Abstract
A new K+-channel blocking peptide identified from the scorpion venom of Tityus cambridgei (Tc1) is composed of 23 amino acid residues linked with three disulfide bridges. Tc1 is the shortest known toxin from scorpion venom that recognizes the Shaker B K+ channels and the voltage-dependent K+ channels in the brain. Synthetic Tc1 was produced using solid-phase synthesis, and its activity was found to be the same as that of native Tc1. The pairings of three disulfide bridges in the synthetic Tc1 were identified by NMR experiments. The NMR solution structures of Tc1 were determined by simulated annealing and energy-minimization calculations using the X-PLOR program. The results showed that Tc1 contains an α-helix and a 310-helix at N-terminal Gly4–Lys10 and a double-stranded β-sheet at Gly13–Ile16 and Arg19–Tyr23, with a type I` β-turn at Asn17–Gly18. Superposition of each structure with the best structure yielded an average root mean square deviation of 0.26 ± 0.05 Å for the backbone atoms and of 1.40 ± 0.23 Å for heavy atoms in residues 2 to 23. The three-dimensional structure of Tc1 was compared with two structurally and functionally related scorpion toxins, charybdotoxin (ChTx) and noxiustoxin (NTx). We concluded that the C-terminal structure is the most important region for the blocking activity of voltage-gated (Kv-type) channels for scorpion K+-channel blockers. We also found that some of the residues in the larger scorpion K+-channel blockers (31 to 40 amino acids) are not involved in K+-channel blocking activity.
Keywords: Scorpion venom, α-KTx, K+-channel blocker, NMR, structure
Ion channels are involved in diverse biological processes and play essential roles in the physiology of all cells. An increasing number of human and animal diseases have been identified as relating to the defective function of ion channels. Scorpion venoms contain various polypeptides with distinct biological functions that particularly affect the permeability of ion channels in cell membranes (Catterall 1980; Valdivia et al. 1992; Garcia et al. 1997). These polypeptides possess the potency to recognize ion channels and receptors in excitable membranes and are classified into four groups on the basis of ion-channel types: (1) group I modulates Na+-channel activity (Possani et al. 1999) and contains peptides of 60 to 70 amino acids linked by four disulfide bridges; (2) group II blocks K+ channels (Miller 1995; Romi-Lebrun et al. 1997) and are short peptides with 31 to 41 amino acid residues with three or four disulfide bonds; (3) group III supposedly inhibits Cl−channels (DeBin et al. 1993) and contains short-chain insect toxin peptides of ∼36 amino acids with four disulfide bonds; and (4) group IV includes peptides that modulate ryanodine-sensitive Ca2+ channels (Valdivia and Possani 1998). It is believed that the toxin has a unique tertiary structure that may provide valuable information for understanding channels. Thus, understanding the structural basis of the specificity of scorpion toxins for these receptors could lead to the design of new ligands with controlled activity and potency with potential for clinical applications.
Scorpion K+-channel blockers of group II, named α-KTx, have been classified into 12 subfamilies (Miller 1995; Tytgat et al. 1999). These K+-channel blockers block two major classes of K+ channels: voltage-gated (Kv-type) and high-conductance Ca2+-activated (BK-type) K+ channels. The three-dimensional structures of several scorpion K+-channel blockers have been determined by NMR spectroscopy; these include charybdotoxin (ChTx; Bontems et al. 1991), iberiotoxin (IbTx; Johnson et al. 1992), noxiustoxin (NTx; Dauplais et al. 1995), PO5-NH2 (Meunier et al. 1993), kaliotoxin (KTx; Fernandez et al. 1994), margatoxin (MgTx; Johnson et al. 1994), and tityustoxin K-α (TsTx-Kα; Ellis et al. 2001). Although the overall fold of these α-KTx toxins is very similar, there are subtle variations among them in amino acid sequence, the size of the β-sheet, the type of β-turn, or the type of α-helix (i.e., α-helix versus 310-helix). These differences in toxin structure affect the placement of side-chain moieties. Thus, the selectivity that various scorpion toxins have for the outer vestibule of different K+ channels is typically quite distinct. Previously, Doyle et al. (1998) applied X-ray crystallographic methods to determine the three-dimensional structure of the KcsA bacterial K+ channel, which may serve as a good model for understanding the binding site of scorpion toxin on Kv-type channels.
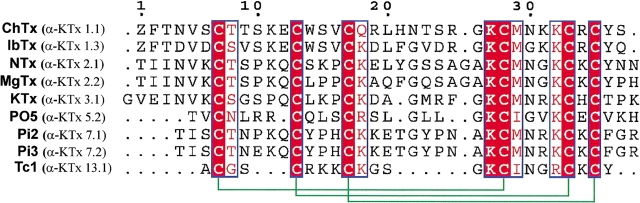
Recently, a new K+-channel blocker was identified from the scorpion venom of Tityus cambridgei (Tc1; Batista et al. 2000). Tc1 contains 23 amino acids linked with three disulfide bridges and is the smallest K+-channel blocker toxin from scorpion venoms. All previously known K+-channel blockers from scorpion venoms are longer than 30 amino acid residues and are classified into 12 subfamilies as described above. Tc1 is classified as the first member of the new subfamily 13. In K+-channel blocking activity, Tc1 recognizes the Shaker B K+ channels with a dissociation constant (Kd) of 65 nM and competes with NTx for binding to the synaptosomal membranes, with an inhibitory concentration 50% (IC50) value in the order of 200 nM (Batista et al. 2000). Tc1 is a highly basic peptide because it contains seven positively charged residues with a pI value of 9.50. The sequence alignment of Tc1 with eight other K+-channel blockers from scorpion toxins is shown in Figure 1 ▶. We found that six cysteine residues (Cys2, Cys5, Cys9, Cys15, Cys20, and Cys22), Gly13, and Lys14 (Tc1 numbering) are conserved, and the C-terminal regions are highly similar among these toxins. In addition, the sequence of Tc1 shows some unique properties. For example, Tc1 possesses Arg at position 19, whereas the corresponding residue in the other toxins is Lys. At position 16, Tc1 has Ile, whereas the other toxins, with the exception of the PO5 peptide, have Met at the corresponding position. Furthermore, Tc1 contains dense positively charged residues at residues 5–10. Unlike other scorpion toxins, Tc1 does not contain either negatively charged residues or proline. These properties make Tc1 an excellent candidate for three-dimensional structure determination and site-directed mutagenesis and for gaining clearer understanding of K+ channels.
Fig. 1.
Sequence alignment of Tc1 with eight other K+-channel blockers was generated using the CLUSTAL-W (Thompson et al. 1994) and ESPript (Gouet et al. 1999) programs. Three-dimensional solution structures for six of them have been determined: They are ChTx (Bontems et al. 1991), IbTx (Johnson and Sugg 1992), NTx (Dauplais et al. 1995), MgTx (Johnson et al. 1994), KTx (Fernandez et al. 1994), and PO5 (Meunier et al. 1993). Residues that are identical are boxed in red; those that are highly conserved are shown in red. The sequence number is based on the sequence of ChTx (top), and the α-KTx name of each peptide (in parentheses) is shown. The three internal disulfide-bridges are indicated with green brackets (bottom).
In this study, synthetic Tc1 was made by conventional solid-phase peptide synthesis and folded into its active conformation. We checked the channel-blocking activity of the synthesized Tc1 and found that both synthetic and native Tc1 possess similar blocking activity against the Shaker K+ channel. Next, we applied circular dichrosim (CD) and NMR techniques to solve the solution structure of Tc1. To further understand the various structure-function relationships among the K+-channel blockers from scorpion venoms, we compared the three-dimensional structure of Tc1 and those of other structurally and functionally related scorpion toxins, ChTx and NTx. We concluded that the C-terminal structure is the most important region for the blocking activity of Kv-type channels for scorpion K+-channel blockers. In addition, we also asserted that some of the residues in the larger scorpion K+-channel blockers, which contain 31 to 40 amino acids, are clearly not involved in K+-channel blocking activity.
Results
Synthetic Tc1 is as potent as the natural scorpion toxin Tc1
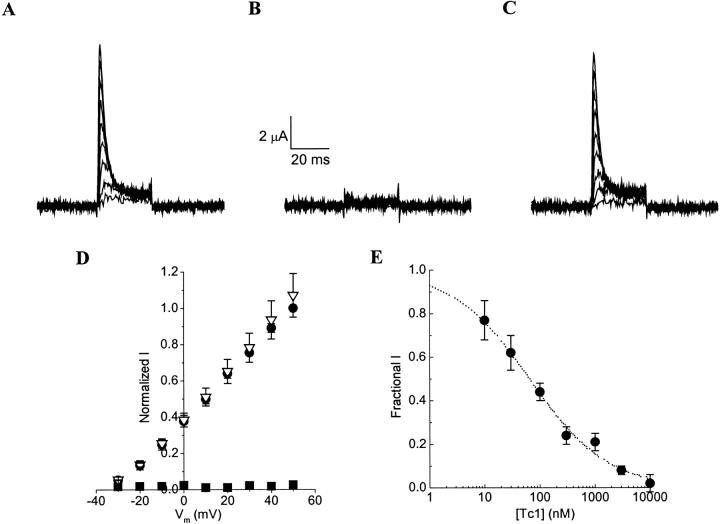
We first examined the effect of the synthetic Tc1 on Shaker GH4 K+ channels expressed in Xenopus ooctyes. Figure 2 ▶ showed that 10 μM of synthetic Tc1 completely blocked outward currents (Fig. 2A,B ▶). The inhibition was reversible on washout (Fig. 2C ▶), but the block was not voltage dependent (Fig. 2D ▶). Figure 2E ▶ shows the dose-response curve for the block of the peak current by synthetic Tc1 at +50 mV. The peak current amplitude in various concentrations of synthetic Tc1 was normalized to that of the control solution and expressed as fractional I. Kd for the synthetic Tc1 block was 65 nM, which is the same as the previously reported value for the scorpion toxin Tc1 block of Shaker B K+ channels transfected in Sf9 cells (Batista et al. 2000). These results indicate that the functional property of the synthetic Tc1 is the same as the natural scorpion toxin Tc1.
Fig. 2.
Block of Shaker GH4 K+ channels by synthetic Tc1. (A) Whole-cell currents through Shaker GH4 K+ channels were recorded in a control solution. (B) Currents were completely eliminated after the addition of 10 μM synthetic Tc1 to the control solution. (C) Currents completely recovered from synthetic Tc1 block after washout in the control solution. (D) Normalized peak current-voltage relationships (n = 4) obtained in control (circles), 10 μM synthetic Tc1 (squares), and washout (triangles). All peak currents were normalized to that recorded at +50 mV in the control solution. (E) Dose-response curves for synthetic Tc1 block of the peak Shaker GH4 current at +50 mV. Dotted line is the best fit of the data (n = 3 to 4) to the Hill equation: 1/{1+[(Tc1)/Kd]h}. Kd = 65 nM is the apparent dissociation constant, and h = 0.6 is the Hill coefficient.
Synthetic Tc1 is a stable K+-channel blocker
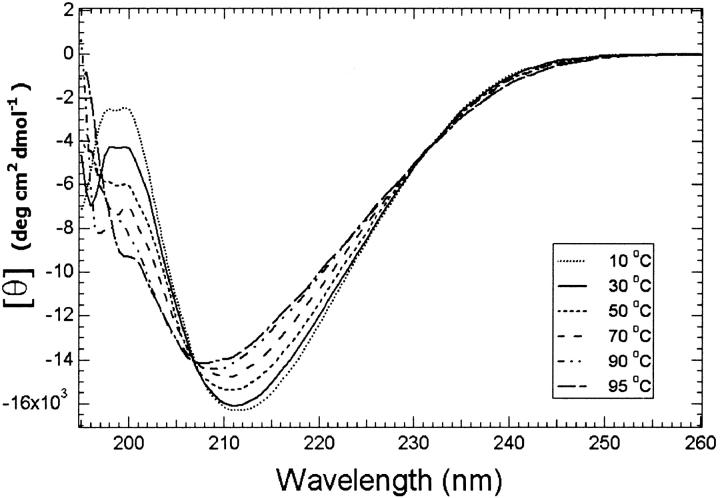
We performed CD experiments of Tc1 at different pH values (3.5 to 7.0) and at various temperatures (0°C to 95°C). The CD spectra were very alike at different pH values, indicating that Tc1 possesses similar conformations at pH ranges of 3.5 to 7.0. The CD spectra at different temperatures (Fig. 3 ▶) displayed a blue-shift of the negative band when the temperature was increased (212 nm at 10°C to 208 nm at 95°C), with an isodichroic point at 207 nm. This indicates that there is a conformational equilibrium for Tc1 at various temperatures. The majority of the secondary structure remained intact even at 95°C, revealing that Tc1 is a thermostable peptide. The contents of the secondary structures of Tc1 estimated using CONTIN-LL, SELCON3, and CDSSTR (Sreerama and Woody 2000) are listed in Table 1.
Fig. 3.
CD spectra of 20 μM Tc1 in 20 mM phosphate buffer at pH 3.0 are shown as a function of temperature. For the sake of clarity, only some of the spectra are shown.
Table 1.
Secondary structure contents of Tc1 estimated from CD spectra acquired at pH 3.0 using three different methods a
| Methods | Temp. | αR | αD | βR | βD | T | U |
| CONTINLL | 25°C | 0.028 | 0.077 | 0.164 | 0.101 | 0.243 | 0.387 |
| 95°C | 0.018 | 0.060 | 0.128 | 0.095 | 0.255 | 0.444 | |
| SELCON3 | 25°C | 0.039 | 0.086 | 0.176 | 0.107 | 0.234 | 0.358 |
| 95°C | 0.030 | 0.079 | 0.182 | 0.106 | 0.229 | 0.374 | |
| CDSSTR | 25°C | 0.013 | 0.082 | 0.209 | 0.111 | 0.242 | 0.343 |
| 95°C | 0.002 | 0.083 | 0.200 | 0.105 | 0.244 | 0.366 |
a (αR) Regular α-helix; (αD) distorted α-helix; (βR) regular β-strand; (βD) distorted β-strand; (T) turns; (U) unordered.
Resonance assignments and secondary structure determination of Tc1
With thermostability and well-dispersed NMR data, Tc1 is an excellent compound for NMR structural studies. Based on the NMR data acquired at different pH values and temperatures, the resonance assignments were accomplished using the standard procedures (Wüthrick 1986). Spin systems were identified based on scalar-correlated spectroscopy (COSY) and total correlation spectroscopy (TOCSY) experiments, and sequential connectivities were obtained from nuclear Overhauser enhancement (NOE) spectroscopy experiments. To obtain as many NOEs as possible, we tested many conditions and finally chose pH 3.0 and pH 7.3 at 275 K for performing our NMR experiments. The representative regions of a NOE spectroscopy spectrum, with partial annotations, are shown in Figure 4 ▶. Interestingly, the amide proton of Cys5 was not observed, presumably because of the fast exchange with water. The amide proton of Arg6 showed a sharper line width compared with that of the other amide protons. These observations indicate that the conformation in the Cys5–Arg6 dipeptide is flexible, and the amide protons of both Cys5 and Arg6 are highly exposed to the solvent. At pH 7.3, the amide protons in the N-terminal Cys2–Cys5 were not detected, suggesting that the N-terminal segment is more flexible than the C terminus. The observed NOE patterns at pH 7.3 were very similar to those at pH 3.0, further supporting the similarity of the structure at both acidic and neutral pH values. Figure 5A ▶ shows the summary of the NMR parameters for Tc1 at pH 3.0. The CαH chemical shift index (CSI; Wishart et al. 1992) indicates that Cys5–Lys10 forms an α-helical structure and that Gly13–Ile16 and Arg19–Tyr23 show β-strand conformations. Based on the α-helical NOEs, we identified an α-helix at the N-terminal Ser4–Lys10, which is in good agreement with CSI results. In Gly11, we did not observe the α-helical NOE of dαN(8, 11), although a medium range NOE of dαN(7, 11) was detected. This indicated that Gly11 likely formed a turn structure. According to the observed long-range NOEs between the two β-strands, among which dαN(15, 21) and dαα(15, 20) could not be accurately assigned because of chemical shift degeneracy at CαH of Cys15 and Cys20, and the deduced hydrogen bonds that were consistent with the β-sheet structure (Fig. 5B ▶), we found that the two β-strands actually formed a double-stranded antiparallel β-sheet, with a β-turn conformation at residues Asn17–Gly18. The observation of medium intensity of the dαN(17, 19) NOE, along with backbone φ, Ψ angles of residues Asn17 and Gly18, calculated based on the derived NMR structures described in the next paragraph, revealed that the Ile16–Arg19 segment is a type I` β-turn. In addition, based on the dββ(i, j) NOEs between the CβH protons of the two cystines forming a disulfide bridge, the connections of three disulfide bridges of the synthetic Tc1 were identified. We observed 17 amide protons possessing medium- or slow- exchange rates at pH 3.0, as shown in Figure 5A ▶. In contrast, only six amide protons at the C terminus (Lys14, Ile16, Gly18, Arg19, Lys21, and Tyr23) showed medium- and slow-exchange rates at pH 7.3. The exchange rate study at pH 3.0 and pH 7.3 proved that the C-terminal β-sheet possesses higher stability than the N-terminal α-helix.
Fig. 4.
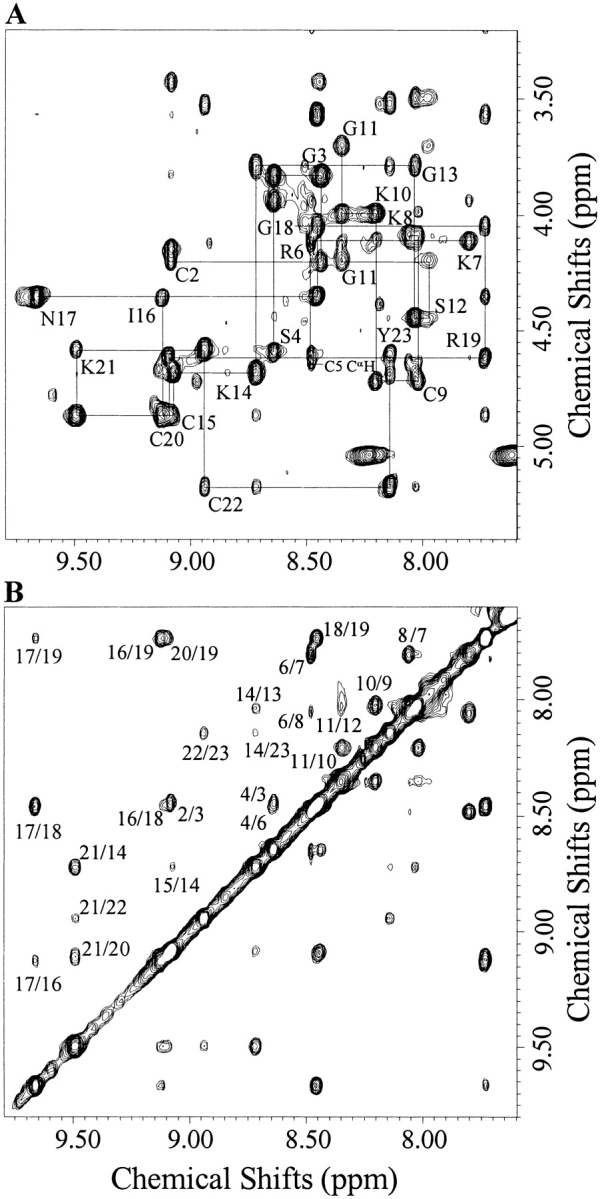
(A) The fingerprint region of a NOE spectroscopy spectrum of Tc1 acquired at 275 K and pH 3.0, with a mixing time of 400 ms. Resonance assignments are indicated with one-letter amino acid codes and residue numbers. (B) The NH/NH region of the same experiment in A. The assignment of each cross-peak is labeled with residue numbers.
Fig. 5.
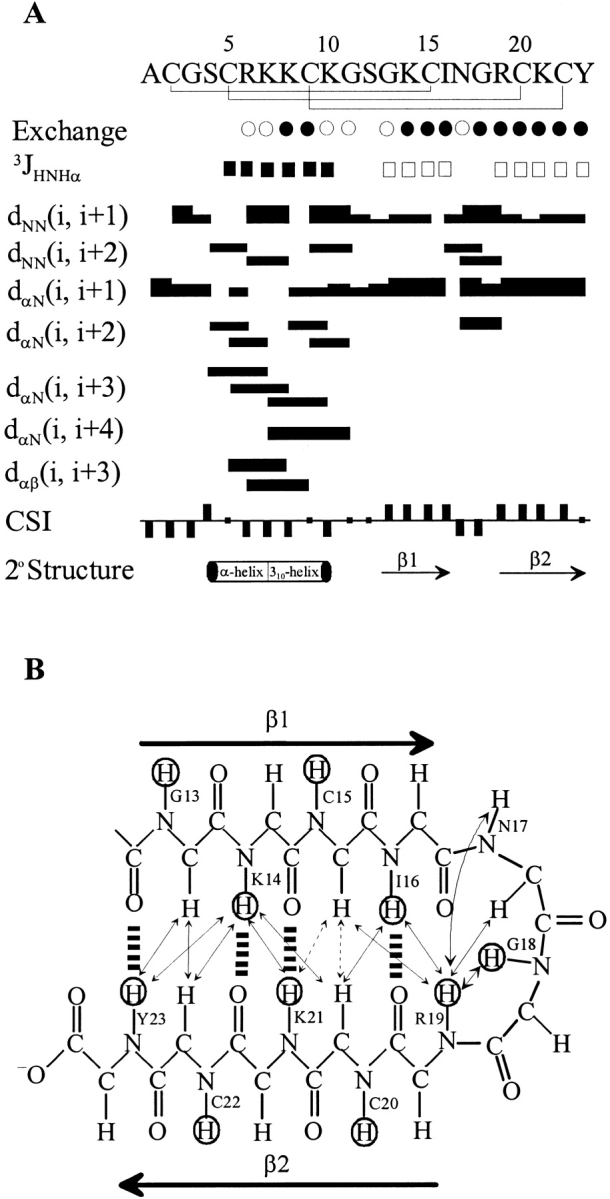
(A) Summary of the amide proton exchange rates, 3JNHα-coupling constants, NOE connectivities, CαH chemical shift index, and the derived secondary structures. Medium-exchange (<3.5 h; open circles) and slow-exchange (>24 h; filled circles) amide protons. 3JNHα coupling constants are <6 Hz (filled squares) and >8 Hz (open squares). Bar thickness indicates the intensity of NOE connectivity, with thicker bars representing stronger NOEs. Negative bars in the chemical shift index indicate upfield shifts of >0.1 ppm of the CαH proton compared with the expected random-coil CαH proton chemical shift. Positive bars indicate downfield shifts of >0.1 ppm of the CαH proton compared with the expected random-coil CαH value. The derived secondary structures based on NMR parameters as described above are shown (bottom). (B) Definition of the β-sheet structure of Tc1 is shown based on the NOEs and amide proton exchange rate. Cross-over NOEs between β-strands are shown in double arrows. Ambiguous cross-over NOEs, because of resonance overlap, are indicated in double arrows with a dashed line. Dashed lines between backbone amide protons and backbone carbonyl oxygens indicate hydrogen bonds consistent with slow-exchanging HN observed in D2O. The amide protons with slow exchange rates are circled.
Three-dimensional solution structure of Tc1
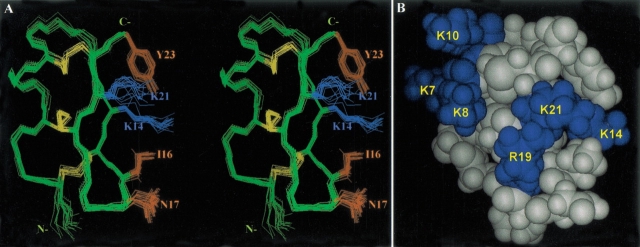
A set of 200 restraints was applied for simulated annealing and energy minimization calculations using the program X-PLOR. Fifteen structures were chosen to represent the ensemble of NMR structures on the basis of the lowest target function and minimal distance and torsional angle restraint violations in the final stage. All of these structures were consistent with both experimental data and standard covalent geometry, and they displayed no violations >0.5 Å for distance restraints. The structure with the lowest final total energy and root mean square deviation (RMSD) of distance restraints was defined as the best structure. Superposition of each structure with the best structure yielded average RMSD of 0.26 ± 0.05 Å for the backbone atoms and 1.40 ± 0.23 Å for heavy atoms in residues 2–23, respectively (Fig. 6A ▶). The structural statistics on the final set of structures are given in Table 2. Analysis of the ensemble of 15 structures using PROCHECK-NMR revealed that 98.8% of the residues lie in the most favored and allowed regions of the Ramachandran φ, Ψ dihedral-angle plot (plot not shown). The distribution of φ and Ψ backbone dihedral angles showed that only three backbone dihedral angles (φ of Cys2, Ψ of Gly11, and φ of Ser12) displayed large deviations, revealing that the backbone conformation is rigid except for the N-terminal Cys2 and the loop region of Gly11 and Ser12. Interestingly, the β-turn of the Asn17–Gly18 dipeptide also contains very stable φ and Ψ dihedral angles. Further, the dihedral angles of three disulfide bonds were found to be rigid, with average angles of −146.10 ± 3.92 (Cys2–Cys15), −107.71 ± 2.27 (Cys5–Cys20), and 115.47 ± 4.18 (Cys9–Cys22). In contrast, the side-chain χ1 dihedral angles in arginines and lysines, with the exception of Lys14, all showed very large deviations. All hydrogen bonds in these 15 structures were located in the N-terminal α-helix, β-sheet, and β-turn regions. In the N-terminal α-helix, we consistently observed a hydrogen bond of Lys7 CO/Lys10 NH, indicating that Lys7–Lys10 forms a 310-helix. In the β-turn region (Ile16–Arg19), as expected, the hydrogen bond of Ile16 CO/Arg19 NH was detected in all the NMR structures. Tc1 is a basic protein with two arginines and five lysine positively charged residues. At neutral pH, it carries an overall positive charge, and the distribution of the charges is clearly distributed into two regions, as shown in the space-filling structure of Tc1 (Fig. 6B ▶).
Fig. 6.
(A) Stereo view of the superimposition of the heavy atoms of 15 NMR structures obtained from simulated annealing and energy minimization calculations. The structures are best fitted to residues 2–23. Side-chains of the five residues (Lys14, Ile16, Asn17, Lys21, and Tyr23), which correspond to the residues for the recognition of a voltage-dependent K+ channel (Shaker B) in ChTx, are shown and labeled. (B) Space-filling model of the best structure of Tc1. The structure is colored according to the charge of the amino acid residues, with positive residues in blue and neutral residues in gray. Two positively charged regions are clearly shown, and the charged residues are labeled in the structure.
Table 2.
Structural statistics on the final set of 15 simulated annealing structures of Tc1
| A. Constraints used | |
| Distance restraints | |
| Intraresidue (|i − j| = 0) | 17 |
| Sequential (|i − j| = 1) | 67 |
| Medium range (|i − j| = 2), (|i − j| = 3) | 34 |
| Long range (|i − j| ≥ 4) | 45 |
| Total distance restraints | 163 |
| Hydrogen bonds | 8 × 2 |
| Dihedral angles | 21 |
| B. Statistics for the final X-PLOR structures | |
| Number of structures in the final set | 15 |
| X-PLOR energy (kcal/mole) | |
| ENOE | 5.40 ± 0.43 |
| Ecdih | 2.02 ± 0.24 |
| Ebond + Eangle + Eimproper | 36.67 ± 1.20 |
| EVDW | 34.39 ± 0.86 |
| NOE violations | |
| Number > 0.5 Å | none |
| Rms deviation (Å) | 0.032 |
| Deviation from idealized covalent geometry | |
| Angle (°) | 0.52 ± 0.01 |
| Impropers (°) | 0.42 ± 0.02 |
| Bonds (Å) | 0.005 |
| Mean global rms deviation (Å) | |
| Backbone (N, Ca, C`) | |
| Residues (2–23) | 0.26 ± 0.05 |
| Heavy atoms | |
| Residues (2–23) | 1.40 ± 0.23 |
| Ramachandran data (%) | |
| Residues in most favored regions | 72.1 |
| Residues in allowed regions | 26.7 |
| Residues in generously allowed regions | 1.2 |
| Residues in disallowed regions | 0.0 |
Discussion
Our studies on the effects of synthetic Tc1 on Shaker GH4 K+ channels indicated that its functional property is the same as the natural scorpion toxin Tc1. We then used CD and NMR techniques to perform our structural study of the synthetic Tc1. CD spectra at different temperatures and pH values showed that Tc1 is a thermostable peptide with a conformation that is independent of pH values in the range of 3.0 to 7.0. The three-dimensional NMR solution structure of Tc1 showed that it is comprised of an α-helix and a 310-helix at N-terminal Gly4–Lys10, a double-stranded antiparallel β-sheet at Gly13–Ile16 and Arg19–Tyr23, with a type I` β-turn at residues Asn17–Gly18. Because the NMR data obtained at pH 3.0 and 275 K were well resolved and showed many medium- and long-range NOEs, high resolutions of Tc1 structures were generated. We found that the overall structures of Tc1 and other α-KTx toxins are similar, although Tc1 only possesses 23 amino acids compared with >30 for the other scorpion toxins.
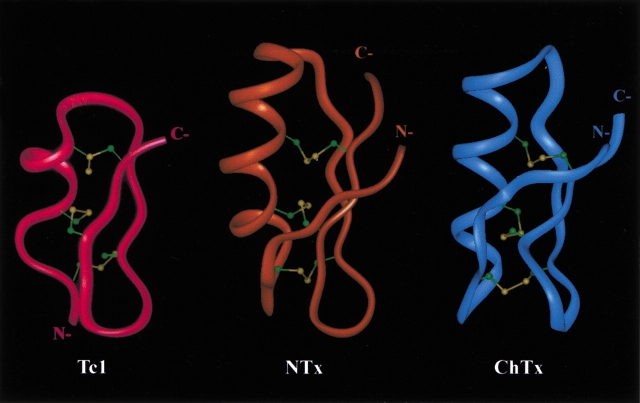
To gain further insight into the structural and functional relationships among the K+-channel blockers from scorpion venoms, we proceeded to a detailed comparison of the three-dimensional structure of Tc1 with two structurally and functionally related scorpion toxins, ChTx (37 amino acids) and NTx (39 amino acids). ChTx is the first member of subfamily 1 of α-KTx and shows a much higher affinity for the Ca2+-activated K+ channels (BK) than for Kv1.3. In contrast, NTx is the first K+-channel blocker isolated from scorpion venoms and displays a strong binding affinity for Kv1.3, whereas it exerts a weaker affinity for BK. Tc1 also has a higher binding affinity for the Shaker B channel than for BK, similar to NTx. In the secondary structure motifs, we found that the N-terminal α-helix, the two C-terminal β-strands, and the β-turn are all located in similar regions based on the sequence alignment of these toxins. However, some variations were observed in the type or length of the secondary elements. For example, Tc1 has a shorter helical conformation, and this helix begins with a regular α-helix and ends with a 310-helix, whereas the helix in NTx begins with a 310-helix and ends with a regular α-helix. Also, because of the presence of a Pro residue in the α-helical region, the α-helix in NTx displays a high degree of curvature. The bending of the α-helix in both Tc1 and ChTx, however, is weak because of the lack of a proline residue in the sequence. Tc1 and NTx have the same type I` β-turn at Asn17–Gly18 and Asn31–Gly32, respectively, whereas ChTx possesses a type I β-turn at Asn30–Lys31. NTx not only contains an extra N-terminal β-strand but also possess longer C-terminal β-strands. Thus, there is a remarkable plasticity within the α/β-scaffold for the α-KTx toxins.
The specificity of scorpion toxins for the various potassium channels has been investigated through the generation of mutants of both receptors and toxins. Mutational analysis of ChTx showed that eight residues (Ser10, Trp14, Arg25, Lys27, Met29, Asn30, Arg34, and Tyr36) are important for the binding of ChTx to BK (Stampe et al. 1994). Five of these eight residues (Lys27, Met29, Asn30, Arg34, and Tyr36) were shown to be critical for the recognition of a voltage-dependent K+ channel (Shaker B) in ChTx (Goldstein et al. 1994). Among these residues, Lys27 is the most important because a mutation of this lysine to arginine destabilizes the toxin by >1000-fold (Miller 1995). The inhibition of the channel permeation is the result of a physical occlusion of the pore-forming region of the channel. Thus, the Lys27 of ChTx is suspected to directly plug into the pore.
The characteristics of five corresponding residues for the recognition of the Shaker B channel in Tc1 (Lys14, Ile16, Asn17, Lys21, and Tyr23) are similar to those in ChTx. However, the properties of other three residues (Ser10, Trp14, and Arg25), which are also important for the binding of ChTx to BK-type channels, were found to be very different in Tc1. The corresponding residues in Tc1 for the first two are Ser4 and Arg6, respectively, and there is no residue occupied for the third. Thus, the different property in these residues offers a possible explanation for the weak affinity of Tc1 to BK-type channels. For the recognition of the Shaker B channel in Tc1, we found that the side-chains of these residues were all exposed to the solvent on the same side (see Fig. 6A ▶). Interestingly, Lys14, which corresponds to Lys27 in ChTx, showed a rigid side-chain conformation and highly protruded into the solvent. Thus, we suggest that Lys14 in Tc1 is the key residue to have electronic interaction with the negative charge in the pore region of the K+ channel.
In addition, IbTx was found to be inactive against the Kv1.3 channel. A sequence comparison between ChTx and IbTx indicates that Asn30 (ChTx numbering) is replaced with Gly in IbTx. Therefore, Asn30 appears to be important for the two types of voltage-dependent channels; in fact, this residue can be found in all the short-chain scorpion toxins that bind Kv1.3 (ChTx, NTx, MgTx, KTx, and Tc1 in Fig. 1 ▶). Two scorpion toxins in subfamily 7, Pi2 and Pi3, have only one amino acid difference at position 7 (a proline for a glutamic acid) in their sequence. However, Pi2 binds the Shaker B K+ channels with a Kd of 8.2 nM, but Pi3 has a much lower affinity of 140 nM (Gomez-Lagunas et al. 1996). The difference in binding affinity supports that the N-terminal residues are part of the domain that recognize Shaker B K+ channels. Interestingly, there is no residue occupied at the corresponding position in Tc1, which has a Kd of 65 nM. Therefore, it is certain that the negative charge at this position disrupts the inhibition of the Shaker B K+ channel.
The surface structures of Tc1, NTx, and ChTx (plot not shown) all indicated that there is a positively charged region at the C terminus and that this region plays an important role for blocking activity. At the N terminus, Tc1 contains a denser positively charged region composed by Arg6, Lys7, Lys8, and Lys10 (Fig. 6B ▶) compared with NTx and ChTx. At present, we do not know whether this region interacts with the K+-channel or whether it plays an important role for activity. We are currently performing mutational studies on Tc1 to further understand its structural-functional relationships.
In addition, Figure 7 ▶ shows the comparison of the ribbon structures of Tc1, NTx, and ChTx. Superposition of backbone atoms (N, Cα, and C`) between Gly13and Tyr23 of Tc1 and between Ala27 and Tyr37 of NTx gave a RMSD of 0.76 Å. However, the RMSD became 0.96 Å between Gly13–Tyr23 of Tc1 and Gly26–Tyr36 of ChTx. Thus, Tc1 is much like NTx, especially at the C-terminal β-sheet and β-turn. Because both Tc1 and NTx contain higher activity for the Shaker B channel, we concluded that the C-terminal structure is the most important region for controling the blocking activity of the Kv-type for scorpion K+-channel blockers. Furthermore, based on the structural data and sequence alignment between Tc1 and NTx, we suggest that for NTx the N-terminal region—including the first β-strand (Thr1-Val5), some residues in the α-helix (Lys11, Gln12, Glu19, and Leu20), some residues in the loop region (Tyr21, Ser23, and Ala25), and some residues in the C-terminal region (Ala27, Asn38, and Asn39)—might not be required for channel-blocking activity. Therefore, we are also in the process of studying the structural and functional relationships of a 24-residue peptide that involves the deletion of the above 15 residues from NTx.
Fig. 7.
Comparison of the ribbon structure of Tc1 and two structurally and functionally related scorpion toxins, NTx and ChTx. Three disulfide bridges are shown as balls and sticks. NTx and ChTx coordinates were obtained from the Protein Data Bank with accession Nos. 1SXM and 2CRD, respectively.
Materials and methods
Chemical synthesis of Tc1
Tc1 was synthesized by solid-phase peptide synthesis using a 433A peptide synthesizer (ABI). Starting with 0.25 mmole (0.238 g) of HMP (p-hydroxymethyl phenoxymethyl polystyrene) resin (1.00 mmole/g), the synthesis was performed using a stepwise Fast Moc protocol. The amino acids were introduced using the manufacturer's prepacked cartridges (1 mmole each). After synthesis, 0.386 g peptide resin was placed in a round-bottom flask containing a micro stirringbar. The cool mixture containing 0.75 g crystalline phenol, 0.25 mL EDT, 0.5 mL thioanisole, 0.5 mL water, and 10 mL TFA was put into the flask and stirred for 2 to 3 h at room temperature. The peptide was further lyophilized. Then the peptide was rinsed with 200 mL cold ether and filtered. The filtered peptide was immediately transferred to 2.9 L of diluted acetic acid solution (5% AcOH in water) and stirred for several minutes. After filtration to remove the resin, the pH of the solution was adjusted to pH 8.6 with ammonium hydroxide. Analytical high performance liquid chromatography (HPLC) was used to monitor the progress of oxidation. The peptide was purified by HPLC using a C18 column (10-μm particle size, 250 × 4.6 mm) with a gradient (5% to 95% buffer B in 30 min) using buffer A (0.1% TFA in water) and buffer B (0.1% TFA in acetonitrile) at a flow rate of 1 mL/min and monitored by absorbance at 214 nm. Mass spectra were determined using a Finnigan LCQ Mass Spectrometer with an electrospray ion source. Unless otherwise stated, all reagents and solvents were obtained commercially as reagent grade and used without further purification.
Whole-cell current recording
Purification of Shaker GH4 K+ channel cDNA and in vitro T7 transcription reactions (mMessage mMachine, Ambion) were performed as previously described (Shieh et al. 1998). Xenopus oocytes were isolated by partial ovariectomy from frogs anaesthetized with 0.1% tricaine (3-aminobenzoic acid ethyl ester). Oocytes were pressure injected with RNA 24 h after defolliculation and used 1 to 3 d after RNA injection. Oocytes were maintained at 18°C in Barth's solution containing (in mM) NaCl (88), KCl (1), NaHCO3 (2.4), Ca(NO3)2 (0.3), CaCl2 (0.41), MgSO4 (0.82), HEPES (15), and gentamicin (20 μg/mL) at pH 7.6.
The block of whole-cell Shaker GH4 K+ channels by synthetic Tc1 was examined at room temperature (21°C to 24°C) using a two-electrode voltage-clamp amplifier (Ca-1 clamp; Dagan). Oocytes were bathed in a control solution containing (in mM) KCl (3), NMG (100), CaCl2 (1), and HEPES (5) at pH 7.4. Both the voltage-sensing and current-injecting electrodes were filled with 3 M KCl (resistance, 0.3 to 1 MΩ). Voltage steps were applied from a holding potential of −90 mV to various test voltages (30 msec) ranging from −30 to +50 mV in 10-mV increments. The command Vm and data acquisition functions were processed using a Pentium computer, a DigiData board, and pClamp6 software (Axon Instruments). The frequency of stimulation was 0.5 Hz, and the sampling rate was 10 kHz. Data were filtered at 2 kHz by an eight-pole low-pass filter (Frequency Devices). Leak current and capacity transients were corrected with a P/4 voltage protocol.
CD experiments
CD experiments were performed using an Aviv CD 202 spectrometer (AVIV) calibrated with (+)-10-camphorsulfonic acid (CSA) at 298 K. In general, a 2-mm pathlength cuvette with 20 μM Tc1 peptide in 20 mM phosphate was used for CD experiments and all the protein solutions were made up to 1 mL. The CD spectra of Tc1 at different temperatures and pH values were recorded. Each piece of the CD data was obtained from an average of three scans with 1-nm bandwidth. The spectra were recorded from 180 nm to 260 nm, at a scanning rate of 38 nm/min with a wavelength step of 0.5 nm and time constant of 100 msec. After background subtraction and smoothing, all the CD data were converted from CD signal (millidegree) into mean residue ellipticity (degcm2dmole−1). The secondary structure content was estimated from the CD spectra according to the methods of CONTIN, SELCON, and CDSSTR (Sreerama and Woody 2000).
NMR experiments
The NMR measurements were performed on a Bruker AMX-500 or AVANCE-600 spectrometer. Samples for NMR experiments contained 0.35 mL of 2 mM Tc1 in 50 mM phosphate buffer at pH 3.0 and 7.3. pH values were measured with a DO microelectronic pH-vision model PHB-9901 pH meter equipped with a 4-mm electrode. All reported pH values were direct readings from the pH meter without correction for isotope effect. To monitor the exchange rates of labile protons, the concentrated sample in water was lyophilized only once and re-dissolved in D2O (99.99% D), and NMR spectra were acquired immediately and thereafter at appropriate time intervals. All chemical shifts were externally referenced to the methyl resonance of 2,2-dimethyl-2-silapentane-5-sulfonate (DSS; 0 ppm). Double-quantum–filtered (DQF)-COSY (Rance et al. 1983), TOCSY (Bax and Davis 1985), and NOE spectroscopy (Kumar et al. 1980) were collected with 512 t1 increments with 2K complex data points. All spectra were recorded in time-proportioned phase sensitive (TPPI) mode (Marion and Wüthrich 1983). Low-temperature studies used a temperature-controlled stream of cooled air using a Bruker BCU refrigeration unit and a B-VT 2000 control unit. Water suppression was achieved by 1.4-sec presaturation at the water frequency, or by the gradient method (Piotto et al. 1992). All spectra were collected with 6024.1- and 7788.16-Hz spectral widths for AMX-500 and Avance-600 respectively.
The data were transferred to an SGI O2 workstation, 200-MHz R5000SC (Silicon Graphics), for all processing and further analysis using the Bruker XWINNMR and AURELIA software packages. All data sets acquired were zero-filled to equal points in both dimensions before further processing. A 60°-shifted skewed sine bell window function was applied in all NOE spectroscopy and TOCSY spectra, and a 20°- or 30°-shifted skewed sine bell function was used for all COSY spectra. To help with resolving spectral overlap, data were collected at different temperatures.
Torsional angle restraints and stereospecific assignment
The 3JNHα-coupling constants were estimated from the residual intensity of the antiphase cross-peak in DQF-COSY spectra recorded in water. φ-Torsional restraints of −130 ± 30° for 3JNHα-coupling constants >8 Hz, and −60±30° for 3JNHα-coupling constants <6 Hz were used for structure calculation. We obtained a total of 15 φ-torsional restraints located in the α-helix and β-sheet regions. The φ-torsional restraints were used for structure generations starting from the early stage when NOE correlations were also consistent. The stereospecific assignments were derived using the method of Hyberts et al. (1987). The 3Jαβ- and 3Jαβ`-coupling constants were estimated as either large or small based on (1) the intensities of the cross-peaks observed in a DQF-COSY spectrum in D2O and a TOCSY spectrum recorded in short mixing time, and (2) the relative intensities of the intraresidue CαH-CβH and NH-CβH NOE cross-peaks. The stereospecific assignment of β-methylene also allowed us to assign the χ1-torsional angle restraints to 60 ± 30°, 180 ± 30°, or −60 ± 30°. To ensure the accuracy of stereospecific assignments, we obtained 6 prochiral assignments (Cys2, Cys9, Asn17, Cys20, Cys22, and Tyr23) with certainty. We found that the stereospecific assignments agreed well with our generated structures in the early stage. Thus, in the later stage of structure generation, we also added χ1 and prochiral assignments as restraints in the structure calculation.
Hydrogen bond and disulfide restraints
The amide-proton exchange rates were identified from residual amide proton signals observed in several TOCSY spectra recorded at 275 K at pH 3.0 and 7.3, respectively. The first spectrum was recorded within 3.5 h after the lyophilized sample was redissolved in D2O. The amide proton exchange rates were categorized into three classes: fast-, medium-, and slow-exchange rates. Hydrogen-bond formation or solvent exclusion from the amide protons was assumed to account for the slow- and medium-exchange-rate amide protons. For better convergence, a number of hydrogen bonds involved in the secondary structure were included as distance restraints in the final stage of structure generation, that is, an O–N distance of between 2.5 and 3.3 Å and O–HN distances of 1.8 and 2.5 Å between NH protons and the backbone carbonyl oxygen atoms were assigned to the slow- and medium-exchanging protons, respectively, in the latter stage of structure determination. In the final stage of structure calculation, the hydrogen bonds between NHi and OCj in the β-sheet structures were included as restraints only if the β-sheet interstrand NHi/NHj, NHi/CαHj+1, and CαHi-1/CαHj1 NOE cross-peaks were observed. The disulfide bonds used in the structure calculation were Cys2 to Cys15, Cys5 to Cys20, and Cys9 to Cys22. Covalent bonds between the sulfur atoms of disulfide bridges were modeled by restraining the distances between the two sulfur atoms to 1.80 to 2.30 Å.
Tertiary structure calculations
Distance restraints of Tc1 were derived primarily from the 200- and/or 400-msec NOESY spectra recorded in aqueous solution at 275 K and pH 3.0. Comparison was made to the 100-msec NOE spectroscopy spectrum to assess possible contributions of the NOEs from spin diffusion. Peak intensities were classified as large, medium, small, and very small, corresponding to upper bound interproton distance restraints of 2.5, 3.5, 4.5, and 6.0 Å, respectively. An additional correction of 1.0 Å was added for methylene and methyl groups. The structure determination was performed using 163 distance restraints, of which 17 were intraresidue, 67 were sequential, 79 were medium- and long-range interproton distances, 16 were hydrogen bonds, 15 were φ-torsional angles, and 6 were χ1-torsion angles. All minimization and dynamical simulated annealing calculations were performed with the program X-PLOR 98 (Brünger 1998) on a SGI O2 workstation. The INSIGHT II (Molecular Simulation Inc.), MOLMOL (Koradi et al. 1996), and GRASP (Nicholls et al. 1991) programs were used to visually observe sets of structures and to calculate and make the electrostatic surface potential of the final three-dimensional models. The distributions of the backbone dihedral angles of the final converged structures were evaluated by the representation of the Ramachandran dihedral pattern, indicating the deviations from the sterically allowed (φ, Ψ) angle limits using PROCHECK-NMR (Laskowski et al. 1996) and MOLMOL.
Data bank accession numbers
The chemical shifts of Tc1 at pH 3.0 and 275 K have been deposited to BioMagResBank (BMRB) under accession No. 5082. The atomic coordinates of the 15 energy-minimized conformers used to represent the solution structure of Tc1 have been deposited in the Brookhaven data bank, together with the complete input of conformational restraints used for the structure calculation under accession No. 1JLZ.
Acknowledgments
We thank N.Y. Su for assistance with NMR experiments and C.H. Hsu for helpful suggestions on this work. We also thank Academia Sinica and the National Science Council, Taipei, Taiwan, R.O.C for support of this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Tc1, a new scorpion toxin from Tityus cambridgei
NTx, noxiustoxin
ChTx, charybdotoxin
KTx, kaliotoxin
MgTx, margatoxin
IbTx, iberiotoxin
TsTx-Kα, tityustoxin K-α
BKca, large-conductance calcium-activated potassium channel
Kv, Shaker-related voltage-gated potassium channel
CD, circular dichroism
NOE, nuclear Overhauser enhancement
DQF-COSY, double-quantum-filtered scalar-correlated spectroscopy
TOCSY, total correlation spectroscopy
RMSD, root mean square deviation
CSI, chemical shift index
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.33402.
References
- Batista, C.V.F., Gomez-Lagunas, F., Lucas, S., and Possani, L.D. 2000. Tc1, from Tityus cambridgei, is the first member of a new subfamily of scorpion toxin that blocks K+-channels. FEBS Lett. 486 117–120. [DOI] [PubMed] [Google Scholar]
- Bax, A. and Davis, D.G. 1985. MLEV-17–based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65 355–360. [Google Scholar]
- Bontems, F., Roumestand, C., Boyot, P., Gilquin, B., Doljansky, Y., Menez, A., and Toma, F. 1991. Three-dimensional structure of natural charybdotoxin in aqueous solution by 1H-NMR: Charybdotoxin possesses a structural motif found in other scorpion toxins. Eur. J. Biochem. 196 19–28. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1998. X-PLOR version 98. Yale University Press, New Haven, CT.
- Catterall, W.A. 1980. Neurotoxins that act on voltage-sensitive sodium channels in excitable membranes. Annu. Rev. Pharmacol. Toxicol. 20 15–43. [DOI] [PubMed] [Google Scholar]
- Dauplais, M., Gilquin, B., Possani, L.D., Gurrola-Briones, G., Roumestand, C., and Menez, A. 1995. Determination of the three-dimensional solution structure of noxiustoxin: Analysis of structural differences with related short-chain scorpion toxins. Biochemistry 34 16563–16573. [DOI] [PubMed] [Google Scholar]
- DeBin, J.A., Maggio, J.E., and Strichartz, G.R. 1993. Purification and characterization of chlorotoxin, a chloride channel ligand from the venom of the scorpion. Am. J. Physiol. 264 361–369. [DOI] [PubMed] [Google Scholar]
- Doyle, D.A., Morais Cabral, J., Pfuetzner, R.A., Kuo, A., Gulbis, J.M., Cohen, S.L., Chait, B.T., and MacKinnon, R. 1998. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 280 69–77. [DOI] [PubMed] [Google Scholar]
- Ellis, K.C., Tenenholz, T.C., Jerng, H., Hayhurst, M., Dudlak, C.S., Gilly, W.F., Blaustein, M.P., and Weber, D.J. 2001. Interaction of a toxin from the scorpion Tityus serrulatus with a cloned K+ channel from squid (sqKv1A). Biochemistry 40 5942–5953. [DOI] [PubMed] [Google Scholar]
- Fernandez, I., Romi, R., Szendeffy, S., Martin-Eauclaire, M.F., Rochat, H., Van Rietschoten, J., Pons, M., and Giralt, E. 1994. Kaliotoxin (1-37) shows structural differences with related potassium channel blockers. Biochemistry 33 14256–14263. [DOI] [PubMed] [Google Scholar]
- Garcia, M.L., Hanner, M., Knaus, H.G., Koch, R., Schmalhofer, W., Slaughter, R.S., and Kaczorowski, G.J. 1997. Pharmacology of potassium channels. Adv. Pharmacol. 39 425–471. [DOI] [PubMed] [Google Scholar]
- Goldstein, S.A., Pheasant, D.J., and Miller, C. 1994. The charybdotoxin receptor of a Shaker K+ channel: Peptide and channel residues mediating molecular recognition. Neuron 12 1377–1388. [DOI] [PubMed] [Google Scholar]
- Gomez-Lagunas, F., Olamendi-Portugal, T., Zamudio, F.Z., and Possani, L.D. 1996. Two novel toxins from the venom of the scorpion Pandinus imperator show that the N-terminal amino acid sequence is important for their affinities towards Shaker. J. Membr. Biol. 152 49–56. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. 1999. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15 305–308. [DOI] [PubMed] [Google Scholar]
- Hyberts, S.G., Marki, W., and Wagner, G. 1987. Stereospecific assignments of side-chain protons and characterization of torsion angles in eglin c. Eur. J. Biochem. 164 625–635. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A. and Sugg, E.E. 1992. Determination of the three-dimensional structure of iberiotoxin in solution by 1H nuclear magnetic resonance spectroscopy. Biochemistry 31 8151–8159. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A., Stevens, S.P., and Williamson, J.M. 1994. Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry 33 15061–15070. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1451–5, 29–32. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Ernst, R.R., and Wüthrich, K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95 1–6. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 8 477–486. [DOI] [PubMed] [Google Scholar]
- Marion, D. and Wüthrich, K. 1983. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113 967–974. [DOI] [PubMed] [Google Scholar]
- Meunier, S., Bernassau, J.M., Sabatier, J.M., Martin-Eauclaire, M.F., Van Rietschoten, J., Cambillau, C., and Darbon, H. 1993. Solution structure of P05-NH2, a scorpion toxin analog with high affinity for the apamin-sensitive potassium channel. Biochemistry 32 11969–11976. [DOI] [PubMed] [Google Scholar]
- Miller, C. 1995. The charybdotoxin family of K+ channel-blocking peptides. Neuron 15 5–10. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 2 661–665. [DOI] [PubMed] [Google Scholar]
- Possani, L.D., Becerril, B., Delepierre, M., and Tytgat, J. 1999. Scorpion toxins specific for Na+-channels. Eur. J. Biochem. 264 287–300. [DOI] [PubMed] [Google Scholar]
- Rance, M., Sorensen, O.W., Bodenhausen, G., Wagner, G., Ernst, R.R., and Wüthrich, K. 1983. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117 479–485. [DOI] [PubMed] [Google Scholar]
- Romi-Lebrun, R., Lebrun, B., Martin-Eauclaire, M.F., Ishiguro, M., Escoubas, P., Wu, F.Q., Hisada, M., Pongs, O., and Nakajima, T. 1997. Purification, characterization, and synthesis of three novel toxins from the Chinese scorpion Buthus martensi, which act on K+ channels. Biochemistry 36 13473–13482. [DOI] [PubMed] [Google Scholar]
- Shieh, R.C., Chang, J.C., and Arreola, J. 1998. Interaction of Ba2+ with the pores of the cloned inward rectifier K+ channels Kir2.1 expressed in Xenopus oocytes. Biophys. J. 75 2313–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama, N. and Woody, R.W. 2000. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287 252–260. [DOI] [PubMed] [Google Scholar]
- Stampe, P., Kolmakova-Partensky, L., and Miller, C. 1994. Intimations of K+ channel structure from a complete functional map of the molecular surface of charybdotoxin. Biochemistry 33 443–450. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl. Acids. Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat, J., Chandy, K.G., Garcia, M.L., Gutman, G.A., Martin-Eauclaire, M.F., van der Walt, J.J., and Possani, L.D. 1999. A unified nomenclature for short-chain peptides isolated from scorpion venoms: α-KTx molecular subfamilies. Trends Pharmacol. Sci. 20 444–447. [DOI] [PubMed] [Google Scholar]
- Valdivia, H.H. and Possani, L.D. 1998. Peptide toxins as probes of ryanodine receptor structure and function. Trends Cardiovasc. Med. 8 111–118. [DOI] [PubMed] [Google Scholar]
- Valdivia, H.H., Martin, B.M., Escobar, L., and Possani, L.D. 1992. Noxiustoxin and leiurutoxin III, two homologous peptide toxins with binding properties to synaptosomal membrane K+ channels. Biochem. Int. 27 953–962. [PubMed] [Google Scholar]
- Wishart, D.S., Sykes, B.D., and Richards, F.M. 1992. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wüthrick, K. 1986. NMR of protein and nucleic acids. Wiley Interscience, New York, NY.