigeon enzyme. At the dimer interface, the pigeon enzyme has several additional charged residues, such as Arg30 (Leu in human ME) and Glu27 (Pro). It may be possible that the destabilization of dimers of the pigeon enzyme at lower pH is due to changes in the ionic interactions in the dimer interface.
The four monomers in the tetramer have essentially the same conformation, and they obey 222 point-group symmetry. This means the monomers have essentially the same environment in the tetramer. Kinetic and chemical modification studies have shown that the pigeon enzyme tetramer has half-of-the-sites reactivity, such that only two of the active sites have high activity (Chang and Hsu 1977; Hsu and Pry 1980; Hsu 1982). An anticooperativity among the four active sites has been proposed (Pry and Hsu 1980). The structure of the oxalate complex here, however, does not provide a strong indication of asymmetry among the four monomers. An oxalate molecule is bound in each of the active sites of the tetramer (Fig. 4A ▶). Further studies are needed to characterize the molecular basis for the half-of-the-sites reactivity of pigeon cytosolic NADP+-dependent malic enzyme.
In summary, we have determined the crystal structure at 2.5-Å resolution of the pigeon cytosolic NADP+-dependent malic enzyme in a quaternary complex with NADP+, Mn2+, and oxalate. The structure revealed the binding modes of the cofactors and the transition-state analog, and suggests that the pigeon enzyme may use the same catalytic mechanism as the human enzyme. Structural comparisons between the human and pigeon enzymes suggest a possible molecular mechanism for the cofactor selectivity of the enzymes, with residue Lys362 playing an important role in this process. Additional residues, such as Ser346, may also help determine the cofactor preference. The structure reveals that the tetramer of the enzyme is fully symmetric, with each monomer having essentially the same conformation and environment in the tetramer. Further biochemical and structural studies are needed to understand the molecular basis for the half-of-the-sites reactivity of the pigeon malic enzyme.
Materials and methods
Expression and purification of pigeon cytosolic NADP+-dependent ME
The expression and purification of pigeon cytosolic NADP+-dependent ME (c-NADP-ME) followed protocols described earlier (Chang et al. 1991; Chou et al. 1997), with modifications in the ion-exchange protocol to follow that used for the human enzyme (Bhargava et al. 1999). Briefly, the c-NADP-ME gene was subcloned into expression vector pET21b (Novagen) and overexpressed in E. coli. The bacterial cells were grown at 37°C until OD of 0.5 was reached. IPTG was added to the concentration of 1 mM to induce protein expression, and the temperature of the culture was lowered to 25°C. The cells were harvested 18 h later, and stored at −80°C.
All the purification steps were carried out at 4°C unless otherwise noted. The bacterial cell pellet was thawed on ice and resuspended in buffer A (30 mM Tris, 3 mM MgCl2, 0.2 mM EDTA, pH 7.5) supplemented with 0.2 mM DTT, 0.5 mM PMSF, and 0.2% NP-40. Lysozyme (Sigma) was added to a final concentration of 0.5 mg/mL, and the cell suspension was sonicated on ice 30 min later. The cell debris was removed by centrifugation, and then the DNA was precipitated by the addition of streptomycin sulfate (Sigma) to a final concentration of 10 mg/mL. After centrifugation, the supernatant was diluted 1:6 with buffer A, and loaded onto a 20 mL DEAE-FF anion exchange column that has be pre-equilibrated in buffer A. The bound protein was eluted with a linear gradient of 0–300 mM NaCl.
The fractions that contain ME activity based on the enzyme assay (see below) were pooled and loaded onto a 5 mL adenosine 2`,5`-bisphosphate agarose column (Sigma). The bound pigeon ME was eluted with buffer A containing 0.23 mM NADP+. The column fractions after affinity purification were concentrated and loaded onto a Sephacryl S-300 gel filtration column (Amersham Pharmacia). The running condition was buffer A supplemented with 250 mM NaCl and 2 mM DTT. The pigeon ME migrated as a tetramer on this column. The column fractions containing the enzyme were pooled, and concentrated to 30 mg/mL protein and stored at −80°C. The protein after purification is more than 98% pure based on Coomassie-stained SDS gels.
Enzyme activity assay
The assay buffer contained 67 mM Tris (pH 7.5), 5 mM l-malate, 4 mM MnCl2, and 0.23 mM NADP+ (Chou et al. 1997). The reaction was initiated by the addition of enzyme, and the increase of absorption at 340 nm (due to the appearance of NADPH) was monitored for 3 min. The amount and the activity of the enzyme are related to the rate of increase in the absorbance.
Crystallization, data colleciton, and data procesing
The initial crystallization condition was identified from sparse-matrix screening using a commercial kit (Hampton Research). Crystals were grown at 4°C using the hanging drop vapor diffusion method. Purified pigeon ME was preincubated on ice at 10 mg/mL concentration in a solution containing 20 mM Tris (pH 7.4), 10 mM oxalate, 5 mM MnCl2, 0.23 mM NADP+, and 2 mM DTT. The reservoir solution contained 100 mM sodium citrate (pH 5.5), 8% PEG 6000, and 1 M LiCl. Crystals generally appeared overnight, and grew to full size in 3 to 4 days. Crystals were flash-frozen in liquid propane for data collection at 100 K. The crystals belong to the space group P1 with unit cell dimensions of a = 124.1 Å, b = 140.9 Å, c = 167.1 Å, α = 90.0°, β = 87.2°, and γ = 75.6°. There are four unique tetramers (16 monomers) in the asymmetric unit (or unit cell) of this crystal. X-ray diffraction data to 2.5-Å resolution were collected at beam line X8C at the National Synchrotron Light Source (NSLS). The X-ray wavelength was 1.171 Å, and the crystal-to-detector distance was 180 mm. The diffraction images were recorded on an ADSC Quantum-4 CCD and processed with the HKL package (Table 1) (Otwinowski and Minor 1997).
Under similar conditions, we also obtained crystals of pigeon ME that belonged to space group C2221, with cell dimensions of a = 139.4 Å, b = 161.7 Å, and c = 199.3 Å. However, these crystals are highly twinned, and structure solution based on a data set collected to 2.5-Å resolution on such a crystal, at the X4A beam line of the NSLS, was not successful.
Structure determination and refinement
The structure of pigeon ME was determined by the combined molecular replacement protocol, with the program COMO (Tong 1996; Jogl et al. 2001). The structure of the human mitochondrial NAD(P)+-dependent malic enzyme in complex with NAD+, Mn2+, and oxalate, in a closed form, was used as the search model (Yang et al. 2000). Reflection data between 10- and 4-Å resolution was used in the calculation, and the orientation and position of the four tetramers were located automatically by the program.
The orientation and position of each of the 16 monomers was then subjected to rigid-body refinement using reflections between 5 and 4-Å resolution, with the program CNS (Brunger et al. 1998). Structure factors were calculated for all reflections to 2.7-Å resolution using the atomic model after rigid-body refinement, and the calculated phases were applied to the observed structure factor amplitudes. The phase information was improved by 16-fold noncrystallographic symmetry (NCS) averaging over the four tetramers with the program DM (CCP4 1994). The resulting electron density map was of excellent quality, clearly showing the differences in amino acid sequences between the human and the pigeon enzymes as well as the NADP+, oxalate, and Mn2+ molecules (Fig. 1 ▶). The atomic model for the monomer of pigeon ME was generated using this electron density map with the program O (Jones et al. 1991). The atomic model for the other 15 monomers in the unit cell was then generated by applying the NCS operations. The structure refinement was carried out with the program CNS (Brunger et al. 1998). NCS restraints on the main chain atoms were applied during the refinement. The crystallographic information is summarized in Table 1.
Acknowledgments
This work was supported by the National Science Foundation (grant number MCB-99–74700 to L.T.). We thank Renu Batra, Gerwald Jogl and Xiao Tao for help with data collection at the synchrotron, Joel Berendzen and Craig Ogata for access to the X8C and X4A beamline, respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
CCD, charge-coupled device
c-NADP-ME, cytosolic NADP+-dependent malic enzyme
DTT, dithiothreitol
IPTG, isopropyl β-D-thio-galactopyranoside
ME, malic enzyme
m-NAD-ME, mitochondrial NAD+-dependent malic enzyme
NCS, noncrystallographic symmetry
NSLS, national synchrotron light source
OD, optical density
PEG, polyethylene glycol
PMSF, phenylmethylsulfonyl fluoride
RMS, root mean square
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.38002.
References
- Bhargava, G., Mui, S., Pav, S., Wu, H., Loeber, G., and Tong, L. 1999. Preliminary crystallographic studies of human mitochondrial NAD(P)+-dependent malic enzyme. J. Struct. Biol. 127: 72–75. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T., and Warren, G.L. 1998. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D54: 905–921. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1987. Ribbon models of macromolecules. J. Mol. Graphics 5: 103–106. [Google Scholar]
- CCP4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50: 760–763. [DOI] [PubMed] [Google Scholar]
- Chang, G.-G. and Hsu, R.Y. 1977. Mechanism of pigeon liver malic enzyme. Kinetics, specificity, and half-site stoichiometry of the alkylation of a cysteinyl residue by the substrate-inhibitor bromopyruvate. Biochemistry 16: 311–320. [DOI] [PubMed] [Google Scholar]
- Chang, G.-G. and Huang, T.-M. 1980. Involvement of tyrosyl residues in the substrate binding of pigeon liver malic enzyme. Biochim. Biophys. Acta 611: 217–226. [DOI] [PubMed] [Google Scholar]
- Chang, G.-G. and Huang, T.-M. 1981. Modification of essential arginine residues of pigeon liver malic enzyme. Biochim. Biophys. Acta 660: 341–347. [DOI] [PubMed] [Google Scholar]
- Chang, G.-G., Huang, T.-M., and Chang, T.-C. 1988. Reversible dissociation of the catalytically active subunits of pigeon liver malic enzyme. Biochem. J. 254: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, G.-G., Wang, J.-K., Huang, T.-M., Lee, H.-J., Chou, W.-Y., and Meng, C.-L. 1991. Purification and characterization of the cytosolic NADP+-dependent malic enzyme from human breast cancer cell line. Eur. J. Biochem. 202: 681–688. [DOI] [PubMed] [Google Scholar]
- Chou, W.-Y., Huang, S.-M., and Chang, G.-G. 1997. Functional roles of the N-terminal amino acid residues in the Mn(II)-L-malate binding and subunit interactions of pigeon liver malic enzyme. Protein Eng. 10: 1205–1211. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Conformational stability of the N-terminal amino acid residues of mutated recombinant pigeon liver malic enzymes. Protein Eng. 11: 371–376. [DOI] [PubMed] [Google Scholar]
- Chou, W.-Y., Liu, M.-Y., Huang, S.-M., and Chang, G.-G. 1996. Involvement of Phe19 in the Mn2+-L-malate binding and the subunit interactions of pigeon liver malic enzyme. Biochemistry 35: 9873–9879. [DOI] [PubMed] [Google Scholar]
- Evans, S.V. 1993. SETOR: Hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11: 134–138. [DOI] [PubMed] [Google Scholar]
- Hsu, R.Y. 1982. Pigeon liver malic enzyme. Mol. Cell. Biochem. 43: 3–26. [DOI] [PubMed] [Google Scholar]
- Hsu, R.Y. and Lardy, H.A. 1967. Pigeon liver malic enzyme. II. Isolation, crystallization, and some properties. J. Biol. Chem. 242: 520–526. [PubMed] [Google Scholar]
- Hsu, R.Y. and Pry, T.A. 1980. Kinetic studies of the malic enzyme from pigeon liver. "Half-of-the-sites" behavior of the enzyme tetramer in catalysis and substrate inhibition. Biochemistry 19: 962–968. [DOI] [PubMed] [Google Scholar]
- Huang, S.-M., Chou, W.-Y., Lin, S.-I., and Chang, G.-G. 1998. Engineering of a stable mutant malic enzyme by introducing an extra ion-pair to the protein. Proteins 31: 61–73. [DOI] [PubMed] [Google Scholar]
- Jogl, G., Tao, X., Xu, Y., and Tong, L. 2001. COMO: A program for combined molecular replacement. Acta Crystallogr. D57: 1127–1134. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kuo, C.-C., Tsai, L.-C., Chin, T.-Y., Chang, G.-G., and Chou, W.-Y. 2000. Lysine residues 162 and 340 are involved in the catalysis and coenzyme binding of NADP+-dependent malic enzyme from pigeon. Biochem. Biophys. Res. Commun. 270: 821–825. [DOI] [PubMed] [Google Scholar]
- Lee, H.-J. and Chang, G.-G. 1990. Quarternary structure of pigeon liver malic enzyme. FEBS Lett. 277: 175–179. [DOI] [PubMed] [Google Scholar]
- Liu, D., Karsten, W.E., and Cook, P.F. 2000. Lysine 199 is the general acid in the NAD-malic enzyme reaction. Biochemistry 39: 11955–11960. [DOI] [PubMed] [Google Scholar]
- Loeber, G., Infante, A.A., Maurer-Fogy, I., Krystek, E., and Dworkin, M.B. 1991. Human NAD+-dependent mitochondrial malic enzyme. J. Biol. Chem. 266: 3016–3021. [PubMed] [Google Scholar]
- Nevaldine, B.H., Bassel, A.R., and Hsu, R.Y. 1974. Mechanism of pigeon liver malic enzyme subunit structure. Biochim. Biophys. Acta 336: 283–293. [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296. [DOI] [PubMed] [Google Scholar]
- Ochoa, S., Mehler, A., and Kornberg, A. 1947. Reversible oxidative decarboxylation of malic acid. J. Biol. Chem. 167: 871–872. [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pry, T.A. and Hsu, R.Y. 1980. Equilibrium substrate binding studies of the malic enzyme of pigeon liver. Equivalence of nucleotide sites and anticooperativity associated with the binding of L-malate to the enzyme-manganese(II)-reduced nicotinamide adenine dinucleotide phosphate ternary complex. Biochemistry 19: 951–962. [DOI] [PubMed] [Google Scholar]
- Rao, S.R., Kamath, B.G., and Bhagwat, A.S. 1991. Chemical modification of the functional arginine residue(s) of malic enzyme from Zea mays. Phytochemistry 30: 431–435. [Google Scholar]
- Satterlee, J. and Hsu, R.Y. 1991. Duck liver malic enzyme: Sequence of a tryptic peptide containing the cysteine residue labeled by the substrate analog bromopyruvate. Biochim. Biophys. Acta 1079: 247–252. [DOI] [PubMed] [Google Scholar]
- Scrutton, N.S., Berry, A., and Perham, R.N. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343: 38–43. [DOI] [PubMed] [Google Scholar]
- Tong, L. 1996. Combined molecular replacement. Acta Crystallogr. A52: 782–784. [Google Scholar]
- Tsai, L.-C., Kuo, C.-C., Chou, W.-Y., Chang, G.-G., and Yuan, H.S. 1999. Crystallization and preliminary X-ray diffraction analysis of malic enzyme from pigeon liver. Acta Crystallogr. D55: 1930–1932. [DOI] [PubMed] [Google Scholar]
- Wei, C.-H., Chou, W.-Y., and Chang, G.-G. 1995. Identification of Asp258 as the metal coordinate of pigeon liver malic enzyme by site-specific mutagenesis. Biochemistry 34: 7949–7954. [DOI] [PubMed] [Google Scholar]
- Wei, C.-H., Chou, W.-Y., Huang, S.-M., Lin, C.-C., and Chang, G.-G. 1994. Affinity cleavage at the putative metal-binding site of pigeon liver malic enzyme by the Fe2+-ascorbate system. Biochemistry 33: 7931–7936. [DOI] [PubMed] [Google Scholar]
- Wierenga, R.K., Terpstra, P., and Hol, W.G.J. 1986. Prediction of the occurrence of the ADP-binding bab-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187: 101–107. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Bhargava, G., Wu, H., Loeber, G., and Tong, L. 1999. Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: A new class of oxidative decarboxylases. Structure 7: 877–889. [PubMed] [Google Scholar]
- Yang, Z., Floyd, D.L., Loeber, G., and Tong, L. 2000. Structure of a closed form of human malic enzyme and implications for catalytic mechanism. Nat. Struct. Biol. 7: 251–257. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Tong, L. 2000. Structural studies of a human malic enzyme. Protein Pept. Lett. 7: 287–296. [Google Scholar]