Abstract
Yersinia pestis, the causative agent of bubonic plague, evades the immune response of the infected organism by using a type III (contact-dependent) secretion system to deliver effector proteins into the cytosol of mammalian cells, where they interfere with signaling pathways that regulate inflammation and cytoskeleton dynamics. The cytotoxic effector YopE functions as a potent GTPase-activating protein (GAP) for Rho family GTP-binding proteins, including RhoA, Rac1, and Cdc42. Down-regulation of these molecular switches results in the loss of cell motility and inhibition of phagocytosis, enabling Y. pestis to thrive on the surface of macrophages. We have determined the crystal structure of the GAP domain of YopE (YopEGAP; residues 90–219) at 2.2-Å resolution. Apart from the fact that it is composed almost entirely of α-helices, YopEGAP shows no obvious structural similarity with eukaryotic RhoGAP domains. Moreover, unlike the catalytically equivalent arginine fingers of the eukaryotic GAPs, which are invariably contained within flexible loops, the critical arginine in YopEGAP (Arg144) is part of an α-helix. The structure of YopEGAP is strikingly similar to the GAP domains from Pseudomonas aeruginosa (ExoSGAP) and Salmonella enterica (SptPGAP), despite the fact that the three amino acid sequences are not highly conserved. A comparison of the YopEGAP structure with those of the Rac1-ExoSGAP and Rac1-SptP complexes indicates that few, if any, significant conformational changes occur in YopEGAP when it interacts with its G protein targets. The structure of YopEGAP may provide an avenue for the development of novel therapeutic agents to combat plague.
Keywords: GAP, GTPase-activating protein, plague, cytotoxin, cytoskeleton, Rho
Manipulation of the actin cytoskeleton in eukaryotic cells is one of the principal virulence strategies used by bacterial pathogens. Some bacteria elicit changes in cytoskeletal dynamics that are intended to promote their uptake by eukaryotic cells, whereas pathogens with an extracellular lifestyle manipulate the actin cytoskeleton for precisely the opposite reason: to avoid engulfment and destruction by macrophages and other professional phagocytes. At least some pathogenic Salmonella spp. evidently use both strategies in succession (Fu and Galan 1999).
Because they are master regulators of actin cytoskeleton dynamics (Van Aelst and D'Souza-Schorey 1997; Hall 1998), the Rho family of small GTPases are frequent targets for bacterial cytotoxins (Aktories 1997; Lerm et al. 2000). Like other G proteins, Rho GTPases cycle between active (GTP-bound) and inactive (GDP-bound) states. They become activated by nucleotide exchange, which is promoted by guanine nucleotide exchange factors. The intrinsic GTPase activity of Rho proteins returns them to their inactive state. Other regulatory factors, called GTPase-activating proteins (GAPs), can accelerate the rate of GTP hydrolysis. The stimulation of Rho GTPases causes membrane ruffling, which leads to internalization of bacteria by macropinocytosis, whereas down-regulation of these molecular switches inhibits phagocytosis (Galan 1999).
The majority of bacterial cytotoxins that target Rho family GTPases appear to be down-regulators. For example, Clostridium botulinum, Staphylococcus aureus, and Bacillus cereus produce C3-like exotoxins that inactivate Rho GTPases by ADP-ribosylation of Asn41 (Lerm et al. 2000). Other pathogenic Clostridia produce large cytotoxins that inactivate Rho GTPases by glycosylation, using either UDP-glucose or UDP-N-acetylglucosamine as cofactors (Lerm et al. 2000). Rho GTPases are also the targets of toxins that are injected into eukaryotic cells by the type III secretion systems of certain bacterial pathogens, including Salmonella enterica, Pseudomonas aeruginosa, and Yersinia spp. Unlike the bacterial enzymes that inactivate Rho GTPases by covalent modification, these injected toxins transiently down-regulate Rho GTPases by mimicking the activity of eukaryotic GAPs. The Yersinia pestis cytotoxin YopE stimulates the GTPase activity of all three Rho family subtypes (RhoA, Rac1, and Cdc42) but has no GAP activity against Ras family members (Black and Bliska 2000; von Pawel-Rammingen et al. 2000). The P. aeruginosa cytotoxins ExoS (Goehring et al. 1999) and ExoT (Krall et al. 2000) and S. enterica SptP (Fu and Galan 1999) also have RhoGAP activity, although the substrate specificity of SptP appears to be somewhat more restricted than that of the others. The GAP activity of YopE is essential for virulence in Y. pestis, the causative agent of plague in humans (Black and Bliska 2000). Therefore, YopE is a valid molecular target for the development of antiplague therapeutics.
Like many proteins that transit type III secretion systems, YopE has a modular structure. Its N-terminal domain (residues 1–89) contains the signals that target the protein for secretion from the bacterium and translocation into eukaryotic cells by the type III secretion machinery in Y. pestis (Sory et al. 1995; Schesser et al. 1996). The C-terminal domain of YopE (residues 90–219) is the seat of the GAP activity (von Pawel-Rammingen et al. 2000). To elucidate the structural basis of its GTPase activity, we have crystallized the GAP domain of Y. pestis YopE (YopEGAP) and solved its structure at 2.2-Å resolution.
Results and Discussion
The structure of YopEGAP
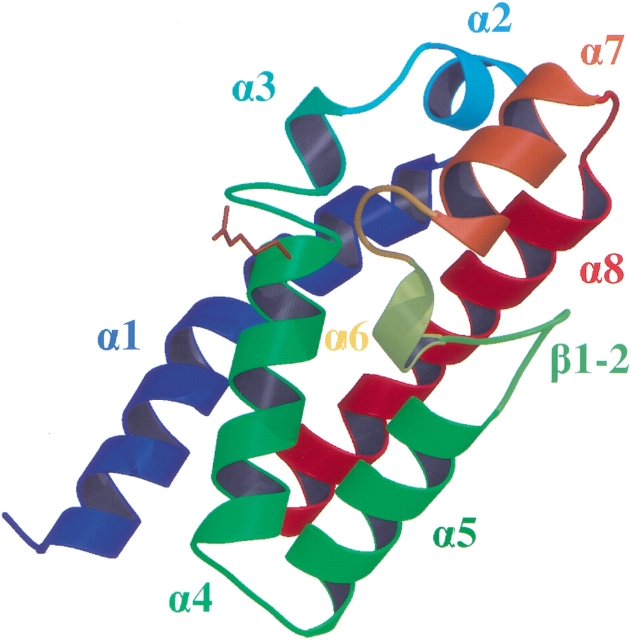
The crystal structure of YopEGAP was solved by the multiwavelength anomalous dispersion (MAD) method, using selenomethionine-substituted protein. The asymmetric unit of the crystal contained two protein monomers that are virtually identical (Cα root mean square deviation, 0.34 Å). The N- and C- termini of YopEGAP are located on the same face of a cylinder, measuring ∼45 × 25 Å, that approximates the shape of the protein. The fold of YopEGAP can be described as an antiparallel four-helix bundle (α1, α4, α5, and α8) that is capped on one end by a convoluted arrangement of four small α-helices (α2, α3, α6, and α7) and one short β-hairpin (β1-2; Fig. 1 ▶). The presence of a proline residue in helix α1 causes it to adopt a kinked conformation. The distribution of atomic thermal displacement parameters (ADPs, B-factors) is fairly even throughout the protein backbone (≤Bcα> = 27; Table 1), with the exception of helices α2 and α3, as well as the first few N-terminal residues, which are not quite as well ordered as the remainder of the structure (≤Bcα> = 38–45).
Fig. 1.
Overall structure of YopEGAP, colored according to secondary structure precession. The critical arginine residue is shown as a stick model.
Table 1.
Data collection and refinement statistics
| Synchrotron | Laboratory source | ||||
| a, b, c (Å); β(°) | 72.51, 72.36, 67.09, 118.17 | 62.69, 71.98, 62.68, 113.06 | |||
| Space group | C2 | C2 | |||
| Resolution (Å) | 30–2.70 | 30–2.25 | |||
| Mosaicity (°) | 1.50 | 1.25 | |||
| Wavelength (Å) | 0.9796a | 0.9795b | 0.9400c | 1.54178 | |
| Completeness (%)a | 85 (74) | 84 (72) | 91 (85) | 98.9 (96.2) | |
| Redundancya | 0.85 (0.78) | 0.84 (0.72) | 0.90 (0.86) | 2.3 (1.8) | |
| Unique reflections | 13302 | 13254 | 14192 | 22062 | |
| I/σIa | 7.9 (1.8) | 8.4 (1.5) | 9.2 (1.9) | 11.2 (3.9) | |
| Rmerge (%)a | 9.1 (61.0) | 10.0 (63.2) | 8.2 (55.0) | 6.0 (22.1) | |
| Anomalous differences (%) | 9.0 | 8.2 | 7.1 | 3.3 | |
| Dispersive differences (%) | 3.3peak−inflection; 5.4peak−remote; 4.1remote−peak | — | |||
| Figure of meritb | 0.53 (0.79) | — | |||
| Rcryst,Rfreeb (%) | 19.8, 23.8 | ||||
| Number of parameters | 7387 | ||||
| Number of restraints | 7258 | ||||
| Parameter/data ratio | 3.96 | ||||
| Root mean square deviation | Bond (Å) | Angle (°) | Dihedral (°) | Planarity (Å) | |
| 0.007 | 1.4 | 16.5 | 0.03 | ||
| Cα | Mainchain | Sidechain | Solvent | ||
| Number of atoms | 241 | 962 | 881 | 67 | |
| Average B-factors (Å2) | 27.3 | 28.2 | 38.4 | 37.0 | |
a Highest resolution shell data is shown in parentheses.
b FOM after density modification with noncrystallographic symmetry averaging is shown in parentheses.
c Randomly selected 8% of the reflections.
The three-dimensional structures of eukaryotic RasGAP, RhoGAP, RanGAP, and ArfGAP have been reported (Rittinger et al. 1997; Scheffzek et al. 1997; Hillig et al. 1999; Mandiyan et al. 1999). Comparison of YopEGAP fold with the structures of these proteins did not reveal any significant similarities. In fact, apart from the other bacterial GAPs, the closest structural relatives of YopEGAP are generic four-helix bundles such as cytochrome b562 and cytochrome c, which yielded DALI scores of 2.9 and 2.8, respectively (Holm and Sander 1993).
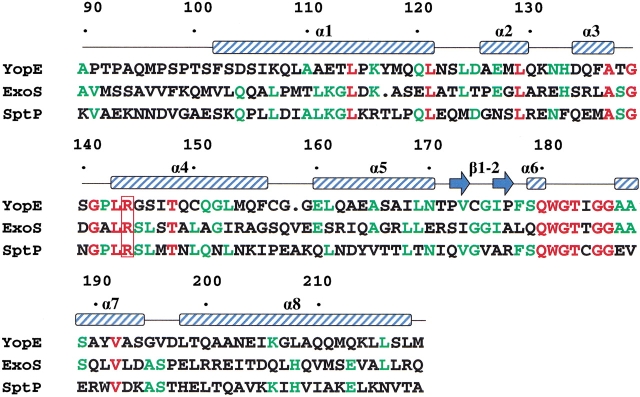
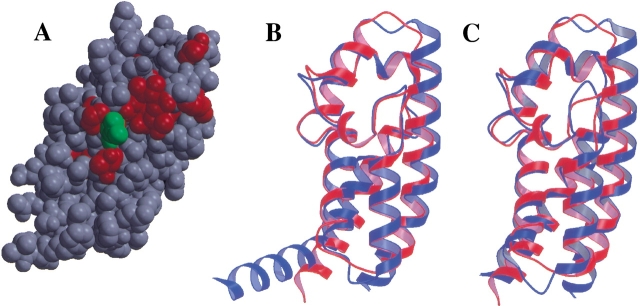
Eukaryotic GAPs that regulate the activity of small GTPases (e.g., RasGAP and RhoGAP) exploit an arginine side-chain, termed the arginine finger, to neutralize the negative charge that develops on the leaving group during GTP hydrolysis (Scheffzek et al. 1997). The arginine finger is contained within a flexible loop in all of these proteins. YopE also contains an arginine residue (Arg 144) that is essential for its GAP activity (Black and Bliska 2000; von Pawel-Rammingen et al. 2000). However, in YopE this arginine side-chain protrudes from the side of an α-helix instead of from within a loop. The critical arginine in YopE, which is located within helix α4, is situated directly underneath a bulge between helices α3 and α4, which is formed by residues 137–141 (Fig. 1 ▶). The key arginine residue and the two glycines associated with this bulge are absolutely conserved in the bacterial GAP domains (Fig. 2 ▶). In close proximity to this bulge is a second protrusion formed by residues 182–186 that connects helices α6 and α7. The residues within and around this protrusion are also strictly conserved in the bacterial GAPs. Collectively, these conserved residues form a patch on the surface of YopEGAP that is likely to play a key role in the recognition of its G protein targets (Fig. 3A ▶).
Fig. 2.
Structure-based sequence alignment of the bacterial GAP domains. Residues that are identical in all three sequences are shown in red; residues identical in two out of three sequences, in green. The critical arginine residue is enclosed by a red box. The positions of α-helices and β-strands are indicated above the sequence. Residues are numbered according to the full-length YopE sequence.
Fig. 3.
Structural alignment of bacterial GAP domains. (A) Locations of strictly conserved residues (red) on the surface of YopEGAP. The critical arginine is in green. (B) YopEGAP (red) aligned with ExoSGAP (blue). (C) YopEGAP (red) aligned with SptPGAP (blue).
Comparison of bacterial GAP domain structures
Salmonella spp. and the opportunistic pathogen P. aeruginosa also inject cytotoxins with functional RhoGAP domains into eukaryotic cells. P. aeruginosa ExoS is a bifunctional toxin: Its N-terminal domain is a RhoGAP, and its C-terminal domain inactivates Ras by ADP ribosylation. ExoT, the other P. aeruginosa cytotoxin with a GAP domain, is very similar to ExoS (Liu et al. 1997). S. enterica SptP also has two functional domains. Its C-terminal domain is a protein tyrosine phosphatase (PTPase) that resembles the PTPase domain of YopH, another cytotoxin produced by Y. pestis. The N-terminal domain of SptP is a RhoGAP. The RhoGAP domain of ExoS and a fragment of SptP that contains both the RhoGAP and PTPase domains have been crystallized in complex with Rac1 (Stebbins and Galan 2000; Würtele et al. 2001a). The ExoSGAP structure has also been determined in the absence of a G protein target (Würtele et al. 2001b). The availability of these structures enabled us to compare and contrast them with the structure of YopEGAP.
Despite a considerable degree of variation in their amino acid sequences (22% identity with ExoSGAP, 29% identity with SptPGAP), the backbone of YopEGAP superimposes remarkably well with the backbones of the two other bacterial GAPs: Alignment of YopEGAP with ExoSGAP (Fig. 3B ▶) and SptPGAP (Fig. 3C ▶) results in Cα root mean square deviations of 1.26 Å and 1.36 Å, respectively. The greatest difference between ExoSGAP and the other two structures occurs near their N termini, where helix α1 in ExoSGAP takes a sharp turn and the corresponding helices of SptPGAP and YopEGAP are kinked but not broken. Helix α1 is also shorter in SptP and YopEGAP than in ExoSGAP. A few other significant differences between the backbone conformations of the three proteins occur in the turns between the helices. Not surprisingly, many of the residues that define the hydrophobic core of YopEGAP are conserved in SptPGAP and ExoSGAP (Leu114, Leu121, Leu129, Ala137, Trp181, Val192). Remarkably, however, despite the fact that the proline associated with the bulge between helices α3 and α4 in YopEGAP and SptPGAP is not conserved in ExoSGAP, the conformation of this bulge is virtually identical in all three proteins. This implies that in ExoS, the bulge is stabilized by interactions with the main body of the protein.
Interaction of YopEGAP with G proteins
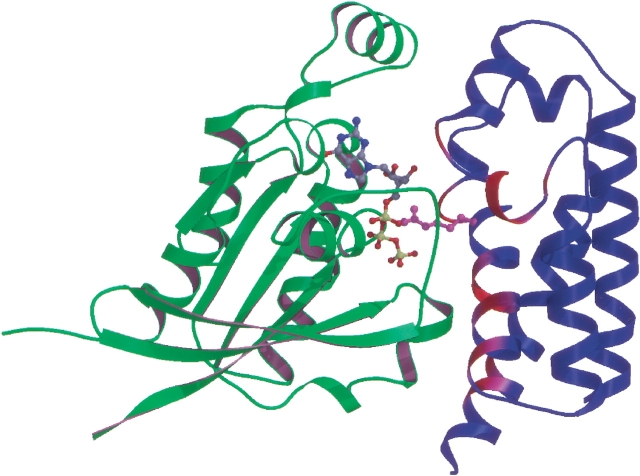
Biochemical experiments revealed slight variations in the specificity of bacterial GAPs for the mammalian G proteins Rac1, Cdc42, and RhoA (Fu and Galan 1999; Goehring et al. 1999; Black and Bliska 2000; Krall et al. 2000; von Pawel-Rammingen et al. 2000). Most notably, ExoSGAP and YopEGAP show similar activity for all three Rho subfamily members, whereas SptPGAP has somewhat lower activity for RhoA (Fu and Galan 1999). Complexes of YopEGAP with Rac1 and Cdc42 bound to a transition-state analog formed by GDP and aluminum fluoride (Ahmadian et al. 1997) were easily purified by gel-filtration chromatography, whereas the corresponding complex of YopEGAP with RhoA was unstable and had to be reconstituted (data not shown). To date, no crystals of these complexes have been obtained despite extensive screening. Lacking structural data, we have drawn on the similarity between YopEGAP and the other bacterial GAPs to construct a model of the YopE-Rac1 complex (Fig. 4 ▶).
Fig. 4.
Model of the YopEGAP-Rac1 complex. YopEGAP and Rac1 are shown in blue and green, respectively. The bound GTP molecule is represented by a ball-and-stick model, colored according to atom type (C, gray; N, blue; O, red; and P, yellow). The critical arginine in YopEGAP is in magenta. Other parts of the YopEGAP structure that are likely to contact Rac1 are depicted in a gradient from magenta to red, with red being most probable.
The polypeptide backbones of the region that contains the critical arginine and the bulge directly above it overlap almost exactly in YopEGAP and the ExoSGAP-Rac1 and SptP-Rac1 complexes (root mean square deviation, 0.3 Å). Therefore, we can conclude that any structural changes that occur in the bacterial GAPs on binding to their G protein targets are restricted to rearrangements of the side-chains rather than the backbone. A potential exception is helix α1, which in our model of YopEGAP-Rac1 complex has to move to optimize contact with the GTPase. Helix α1 is significantly bent by Pro115, which is present only in YopEGAP. This bend is very likely to influence the energetics of any conformational rearrangements involving this α-helix and may contribute to the thermodynamic parameters and selectivity of the interaction between YopE and its targets.
As observed for SptPGAP and ExoSGAP, our model of the YopEGAP-Rac1 complex indicates that interactions between the two proteins are limited to three distinct regions of the GAP structure: residues Ile106, Leu109, Thr138, Gly139, Ser140, and Gln149, contacting Switch II region of the G protein; the key arginine and the bulge residues Thr183, Ile184, and Gly185, contacting GTP and both of the switch regions; and residues Thr148, Gln151, Gln155, Pro177, Ser179, and Gln180, contacting Switch I and the bound nucleotide. However, not all of these residues are conserved in the three bacterial GAPs (Fig. 2 ▶). The conserved residues (Thr/Ser138, Gly139, Thr148, Gln180, Thr183, and Gly185), which lie in close proximity to the critical arginine (Agr144) on the surface of YopE, recognize the bound nucleotide triphosphate and/or highly conserved features of the Rho GTPase. The other contacts between Rac1 and the three bacterial GAPs are slightly different in each case. Either directly or indirectly, these mutations must be responsible for the observed differences in specificity and catalytic efficiency of the bacterial GAPs. Further studies, especially the direct calorimetric measurements of stability constants and catalytic activities of the GTPase-bacterial GAP domain complexes, will be required to reach more definitive conclusions.
Conclusions
One of the strategies most frequently used by bacterial pathogens to avoid phagocytosis and destruction by macrophages is down-regulation of Rho GTPases. Yersinia spp. and at least two other bacterial pathogens have evolved a remarkable way of accomplishing this feat: by injecting a cytotoxin into eukaryotic cells that mimics the activity of eukaryotic GAPs. Like their eukaryotic counterparts, the bacterial GAPs use a conserved arginine side-chain (Arg144 in Y. pestis YopE) to catalyze GTP hydrolysis. In practically all other respects, however, they are quite unlike the eukaryotic GAPs. Accordingly, the bacterial GAPs most likely arose by convergent evolution with their eukaryotic functional analogs. The amino acid sequences of the bacterial GAPs appear to be diverging at a rapid rate, as evidenced by their dissimilarity, but their tertiary structures are remarkably well conserved.
The similarity among the crystal structures of YopEGAP, ExoSGAP, and SptP implies that these three proteins use the same mechanism for stimulating GTP hydrolysis by Rho family G proteins. It seems likely that slight variations in the global conformations of the proteins are responsible for the differing activities and stabilities of the various Rho-GTPase complexes with bacterial GAPs.
Materials and methods
Cloning, expression, and purification of YopEGAP
The open reading frame encoding YopEGAP (residues 90–219 of YopE) was amplified from Y. pestis genomic DNA (strain 195/P) by polymerase chain reaction (PCR) using the following oligonucleotide primers: 5`-GAG AAC CTG TAC TTC CAG GGT GCA CCC ACA CCT GCA CAA ATG CCA AG-3` and 5`-ATT AGT GAT GAT GGT GGT GAT GCA TCA ATG ACA GTA ATT TCT GCA TCT GTT G-3`. This PCR amplicon was subsequently used as the template for a second PCR with the following primers: 5`-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTC GGA GAA CCT GTA CTT CCA G-3` and 5`-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTT ATT AGT GAT GAT GGT GGT GAT G-3`. The amplicon from the second PCR was inserted by recombinational cloning into the entry vector pDONR201 (Invitrogen) to create pKM946, and the nucleotide sequence of the entire insert was confirmed experimentally. The open reading frame encoding YopEGAP, now bracketed by a hexahistidine tag on its C terminus and a recognition site (ENLYFQG) for tobacco etch virus (TEV) protease on its N terminus, was moved by recombinational cloning from pKM946 into the destination vector pKM596 (Evdokimov et al. 2000) to construct pKM948. pKM596, a derivative of pMal-C2 (New England Biolabs), was designed to produce recombinant proteins as in-frame fusions to the C terminus of Escherichia coli maltose-binding protein (MBP). Therefore, pKM948 directed the expression of YopEGAP in the form of an affinity sandwich, with MBP fused to its N terminus and a hexahistidine tag joined to its C terminus. The MBP moiety could be removed by cleaving the fusion protein with TEV protease at a designed site in the linker to yield a recombinant YopEGAP with a single non-native glycine residue on its N terminus and a hexahistidine tag on its C terminus.
The MBP-YopEGAP-His6 fusion protein was overproduced in E. coli BL21(DE3) containing an auxiliary plasmid, pRK603, that produces the catalytic domain of TEV protease (Kapust and Waugh 2000). The fusion protein was cleaved in vivo to generate YopEGAP with a C-terminal hexahistidine tag. Although the MBP moiety was not exploited for affinity chromatography, it probably contributed to the high yield of soluble YopEGAP that was obtained under these conditions. Recombinant cells were grown to mid-log phase (A600 = 0.5) at 37°C in Luria broth (Miller 1972) containing 100 μg/mL ampicillin, 30 μg/mL kanamycin, and 0.2% glucose. Overproduction of MBP-YopEGAP-His6 was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM for 4 h at 25°C. The cells were pelleted by centrifugation and stored at −80°C.
E. coli cell paste was suspended in ice-cold 50 mM sodium phosphate (pH 8) and 300 mM NaCl (Buffer A) containing "complete" protease inhibitor cocktail (Roche Molecular Biochemicals) and disrupted with an APV Gaulin Model G1000 homogenizer at 10,000 psi. The homogenate was centrifuged at 20,000g for 30 min at 4°C to remove insoluble material. The supernatant was filtered through a 0.45-μm cellulose acetate membrane and applied to a 50-mL Ni-NTA Superflow affinity column (Qiagen) equilibrated in buffer A. The column was washed with 5 volumes of equilibration buffer and then with 5 volumes of buffer A containing 25 mM imidazole to remove nonspecifically bound proteins. Elution was performed with a linear gradient from 25 to 250 mM imidazole in buffer A. Fractions containing recombinant YopEGAP-His6 were pooled, and dithiothreitol (DTT) and ethylenediaminetetraacetic acid (EDTA) were added to final concentrations of 5 mM and 1 mM, respectively. The sample was concentrated by diafiltration and fractionated on a HiPrep 26/60 Sephacryl S-100 HR column (Amersham Pharmacia Biotech) equilibrated in buffer A containing 1 mM EDTA and 5 mM DTT. Fractions containing YopEGAP-His6 were pooled; dialyzed against a 25 mM HEPES (pH 7), 30 mM NaCl, and 5 mM DTT buffer; and concentrated to 14 mg/mL (determined spectrophotometrically using the theoretical A280 = 8370 cm−1/M). Aliquots were flash-frozen with liquid nitrogen and stored at −80°C until use. The final product was judged to be >95% pure, on the basis of silver staining after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (data not shown). The molecular weight was confirmed by electrospray mass spectrometry.
Selenomethionine-YopEGAP was produced using the saturation of the methionine biosynthetic pathway protocol (Doublié 1997) and purified in essentially the same fashion as the native YopEGAP, except that the protein was maintained in 10 mM DTT at all the stages after immobilized metal affinity chromatography. Electrospray mass spectrometry showed that the efficiency of selenomethionine incorporation was >98%.
Crystallization and data collection
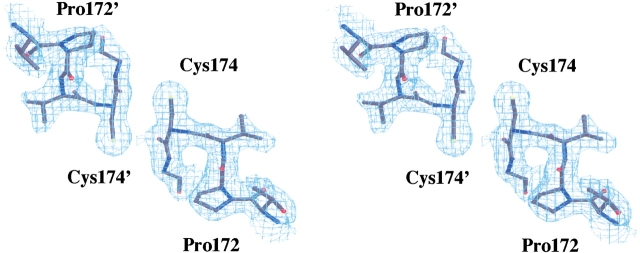
YopEGAP crystals were grown by vapor diffusion in VDX 24-well plates containing 1 mL of precipitant solution per well. Initial crystallization trials were performed with sparse-matrix kits (Jancarik and Kim 1991) obtained from Hampton Research and Emerald BioStructures. YopEGAP crystallized under several neutral and alkaline conditions with ammonium sulfate as the precipitant. Small crystals formed in the presence of 2.0 M ammonium sulfate, but they never grew large enough for data collection. The optimum crystallization conditions were 1.2 to 1.6 M ammonium sulfate, 100 mM HEPES (pH 7.0; or Bicine at pH 9.0), and 100 to 200 mM potassium nitrate. Diffraction quality crystals were obtained under these conditions by streak-seeding 3μL : 3μL (reservoir:protein) drops after 20 h of equilibration at 18°C with the stock crystals grown from 2.0 M ammonium sulfate. The resulting crystals grew to a maximum size of 0.7 × 0.2 × 0.2 mm within 30 to 40 h and started to decay almost immediately after that. The reason for this rapid crystal decay is not completely clear; it may be due to either a transition from one crystal form into another or to oxidative crosslinking (the structure showed that cysteine residues from separate protein monomers can come within bonding distance in the crystal; Fig. 5 ▶). The YopEGAP crystals were of two types, both belonging to the C2 space group with somewhat different cell dimensions. It was impossible to distinguish visually which form appeared in any given crystallization experiment because the two crystal habits were nearly identical. Both crystal forms were found to be extremely sensitive to temperature variations, because as little as a 2° increase in ambient temperature was enough to dissolve or irreversibly damage them.
Fig. 5.
Stereo diagram of the final electron density (│3Fo–2Fc│, 1.3σ) contoured around the region where the Cys174 residues of noncrystallographically symmetrical monomers (equivalent residues labeled with apostrophes) come in close contact.
Crystals of selenomethionine-substituted YopEGAP were grown under the same conditions: first by microseeding with native YopEGAP crystals and then by using the resulting crystals for streak seeding. For data collection, the crystals either had to be frozen immediately after they reached the optimum size or grown fresh every time. In general, crystals of the selenomethionine-substituted YopEGAP diffracted to higher resolution and had lower mosaicity than did the native protein crystals.
Before data collection, protein crystals measuring 0.2 × 0.1 × 0.1 mm were dipped into artificial mother liquor containing 16% glycerol, 1.55 M ammonium sulfate, 200 mM potassium nitrate, and 100 mM Na-Bicine (pH 9.0); mounted immediately in a monofilament loop; and flash-frozen in a cryogenic nitrogen stream (Oxford Cryostream) at 100 K. Cryogenic single-crystal MAD data were collected around the K edge absorption of selenium using a Brandeis CCD detector at the National Synchrotron Light Source beamline X12C. MAD wavelengths were selected on the basis of the X-ray fluorescence spectrum. Data were collected in 150-sec, 1.0° oscillation steps for a total of 120° for each wavelength. All of the YopEGAP crystals that were brought to the synchrotron were anisotropic; the maximum resolution of diffraction was direction dependent and varied from 2.4 to 3.0 Å. An acceptable compromise between resolution, data quality, and completeness was achieved by using the reflections in the 30 to 2.7 Å range. The data sets were processed using the HKL package (Otwinowski and Minor 1997). The essential statistics are given in Table 1. Better data were subsequently collected using freshly grown crystals of selenomethionine-substituted YopEGAP and a laboratory X-ray source. When flash frozen, these crystals diffracted isotropically to 2.2 Å, and a data set was collected using a MAR-345 image plate mounted on a Rigaku X-ray generator equipped with Osmic multilayer focusing mirrors.
Structure determination and refinement
The unit cell dimensions (Table 1) indicated that the asymmetric unit of the crystal contained two protein monomers. Using MAD data in the 30 to 2.8 Å range, SOLVE (Terwilliger and Berendzen 1999) located 10 selenium atoms that fell into two groups that were related by a clear noncrystallographic twofold axis. The first experimental electron density maps were of relatively poor quality. Nevertheless, maximum-likelihood phase refinement with SHARP (de La Fortelle and Bricogne 1997), followed by density modification and noncrystallographic symmetry averaging in DM (Cowtan 1994), yielded interpretable electron density maps. The first model of the protein was built into the experimental density with the program O (Jones et al. 1991), relying on the positions of methionine residues for sequence assignment. After several rounds of noncrystallographic symmetry-constrained refinement (SHELXL-97; Sheldrick and Schneider 1997) against the most complete MAD data set, the model was applied to the 2.2-Å data collected in the lab. The orientation of the two protein monomers in the unit cell was determined using AMoRe (Navaza 1994). After several cycles of manual rebuilding in O, interspersed with conjugated-gradient least-squares refinement in SHELXL, the model contained 241 amino acid residues (120 residues in each of the two monomers, and one residue of the His-tag in the second monomer). Residues 90–99 (including selenomethionine 96), as well as most of the His-tag, were not visible in the final map, most likely because they were disordered. Finally, 67 water molecules were added to the model on the basis of difference density maps and standard water coordination criteria. A representative section of the final electron density map is shown in Figure 5 ▶. The essential refinement parameters and model quality indicators are given in Table 1. The atomic coordinates and the structure factors for the refined YopEGAP were deposited in the Protein Data Bank (Berman et al. 2000) under reference code 1HY5.
YopEGAP-Rac1 complex model
A model of the YopEGAP-Rac1 complex was constructed by least-squares alignment of the Cα atom positions in the YopEGAP monomer on to either SptPGAP or ExoSGAP in their complexes with Rac1 using the program LSQMAN (Kleywegt and Jones 1994), followed by manual adjustment of the side-chains wherever necessary.
Acknowledgments
We thank Dr. Patricia Worsham of the United States Army Research Institute of Infectious Diseases for her gift of Y. pestis genomic DNA (strain 195B), as well as Drs. Robert Sweet and Anand Saxena of the NSLS beamline X12C for making the facility available to us for data collection.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
GAP, GTPase activating protein
TEV, tobacco etch virus
IPTG, isopropyl-β-d-thiogalactopyranoside
DTT, dithiothreitol
EDTA, ethylenediaminetetraacetic acid
CCD, charge-coupled device
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.34102.
References
- Ahmadian, M.R., Mittal, R., Hall, A., and Wittinghofer, A. 1997. Aluminum fluoride associates with small guanine nucleotide binding proteins. FEBS Lett. 408 315–318. [DOI] [PubMed] [Google Scholar]
- Aktories, K. 1997. Rho proteins: Targets for bacterial toxins. Trends Microbiol. 5 282–288. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The protein data bank. Nucl. Acid. Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.S. and Bliska, J.B. 2000. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol. 37 515–527. [DOI] [PubMed] [Google Scholar]
- Cowtan, K. 1994. "dm": An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31 34–38. [Google Scholar]
- Doublié, S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276 523–530. [PubMed] [Google Scholar]
- Evdokimov, A.G., Anderson, D.E., Routzahn, K.M., and Waugh, D.S. 2000 Overproduction, purification, crystallization and preliminary X-ray diffraction analysis of YopM, an essential virulence factor extruded by the plague bacterium Yersinia pestis.Acta Crystallogr. D Biol. Crystallogr. 56 1676–1679. [DOI] [PubMed]
- de La Fortelle, E. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods.Methods Enzymol. 276 472–494. [DOI] [PubMed] [Google Scholar]
- Fu, Y. and Galan, J.E. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401 293–297. [DOI] [PubMed] [Google Scholar]
- Galan, J.E. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 2 46–50. [DOI] [PubMed] [Google Scholar]
- Goehring, U.M., Schmidt, G., Pederson, K.J., Aktories, K., and Barbieri, J.T. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274 36369–36372. [DOI] [PubMed] [Google Scholar]
- Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279 509–514. [DOI] [PubMed] [Google Scholar]
- Hillig, R.C., Renault, L., Vetter, I.R., Drell, T., Wittinghofer, A. and Becker, J. 1999. The crystal structure of rna1p: A new fold for a GTPase-activating protein. Mol. Cell 3 781–785. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–138. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. and Kim, S.H. 1991. Sparse matrix sampling: A screening method for crystallization of proteins. J. Appl. Cryst. 24 409–411. [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kapust, R.B. and Waugh, D.S. 2000. Conrolled intracellular processing of fusion proteins by TEV protease. Protein Expr. Purif. 19 312–318. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J. and Jones, T.A. 1994. A super position. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31 9–14. [Google Scholar]
- Krall, R., Schmidt, G., Aktories, K., and Barbieri, J.T. 2000. Pseudomonas aeruginosa ExoT is a Rho GTPase activating protein. Infect. Immun. 68 6066–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerm, M., Schmidt, G., and Aktories, K. 2000. Bacterial protein toxins targeting Rho GTPases. FEMS Microbiol. Lett. 188 1–6. [DOI] [PubMed] [Google Scholar]
- Liu, S. Yahr, T.L., Frank, D.W., and Barbieri, J.T 1997. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa.J. Bacteriol. 179 1609–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandiyan, V., Andreev, J., Schlessinger, J. and Hubbard, S.R. 1999. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein beta. EMBO J. 18 6890–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode Methods Enzymol. 276A 307–326. [DOI] [PubMed] [Google Scholar]
- Rittinger, K., Walker, P.A., Eccleston, J.F., Nurmahomed, K., Owen, D., Laue, E., Gamblin S.J., and Smerdon, S.J. 1997. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature 388 693–697. [DOI] [PubMed] [Google Scholar]
- Scheffzek, K., Ahmadian, M.R., Kabsch, W., Wiesmuller, L., Lautwein, A., Schmitz, F., and Wittinghofer, A. 1997. Three-dimensional view of Ras:Ras-GAP communication: Structural basis for the mechanism of GTPase activation and its block in oncogenic Ras mutants. Science 277 333–338. [DOI] [PubMed] [Google Scholar]
- Schesser, B., Frithz-Lindsten, E., and Wolf-Watz, H. 1996. Deliniation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178 7227–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick, G.M. and Schneider T.R. 1997. SHELXL: High resolution refinement. Methods Enzymol. 277 319–343. [PubMed] [Google Scholar]
- Sory, M.-P., Boland, A., Lambermont, I., and Cornelis, G.R. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. 92 11998–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, C.E. and Galan, J.E. 2000. Modulation of host signaling by a bacterial mimic: Structure of the Salmonella effector SptP bound to Rac1. Mol. Cell 6 1449–1460. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated structure solution for MIR and MAD. Acta Crystallogr. D Biol Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst, L. and D'Souza-Schorey, C. 1997. Rho GTPases and signaling networks. Genes & Dev. 11 2295–2322. [DOI] [PubMed] [Google Scholar]
- Von Pawel-Rammingen, U., Telepnev, M.V., Schmidt, G., Aktories, K., Wolf-Watz, H., and Rosqvist, R. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: A mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36 737–748. [DOI] [PubMed] [Google Scholar]
- Würtele, M., Wolf, E., Pederson, K.J., Buchwald, G., Ahmadian, M.R., Barbieri, J.T., and Wittinghofer, A. 2001a. How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nat. Struct. Biol. 8 23–26. [DOI] [PubMed] [Google Scholar]
- Würtele, M., Renault, L., Barbieri, J.T., Wittinghofer, A., and Wolf, E. 2001. Structure of the ExoS GTPase activating domain. FEBS Letts. 491 26–29. [DOI] [PubMed] [Google Scholar]