Abstract
We describe a new approach for labeling of unique sequences within dsDNA under nondenaturing conditions. The method is based on the site-specific formation of vicinal nicks, which are created by nicking endonucleases (NEases) at specified DNA sites on the same strand within dsDNA. The oligomeric segment flanked by both nicks is then substituted, in a strand displacement reaction, by an oligonucleotide probe that becomes covalently attached to the target site upon subsequent ligation. Monitoring probe hybridization and ligation reactions by electrophoretic mobility retardation assay, we show that selected target sites can be quantitatively labeled with excellent sequence specificity. In these experiments, predominantly probes carrying a target-independent 3′ terminal sequence were employed. At target labeling, thus a branched DNA structure known as 3′-flap DNA is obtained. The single-stranded terminus in 3′-flap DNA is then utilized to prime the replication of an externally supplied ssDNA circle in a rolling circle amplification (RCA) reaction. In model experiments with samples comprised of genomic λ-DNA and human herpes virus 6 type B (HHV-6B) DNA, we have used our labeling method in combination with surface RCA as reporter system to achieve both high sequence specificity of dsDNA targeting and high sensitivity of detection. The method can find applications in sensitive and specific detection of viral duplex DNA.
INTRODUCTION
Considerable interest in methodologies capable of labeling and detecting specific double-stranded (ds) DNA sequences without requiring DNA denaturation has arisen in recent years, as it has been recognized that such approaches may open up new avenues for functional studies of DNA-modifying enzymes, for studies of intracellular DNA trafficking, genomic analysis, medical diagnostics or pathogen identification. Direct dsDNA labeling technologies may provide with a significant improvement in the specificity of DNA targeting, may result in reduced assay time and cost, may be easily implemented in miniature diagnostic devices, and may lead to cellular diagnostic assays (1).
A number of different approaches for sequence-specific dsDNA labeling have been proposed. In a number of methods, DNA molecules are tagged sequence-specifically at short sequence motifs (4–8 bp), which occur quite frequently within genomic DNA. Some of them apply site-specific binding of synthetic molecules [such as peptide nucleic acids (PNAs) (2–4) or hairpin polyamides (5,6)] or of a restriction endonuclease to directly label cognate dsDNA sequences (7). In contrast to these noncovalent dsDNA-labeling strategies, reporter groups such as fluorophores can be incorporated covalently into dsDNA at target sequences by using a methyltransferase in the presence of a chemically modified cofactor (8,9) or by using a nicking endonuclease followed by nick translation (10,11). The interrogation of such recurrent sequence sites has been successfully employed in combination with DNA stretching techniques and fluorescent single molecule detection for novel DNA-mapping technology (3,4,10,11). It has been projected that this technology may be especially useful in the rapid identification of microbial pathogens. However, this technology is not without limitations, as sites must be accurately labeled and labeling positions precisely determined in order to compare calculated (virtual) and experimental sequence motif maps. Being incompatible with other detection schemes, the developed approaches moreover require sophisticated instrumentation for single molecule detection.
We and others have been pursuing alternative approaches to label dsDNA sequence-specifically at sites ranging from 15 to 30 bp. In a number of methods, a single oligonucleotide probe is hybridized to its complementary target sequence at an internal DNA segment and, following hybridization of its termini to a scaffold oligonucleotide, circularized into a padlock-like complex by ligation (12–16). In another approach, a split protein system is reassembled in the presence of a cognate dsDNA target sequence (17,18). Provided that labeling is extremely sequence-specific and occurs with high efficiency, such approaches may allow labeling and detection of a single (or few) unique sequence(s) within a genome yielding, for instance, new assays for genome identification. We have developed an exceedingly specific approach of this kind based on local opening of a dsDNA segment (∼20 bp) by homopyrimide PNAs and subsequent circular probe assembly (19,20). High sensitivity of detection is achieved using the circularized probe as a template for rolling-circle amplification (RCA) (21–24). Recently, we have successfully applied this methodology on surface-immobilized cells for bacterial detection (25,26).
Here, we propose another method for highly sequence-specific dsDNA labeling of unique genomic DNA sequences under nondenaturing conditions, which does not involve PNAs. This method generates stable DNA tagging and is potentially versatile to either be used with single molecule detection or in combination with signal amplification technology. We demonstrate that in two steps comprising cooperative actions of nicking endonucleases, probe oligonucleotides, and a DNA ligase, selected dsDNA sites can be labeled in high yield and with excellent sequence specificity. Labeled DNA samples are detected, in a heterogeneous assay format, through RCA performed in the presence of an amplicon-complementary PNA beacon (27–29). In proof-of-concept experiments we have applied our approach for the specific labeling and detection of target sites present in genomic DNA of dsDNA viruses.
MATERIALS AND METHODS
Materials (chemicals, DNAs and enzymes)
Succinic anhydride, 1-methyl-2-pyrrolidinone, 20× SSC buffer and betaine were obtained from Sigma–Aldrich. Poly-l-lysine microscope slides were from PolySciences. Oligodeoxyribonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). Except for probes P*-45k-3′ and P*-18k (HPLC-purified) and CO-1 (PAGE-purified), oligonucleotides were purchased without purification. Lambda DNA was obtained from New England BioLabs (Berverly, MA, USA), and human herpes virus 6 type B viral DNA was obtained from Advanced Biotechnologies Inc (Columbia, MD, USA). DNA concentrations were determined on a NanoDrop ND-1000 spectrophotometer. CircLigase and Ampligase were obtained from Epicentre (Madison, WI, USA). All other enzymes were purchased from New England BioLabs (Berverly, MA, USA). PNA beacon, Flu-Glu-AAGGCTAGGAA-K-K(Dabcyl)-NH2, was synthesized as described in ref. 29.
Identification of potential target sites
Genome sequences of dsDNA viruses (http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/10239.html) were pasted into RestrictionMapper (http://www.restrictionmapper.org/), and virtually digested by selecting the restriction endonuclease analogs of nicking endonucleases. From these digests, fragments with appropriate length were selected. Sequences with two nicks at opposing strands were then discarded.
PCR
Reactions were performed in 1× ThermoPol buffer (New England Biolabs) containing 200 μM of each dNTP, 0.5 μM each of a corresponding primer pair F1 and R1 (see Supplementary Table 1) and 0.02 U/μl of Taq DNA polymerase. Amplification was carried out with an initial denaturation step at 94°C for 60 s, followed by 37 cycles of denaturation at 94°C for 30 s, primer annealing at 56°C for 30 s and extension at 72°C for 30 s. The last cycle was followed by an extension step at 72°C for 2 min. Like all other DNA samples that were obtained by enzymatic reactions (see below), PCR amplicons were purified by a standard workup procedure, i.e. extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and chloroform/isoamyl alcohol (24:1), precipitated by the addition of 2 volumes of cold ethanol and centrifugation and dissolved in 1× TE buffer [10 mM Tris–HCl (pH 7.4), 0.1 mM EDTA].
Assembly PCR
Assembly PCR was performed by conducting first extension reactions with hybridized, overlapping oligonucleotides in the absence of primers, followed by PCR amplification in the presence of primers. Extension reactions were performed in 1 × Pfu reaction buffer (Stratagene) containing 20 nM of oligonucleotides A-D (see Supplementary Table 1 for sequences), 200 μM dNTPs and 0.025 U/μl of cloned Pfu DNA polymerase (Stratagene) at the following conditions: one step at 94°C for 60 s, followed by 10 cycles of denaturation at 94°C for 30 s, annealing at 35°C for 30 s and extension at 72°C for 30 s. The last cycle was followed by an extension step at 72°C for 2 min. PCR amplification reactions were performed in 1× ThermoPol polymerase buffer (NEB) containing 200 μM dNTPs, 0.5 μM each of primers P1 and P2, 5 μl of extension reaction product and 0.02 U/μl of Taq DNA polymerase. PCR conditions were: one step at 94°C for 60 s, followed by 28 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s and extension at 72°C for 30 s. The last cycle was followed by an extension step at 72°C for 2 min.
Preparation of ssDNA circles
Oligonucleotide CO-1 (20 pmol, see Supplementary Table 1) was incubated for 1 h at 60°C with 5 U/μl of CircLigase in 1× reaction buffer [50 mM MOPS (pH 7.5), 10 mM KCl, 5 mM MgCl2 and 1 mM DTT] supplemented with 0.05 mM ATP and 2.5 mM MnCl2. Remaining linear molecules were removed by incubation of reaction products for 15 min at 37°C with 60 U of Exonuclease I.
Treatment of dsDNA with nicking endonucleases
Duplex DNA samples (λ-DNA, HHV-6B viral DNA or PCR products) were incubated at 55°C with Nt.BstNBI or sequentially incubated with Nb.BsmI and Nb.BsrDI at 65°C in 1× reaction buffer recommended by the supplier. Typically, incubation was performed for 2–4 h with 10–40 U of nicking endonuclease.
Strand displacement probe hybridization and ligation
In a typical procedure, nicked dsDNA samples (40–60 ng per PCR product) were incubated for 10 min at 50°C in the presence of one or several corresponding probe oligonucleotides (50–100 M excess over dsDNA target) in 30-μl buffer containing 10 mM Tris–HCl (pH 7.4) and 0.1 mM EDTA, followed by cooling to 16°C at a rate of 1°C/min. Subsequently, 5 μl of 10× ligation buffer and 14 μl of H2O were added. Samples were then equilibrated at the specified ligation temperature for 5 min prior to addition of 1 μl of DNA ligase (T4 DNA ligase or Ampligase). Ligation reactions were performed for 1 h at 16°C (T4 DNA ligase) or at 65°C (Ampligase) and stopped by addition of 2 μl of 0.5 M EDTA. Samples to be used for RCA were additionally incubated in 1× NEBuffer 2 (New England BioLabs) with RecJf (typically 60–150 U) for 16 h at 37°C.
Gel electrophoresis
Samples were generally analyzed on 7.5% polyacrylamide gels (29:1 acrylamide/bis-acrylamide) containing 3 M urea. Samples obtained after incubation with NEase were analyzed in the presence of 7 M urea to differentiate intact and singly nicked dsDNA fragments (30). Gels were run for 2–3 h at 12.5 V/cm (ambient temperature) in 1× TBE buffer (90 mM Tris–borate, 2 mM EDTA, pH 8.0), stained with ethidium bromide or SYBR Gold (ssDNA samples), illuminated at 302 nm and scanned with a CCD camera. Quantification was performed using the IS-1000 digital imaging system (Alpha Innotech Corporation, CA).
RCA
Reactions were performed at 31°C in 1× ThermoPol buffer (NEB) containing 1 pmol ssDNA circle, 0.25 μg/μl BSA, 0.5 mM of each dNTP, 0.2 μM PNA beacon and 0.4 U/μl units of phi29 DNA polymerase. In real-time RCA experiments, samples were excited at 499 nm, and the emission was measured at 525 nm.
Immobilization of DNA samples onto glass slides
Following probe attachment and RecJf incubation, samples were suspended in buffer containing 3× SSC (45 mM sodium citrate pH 7.0, 450 mM NaCl) supplemented with 1.5 M betaine (31), and spotted onto poly-lysine coated glass slides (∼50 nl per spot). Spotted DNA was crosslinked to slides by UV irradiation with a total energy of 60 mJ in a Stratalinker 2400 UV Crosslinker (Stratagene). After DNA immobilization, glass slides were blocked with succinic anhydride (32). Slides were then washed by immersion twice in 1× SSC and twice in 1× TE buffer. After each washing step, slides were left to dry at room temperature.
Incubations on slides and analysis of slides
Strand displacement probe hybridization, ligation and RCA reactions were performed by placing the corresponding components in 40 μl buffer on the center of the slide. The slide was then covered by a 24 × 50 microscope cover glass, sealed with rubber cement and incubated in a humidifying chamber essentially under conditions described above. RCA reactions were performed overnight. After each reaction, the rubber cement was gently peeled off, and slides were washed by immersion twice in 1× SSC and twice in 1× TE buffer. Following RCA, slides were washed four times in 1× TE buffer and mounted with Vectashield mounting medium. Detection of fluorescence signal was performed on a Nikon Eclipse 80i inverted microscope equipped with an epifluorescence system X-Cite 120 and a Nikon Cool Snap, HQ black and white digital camera (12 bit, 20 MHz), and analyzed using IPLab 3.7 software (Scanalytics). Fluorescence signals were pseudocolored in green.
RESULTS AND DISCUSSION
Outline of the assay
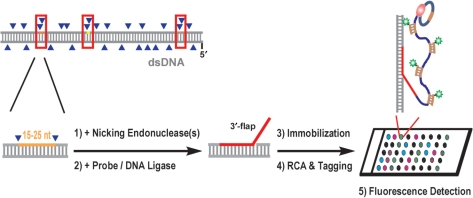
Figure 1 shows the principle of the assay. In the initial step, site-specific nicks are introduced into dsDNA by its treatment with one or two nicking endonucleases (NEases). Although currently only a very limited set of NEases is commercially available, we have found that even short genomes <200 kb (i.e. dsDNA viral genomes) contain a significant number of locations, at which two of the resulting nicks are positioned in close vicinity (≤25 bp) on the same DNA strand (see Table 1 for examples). These regions between vicinal nicks serve in our procedure as target sites for probe oligonucleotide binding via strand displacement hybridization. Through subsequent ligation, hybridized probe oligonucleotides become covalently linked to the dsDNA at the selected target sites. The use of a probe with a target-independent segment at one terminus results in the formation of a bifurcated DNA structure, so-called flap DNA. A structure with a 3′-flap may serve afterward as a priming site for an externally added ssDNA circle, thus initiating RCA in the presence of an appropriate DNA polymerase and dNTPs. The resulting ssDNA amplicon can be detected via hybridization with a complementary fluorophore-labeled probe such as a PNA beacon. We employed this strategy for fluorescence detection of probe-labeled dsDNA following DNA immobilization onto glass slides. As described below, we first examined and optimized individual reactions stepwise in solution before advancing to the heterogeneous assay format.
Figure 1.
Schematic overview of the method. In this study, mainly probe oligonucleotides with an additional, target-independent 3′-terminal section were employed, leading to 3′-flap DNA upon probe hybridization and ligation. Linear rolling-circle amplification (RCA) was used as a reporter system to fluorescently detect probe incorporation events.
Table 1.
Representative targets for the proposed assay within some virus dsDNAsa
| Genomeb | Position (nt) | Length (nt) | NEase(s) | Target sitec (5′→3′) |
|---|---|---|---|---|
| Lambda | 33779-33791 | 13 | Nt.BstNBI | TTCAGAGTCTGAC |
| 26151-26166 | 16 | Nb.BsmI/Nt.BstNBI | CATTCTTGAGTCCAAT | |
| 10841-10857 | 17 | Nt.BstNBI/Nt.AlwI | CGCCGAAGGAGTCCTTC | |
| 44815-44831 | 17 | Nt.BstNBI | ATCGTGAAGAGTCGGCG | |
| 37289-37306 | 18 | Nb.BsrDI/Nt.AlwI | CATTGCATGGGATCATTG | |
| 48483-48502 | 20 | Nt.AlwI | CGTAACCTGTCGGATCACCG | |
| EBV | 50324-50337 | 14 | Nb.BsmI/Nb.BsrDI | CATTGCCCCCAATG |
| 90615-90631 | 17 | Nb.BsmI/Nb.BbvCI | CATTCTCAGGAGCAGGC | |
| HHV-6A | 17380-17397 | 18 | Nb.BsmI/Nb.BsrDI | CATTCCGAAAGTTTTATA |
| 21797-21814 | 18 | Nb.BsrDI | CATTGCCTTTGAACTCTT | |
| HHV-6B | 18321-18338 | 18 | Nb.BsmI/Nb.BsrDI | CATTCCGAAAGTTTTATG |
| 36668-36686 | 19 | Nb.BsmI/Nb.BsrDI | CATTCAACAAACGATGTAT | |
| 125843-125862 | 20 | Nb.BsmI | CATTCGGATCTTGCCTTTGG | |
| 18687-18710 | 24 | Nb.BtsI/Nb.BsrDI | CACTGCCTTATAAAACAGTATGAG | |
| HCMV | 148206-148219 | 14 | Nb.BtsI | CACTGCTCGTCGGT |
| 114721-114738 | 18 | Nb.BsmI/Nb.BsrDI | CATTCTGCCGCTCTTTAT | |
| 202043-202060 | 18 | Nb.BtsI/Nb.BsrDI | CATTGCCCTTCTGGAGCA |
aOnly sites with 13–24 nt between two nicks were selected. Except for lambda, only those sites are shown that can be obtained by using NEases with a recognition sequence of ≥6 bp.
bEBV, Epstein Barr Virus; HHV-6, human herpes virus 6; HCMV, human cytomegalovirus.
cPartial and complete recognition sequences for NEases are shown in italics or in bold, respectively.
Strand displacement probe hybridization and ligation reactions
It was far from being obvious that the strand displacement reaction in our case could be efficient and highly sequence-specific. Indeed, after nicking we deal with a relatively long DNA oligonucleotide hybridized with its complementary partner, which forms a very stable duplex. Moreover, in nicking sites stacking interactions are preserved leading to additional stabilization of the complex (33) that could make the strand exchange with oligonucleotides in solution even more problematic. Yet, our optimism was based on the data of Lyamichev and collaborators (34), which indicated that in the presence of excess displacing oligonucleotide the strand exchange occurs very efficiently even at low temperatures via a branch-migration mechanism. It should be noted, however, that these data were obtained in the absence of the stabilizing effect of stacking.
We therefore performed experiments to evaluate the efficiency and sequence specificity of strand displacement probe hybridization and ligation reactions on nicked DNA substrates under various conditions. To this end, we prepared appropriate dsDNA fragments by PCR. We reasoned that typical PCR amplicons, in contrast to long dsDNA molecules (kb scale), could be analyzed with respect to reaction products by electrophoretic mobility retardation assay, in case probe oligonucleotides resulting in flap DNA structures are employed. In the first set of experiments, we prepared a 170-bp-long PCR product containing a suitable labeling site encountered within λ-DNA that requires only treatment by the nicking endonuclease Nt.BstNBI (wt in Figure 2A). For labeling reactions, we initially employed probe oligonucleotide P-45k-5′F containing a short additional segment of five thymines and a biotin label at the 5′-terminus (see Table 2 for probe oligonucleotide sequences). To establish suitable conditions for strand displacement, the nicked PCR fragment was incubated, in buffer of low ionic strength, with molar excess of probe at different reaction temperatures, followed by ligation with T4 DNA ligase at 16°C. Nearly quantitative formation of 5′-flap product with reduced electrophoretic mobility was obtained, when the hybridization step was carried out at 50–60°C (see Supplementary Figure 1, lanes 5 and 6). Addition of streptavidin led to a further decrease in the mobility of this product, as anticipated for a biotin-labeled branched DNA structure (lane 8).
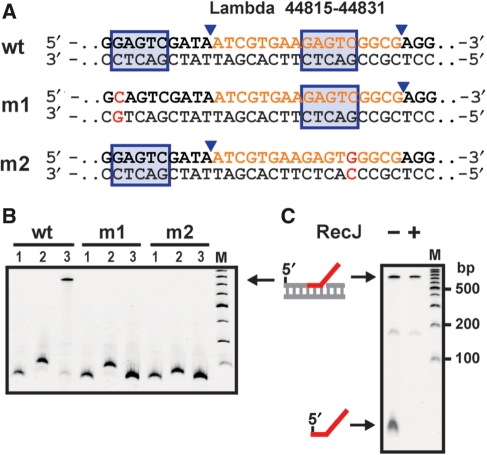
Figure 2.
Vicinal nicks are prerequisite for target labeling. (A) Partial sequences of different dsDNA substrates prepared by assembly PCR. The recognition sequence and cleavage site for NEase Nt.BstNBI present in the selected wild-type and prepared mutant λ-DNA sequences are marked. (B) Probe hybridization and ligation of nicked substrates. PCR amplicons (170 bp, lanes 1) were treated with Nt.BstNBI, leading to doubly nicked wt and singly nicked m1 and m2 fragments (lanes 2). Samples were then incubated with probe P*-45k-3′F for 10 min at 50°C, followed by cooling and incubation with T4 DNA ligase for 1 h at 16°C. Here and below, M denotes a 100-bp DNA ladder (NEB). (C) Removal of excess probe oligonucleotide. Probe-labeled wt DNA sample before and after incubation with RecJf, a 5′→ 3′ ssDNA-specific exonuclease.
Table 2.
Probe oligonucleotides
| Oligo | Sequencea (5′→3′) |
|---|---|
| Lambda | |
| P-45k-5′F | biotin-TTTTTATCGTGAAGAGTCGGCG |
| P*-45k-3′F | P-ATCGTGAAGAGTCGGCGTTTTTCAGACAGCAGAGTGAACA*A*G |
| HHV-6B | |
| P-18k | P-CATTCCGAAAGTTTTATGTTTTTCAGACAGCAGAGTGAACAAG |
| P*-18k | P-CATTCCGAAAGTTTTATGTTTTTCAGACAGCAGAGTGAACA*A*G |
| P-37k | P-CATTCAACAAACGATGTATTTTTTCAGACAGCAGAGTGAACAAG |
| P-126k | P-CATTCGGATCTTGCCTTTGGTTTTTCAGACAGCAGAGTGAACAAG |
P, phosphate; asterisk indicates phosphorothioate. Target-complementary segments are underlined and priming segments are italicized.
To verify that probe labeling requires the occurrence of vicinal nicks, we generated corresponding dsDNA fragments carrying a single mutation in the upstream or downstream recognition sequence for Nt.BstNBI (Figure 2A, m1 and m2). Labeling reactions were now performed with probe oligonucleotide P*-45k-3′F featuring an extra segment of 25 nt at the 3′-terminus, of which 20 nt were intended to serve as a priming site in RCA reactions at a later stage of our protocol. As expected, successful labeling occurred with the nicked wt fragment, but not with the nicked mutant fragments (Figure 2B, lane 3). Of note, due to the considerably longer nontarget tail in this probe compared to the previously used probe, the obtained 3′-flap DNA fragment moved much slower.
Detection of 3′-flap DNA by RCA
Following strand displacement probe hybridization and ligation reactions, the DNA samples (wt, m1 and m2) were treated with RecJf to remove probe molecules not covalently linked to the dsDNA target (Figure 2C). Samples were then subjected to RCA performed with phi29 DNA polymerase and a supplied ssDNA circle. Before embarking on performing RCA with immobilized DNA samples, which should provide with a high sensitivity of detection by fluorescence microscopy, we analyzed RCA reactions in solution. In this homogeneous format, RCA can be monitored in real-time, thus allowing to rapidly examine the performance of RCA with 3′-flap DNA substrate under various conditions. For fluorescent detection, we investigated the use of PNA beacons, which hitherto had not been used for the detection of ssDNA amplicon generated during linear RCA. It is also worth mentioning that phi29 DNA polymerase has a strong 3′ exonucleolytic activity (35). For RCA reactions with circularized probes as template, this enzymatic activity does not create a problem since primers are supplied in large excess. By contrast, in our case, the number of primers for RCA is strictly limited by the number of target sites and therefore any substantial primer degradation will decrease the sensitivity of our assay. To avoid primer degradation, we obtained probe oligonucleotide P*-45k-3′F with phosphorothioate linkages in the two 3′-terminal nucleotides. This modification makes the probe resistant to the 3′-exonuclease activity of phi29 DNA polymerase but does not interfere with enzymatic polymerization activity (36,37).
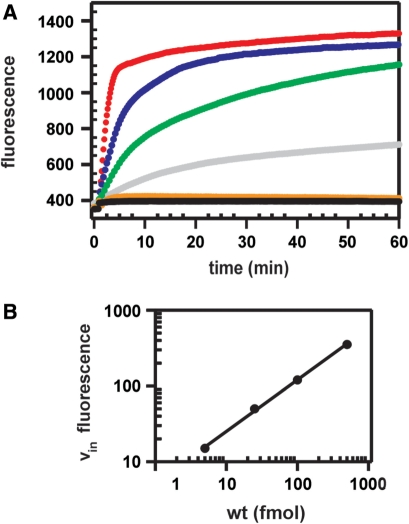
RCA reactions were carried out with various inputs of DNA samples and a fixed amount of ssDNA circle. At input of samples containing labeled wt a significant increase in fluorescence was observed in real-time (Figure 3A). As expected in case of linear RCA, the initial speed of fluorescence signal increases proportional to the number of input molecules (Figure 3B). By contrast, no fluorescent signal could be detected for the analogously treated m1 and m2 samples, compliant with the absence of 3′-flap DNA product in those samples, as assessed by electrophoretic mobility retardation assay. Note that similar data with regard to the sensitivity and quantification have been previously obtained with circularized padlock probes as analytes and 2′-O-Me-RNA molecular beacons (38). Thus, our data demonstrate that the 3′ terminus in flap DNA serves as a very effective primer during RCA and that a PNA beacon makes it possible to detect that amplicon.
Figure 3.
Real-time RCA. (A) Measured fluorescence from PNA beacon Flu-Glu-AAGGCTAGGAA-K-K(Dabcyl)-NH2 plotted against the time of RCA reactions containing 500 (red), 100 (blue), 25 (green) or 5 (gray) fmol wt target or 500 fmol each m1 (black) or m2 target (orange), respectively. Inputs comprised aliquots of samples shown in Figure 2 after RecJf treatment. Phi29 DNA polymerase was added at t = 1 min. (B) Plot of the initial speed of the fluorescence increase (vin) as a function of input wt 3′-flap dsDNA present in RCA reactions.
Fluorescent detection of immobilized lambda DNA
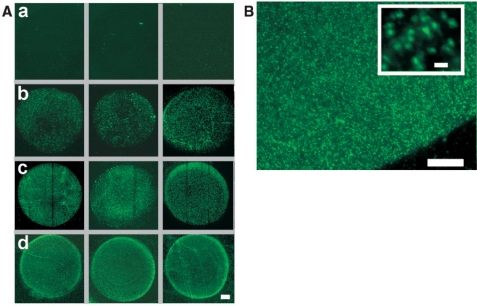
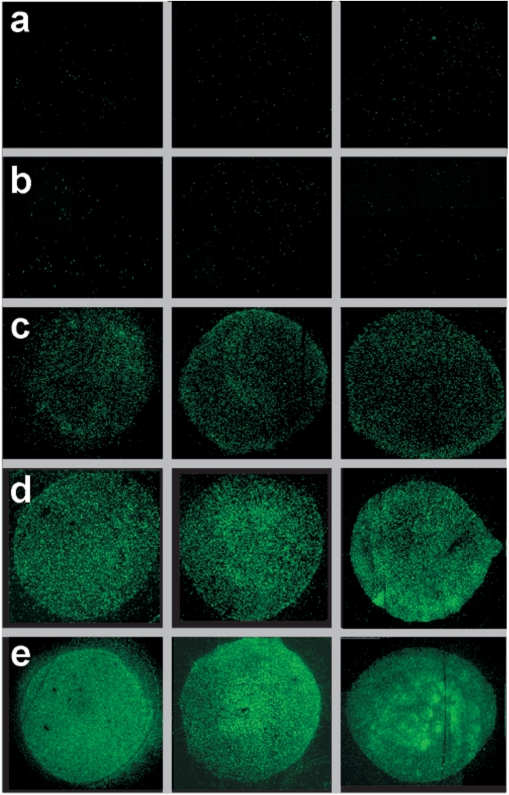
Immobilized RCA products labeled with a detection oligonucleotide can be individually visualized in a straightforward manner by fluorescence microscopy (21). Having established the protocol of our approach in solution, we moved forward by incorporating an immobilization step into our procedure. In addition, DNA samples now consisted of genomic λ-DNA in order to investigate the workability of our assay on a full viral genome. Labeled samples were spotted and immobilized together with control samples onto polylysine-coated glass slides, which were subsequently blocked in succinic anhydride to inactivate free lysine groups. Representative results obtained after RCA in the presence of the PNA beacon for samples immobilized at apparently most favorable conditions (see Materials and methods section) are shown in Figure 4A. Control samples of λ-DNA, in which either incubation with Nt.BstNBI or incubation with probe P*-45k-3′F were omitted, did not yield any detectable fluorescence above background (see row a). By contrast, a clear fluorescence signal was detected with samples, for which all steps were performed appropriately. With inputs between 0.1 and 10 amol (6 × 104 to 6 × 106 copies), the signal intensity increased roughly proportional to the number of spotted molecules (rows b–d). On average, the observed fluorescent objects were about 1 μm in size (Figure 4B), in agreement with previously measured fluorophore-tagged RCA products (39,40). In addition to the condensed fluorescent objects, we observed in some experiments various elongated molecules, which were several micrometers long. The occurrence of such unfolded or partly folded amplification products has been previously reported for RCA reactions performed on polylysine-coated glass slides (21). We conclude that in λ-DNA, our first model system, we successfully incorporated a probe oligonucleotide into the chosen target site and detected it with high sensitivity following sample immobilization and RCA.
Figure 4.
Detection of genomic λ-DNA. All steps of the assay were performed in solution with the exception of RCA, which was conducted on glass slides following surface-immobilization of samples. (A) Fluorescent images of signals generated by RCA. (a) λ-DNA sample (10 amol), for which the nicking step was omitted. (b–d) Probe-labeled λ-DNA samples. Spotted DNA amounts were: 0.1 amol (b), 1 amol (c) or 10 amol (d). Scale bar, 50 μm. (B) Partial image of the central spot in panel d, acquired at higher magnification (100×). Scale bar, 20 μm. Inset: enlarged view of an image section with reduced signal density. Scale bar, 2 μm.
Sequence specificity of the assay
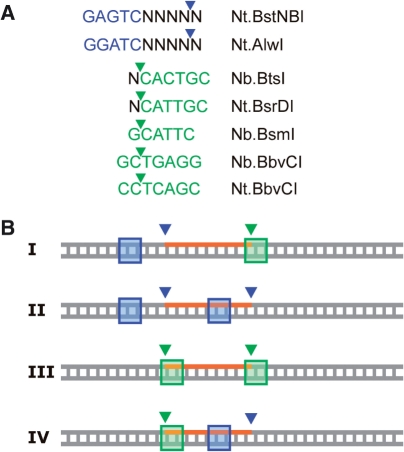
One important factor concerning the specificity of our method is the configuration of the sequence recognition motifs pertaining to the cleavage positions. With the few NEases existing at present (Figure 5A), target sites can be divided into four different configurations. Configuration I, in which both recognition sequences lie completely or largely outside of the cleaved DNA segment, is evidently superior over other configurations in terms of obtaining high sequence specificity of probe labeling, because possible off-target sites with vicinal nicks of the same NEases will likely differ significantly in the cleaved segment (and thus probe sequence). On the contrary, in configuration IV the recognition sequences comprise a substantial part of the cleaved segment. Thus, a site with this configuration seems only appropriate for labeling, if off-target sites can be excluded or if probe labeling is performed under precisely controlled conditions.
Figure 5.
(A) Recognition sequences and cleavage positions of currently available NEases. (B) Possible configurations of NEase sequence recognition motifs at sites with vicinal nicks. Overlaps between these motifs and the cleaved DNA segment are minimal or absent in configuration I, moderate in II and III and extensive in IV.
To investigate the exemplary sequence specificity of probe labeling, we selected three target sites present in human herpes virus 6 type B (HHV-6B) generated by NEases Nb.BsmI and Nb.BrsDI (Figure 6A). These sites, representing intermediate configuration III, constitute a good test for our assay since they are nearly identical in length and contain the same 5-bp-long sequence at the 5′-terminus of the cleaved segment. By PCR, dsDNA fragments of different length carrying each one centrally located target site were prepared. Incubation of each fragment with Nb.BsmI and Nb.BrsDI resulted in the formation of doubly nicked DNA molecules in essentially quantitative yield (Supplementary Figure 2A). To correctly assess the sequence specificity in labeling reactions with pooled amplicons, we first generated deliberately all possible incorrect ligation products by performing ligation with T4 DNA ligase under low fidelity conditions. We also prepared corresponding gapped DNA fragments (41), which may be formed in probe labeling reactions to some extent, if dissociated cleaved segments do not rehybridize and if incorrect probes are not ligated into target sites. The various products were then analyzed concurrently with correct ligation products. Under the employed gel electrophoretic conditions, gapped DNA had a higher electrophoretic mobility and incorrect ligation products had markedly reduced mobility in comparison with the three correct ligation products (Supplementary Figure 2B).
Figure 6.
Sequence specificity of the assay at elevated ligation temperature. (A) Target sequences present in human herpes virus 6 type B (HHV-6B) at nicking with Nb.BsmI and Nb.BsrDI. (B) Individual or combined labeling of HHV-6B target sites. All three sites are amplified within corresponding PCR products, which differ in length to be distinguished in electrophoresis. Ligation reactions were performed for 1 h at 65°C with 5 U of Ampligase.
The fact that the different classes of potential reaction products can be spatially well separated on a gel made it possible to monitor probe hybridization and ligation reactions of all three sites simultaneously. Because the thermophilic DNA ligase Ampligase is commonly employed for achieving high sequence discrimination with probe oligonucleotides (42), we performed experiments with this enzyme to investigate the sequence specificity of target labeling. When incubations were carried out at 65°C, target sites proved to be labeled with high yield (84–93%), individually or in any combination with the correct probe oligonucleotide(s) (Figure 6B). Due to the fact that we employed unpurified probe oligonucleotides, weak bands resulting from truncated oligomers were observed below correct ligation products in any ligation reaction. Interestingly, the sites that were not targeted in a reaction were sealed again into intact dsDNA. Only in some ligation samples, weak bands corresponding to gapped DNA could be detected. Most importantly, no incorrect probe labeling was observed in any of the labeling reactions. Theses data clearly show that sites of vicinal nicks can be labeled with high sequence specificity by probe oligonucleotides, notwithstanding considerable sequence homology between probes at the site of ligation.
Labeling of human herpes virus 6 type B (HHV-6B) genomic DNA
As previously, all steps with the exception of the final amplification step were performed in solution. Strand displacement hybridization was performed in the presence of phosphorothioate-containing probe P*-18k, and ligation was carried out with Ampligase at 65°C. Identical reactions were conducted with either HHV-6B or lambda dsDNA as input. Other controls consisted in omission of one NEase or other components in the labeling of HHV-6B. Figure 7 shows representative images of observed fluorescent signals at the end of the procedure. Clear signals were obtained with immobilized samples containing nicked, probe-labeled HHV-6B DNA at inputs of 7 amol (row d) and 70 amol (row e). At lower input of 0.7 amol, the observed fluorescence signal was relatively weak (row c), albeit still considerably above background signal obtained with control samples at high input (rows a and b). It should be noted that the observed background was slightly higher in those control samples than in previous controls for the detection of lambda genomic DNA. Most probably, incomplete digestion of free oligonucleotide probe molecules caused a higher background.
Figure 7.
Detection of genomic HHV-6B DNA. Fluorescent images of signals generated by RCA. (a) HHV-6B DNA sample (70 amol), for which the nicking step was omitted. (b) λ-DNA sample (150 amol) treated analogous to the HHV-6B sample shown in row e. (c–e) HHV-6B DNA samples after labeling procedure with probe P*-18k. Spotted DNA amounts were: 0.7 amol (c), 7 amol (d) or 70 amol (e).
CONCLUSION
We have proposed and validated a new method for efficient and highly sequence-specific targeting of appropriate sequences within dsDNA. In combination with surface RCA, the method makes possible the specific and sensitive detection of viral dsDNA, as demonstrated for two model genomes. We chose this solid-support detection format for our proof-of-principle experiments because it allowed us to simultaneously analyze labeled samples and appropriate controls under identical amplification conditions. However, with regard to the practical utility of the assay for viral dsDNA detection, other detection formats may prove to be advantageous. These may consist in the use of other reporter systems [e.g. Invader-type amplification reactions (43,44)] or in the use of single molecule detection methods similar to those used by others in conjunction with other labeling techniques (3,4,10,11). Finally, we should note that the proposed labeling method may be useful in other areas of research such as DNA repair, as it provides with a convenient procedure for the site-specific incorporation of modified nucleic acids within long dsDNA duplexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Maria Valencia-Burton for technical assistance and Ron McCullough for discussion. Funding to pay the Open Access publication charges for this article was provided by the Wallace H. Coulter Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ghosh I, Stains CI, Ooi AT, Segal DJ. Direct detection of double-stranded DNA: molecular methods and applications for DNA diagnostics. Mol. Biosyst. 2006;2:551–560. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 2.Zelphati O, Liang X, Hobart P, Felgner PL. Gene chemistry: functionally and conformationally intact fluorescent plasmid DNA. Hum. Gene Ther. 1999;10:15–24. doi: 10.1089/10430349950019156. [DOI] [PubMed] [Google Scholar]
- 3.Chan EY, Goncalves NM, Haeusler RA, Hatch AJ, Larson JW, Maletta AM, Yantz GR, Carstea ED, Fuchs M, Wong GG, et al. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Res. 2004;14:1137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips KM, Larson JW, Yantz GR, D’Antoni CM, Gallo MV, Gillis KA, Goncalves NM, Neely LA, Gullans SR, Gilmanshin R. Application of single molecule technology to rapidly map long DNA and study the conformation of stretched DNA. Nucleic Acids Res. 2005;33:5829–5837. doi: 10.1093/nar/gki895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fechter EJ, Olenyuk B, Dervan PB. Sequence-specific fluorescence detection of DNA by polyamide-thiazole orange conjugates. J. Am. Chem. Soc. 2005;127:16685–16691. doi: 10.1021/ja054650k. [DOI] [PubMed] [Google Scholar]
- 6.Chenoweth DM, Viger A, Dervan PB. Fluorescent sequence-specific dsDNA binding oligomers. J. Am. Chem. Soc. 2007;129:2216–2217. doi: 10.1021/ja0682576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lushnikov AY, Potaman VN, Lyubchenko YL. Site-specific labeling of supercoiled DNA. Nucleic Acids Res. 2006;34:e111. doi: 10.1093/nar/gkl642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pljevaljcic G, Schmidt F, Scheidig AJ, Lurz R, Weinhold E. Quantitative labeling of long plasmid DNA with nanometer precision. Chembiochem. 2007;8:1516–1519. doi: 10.1002/cbic.200700294. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt FH, Huben M, Gider B, Renault F, Teulade-Fichou MP, Weinhold E. Sequence-specific Methyltransferase-Induced Labelling (SMILing) of plasmid DNA for studying cell transfection. Bioorg. Med. Chem. 2008;16:40–48. doi: 10.1016/j.bmc.2007.04.054. [DOI] [PubMed] [Google Scholar]
- 10.Xiao M, Phong A, Ha C, Chan TF, Cai D, Leung L, Wan E, Kistler AL, DeRisi JL, Selvin PR, et al. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 2007;35:e16. doi: 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo K, Dhingra DM, Odijk T, de Pablo JJ, Graham MD, Runnheim R, Forrest D, Schwartz DC. A single-molecule barcoding system using nanoslits for DNA analysis. Proc. Natl Acad. Sci. USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potaman VN, Lushnikov AY, Sinden RR, Lyubchenko YL. Site-specific labeling of supercoiled DNA at the A+T rich sequences. Biochemistry. 2002;41:13198–13206. doi: 10.1021/bi026402w. [DOI] [PubMed] [Google Scholar]
- 13.Roulon T, Coulaud D, Delain E, Le Cam E, Helene C, Escude C. Padlock oligonucleotides as a tool for labeling superhelical DNA. Nucleic Acids Res. 2002;30:E12. doi: 10.1093/nar/30.3.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geron-Landre B, Roulon T, Desbiolles P, Escude C. Sequence-specific fluorescent labeling of double-stranded DNA observed at the single molecule level. Nucleic Acids Res. 2003;31:e125. doi: 10.1093/nar/gng125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potaman VN. Applications of triple-stranded nucleic acid structures to DNA purification, detection and analysis. Expert Rev. Mol. Diagn. 2003;3:481–496. doi: 10.1586/14737159.3.4.481. [DOI] [PubMed] [Google Scholar]
- 16.Shigemori Y, Haruta H, Okada T, Oishi M. Marking of specific sequences in double-stranded DNA molecules–SNP detection and direct observation. Genome Res. 2004;14:2478–2485. doi: 10.1101/gr.2789604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. DNA sequence-enabled reassembly of the green fluorescent protein. J. Am. Chem. Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 18.Ooi AT, Stains CI, Ghosh I, Segal DJ. Sequence-enabled reassembly of beta-lactamase (SEER-LAC): a sensitive method for the detection of double-stranded DNA. Biochemistry. 2006;45:3620–3625. doi: 10.1021/bi0517032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukanov NO, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. PD-loop: a complex of duplex DNA with an oligonucleotide. Proc. Natl Acad. Sci. USA. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn H, Demidov VV, Frank-Kamenetskii MD. Topological links between duplex DNA and a circular DNA single strand. Angew. Chem. Int. Ed. 1999;38:1446–1449. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1446::AID-ANIE1446>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 22.Zhang DY, Brandwein M, Hsuih TC, Li H. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn H, Demidov VV, Frank-Kamenetskii MD. Rolling-circle amplification under topological constraints. Nucleic Acids Res. 2002;30:574–580. doi: 10.1093/nar/30.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Wu J, Ye F, Feng T, Lee I, Yin B. Amplification of circularizable probes for the detection of target nucleic acids and proteins. Clin. Chim. Acta. 2006;363:61–70. doi: 10.1016/j.cccn.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 25.Smolina I, Lee C, Frank-Kamenetskii M. Detection of low-copy-number genomic DNA sequences in individual bacterial cells by using peptide nucleic acid-assisted rolling-circle amplification and fluorescence in situ hybridization. Appl. Environ. Microbiol. 2007;73:2324–2328. doi: 10.1128/AEM.02038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smolina IV, Kuhn H, Lee C, Frank-Kamenetskii MD. Fluorescence-based detection of short DNA sequences under non-denaturing conditions. Bioorg. Med. Chem. 2008;16:84–93. doi: 10.1016/j.bmc.2007.04.063. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn H, Demidov VV, Gildea BD, Fiandaca MJ, Coull JC, Frank-Kamenetskii MD. PNA beacons for duplex DNA. Antisense Nucleic Acid Drug Dev. 2001;11:265–270. doi: 10.1089/108729001317022269. [DOI] [PubMed] [Google Scholar]
- 28.Seitz O, Kohler O. Convergent strategies for the attachment of fluorescing reporter groups to peptide nucleic acids in solution and on solid phase. Chemistry. 2001;7:3911–3925. doi: 10.1002/1521-3765(20010917)7:18<3911::aid-chem3911>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn H, Protozanova E, Demidov VV. Monitoring of single nicks in duplex DNA by gel electrophoretic mobility-shift assay. Electrophoresis. 2002;23:2384–2387. doi: 10.1002/1522-2683(200208)23:15<2384::AID-ELPS2384>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 31.Diehl F, Grahlmann S, Beier M, Hoheisel JD. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 2001;29:E38. doi: 10.1093/nar/29.7.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 33.Kotler LE, Zevin-Sonkin D, Sobolev IA, Beskin AD, Ulanovsky LE. DNA sequencing: modular primers assembled from a library of hexamers or pentamers. Proc. Natl Acad. Sci. USA. 1993;90:4241–4245. doi: 10.1073/pnas.90.9.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynaldo LP, Vologodskii AV, Neri BP, Lyamichev VI. The kinetics of oligonucleotide replacements. J. Mol. Biol. 2000;297:511–520. doi: 10.1006/jmbi.2000.3573. [DOI] [PubMed] [Google Scholar]
- 35.de Vega M, Lazaro JM, Salas M, Blanco L. Mutational analysis of phi29 DNA polymerase residues acting as ssDNA ligands for 3′–5′ exonucleolysis. J. Mol. Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- 36.Skerra A. Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res. 1992;20:3551–3554. doi: 10.1093/nar/20.14.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsson M, Gullberg M, Dahl F, Szuhai K, Raap AK. Real-time monitoring of rolling-circle amplification using a modified molecular beacon design. Nucleic Acids Res. 2002;30:e66. doi: 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melin J, Jarvius J, Goransson J, Nilsson M. Homogeneous amplified single-molecule detection: characterization of key parameters. Anal. Biochem. 2007;368:230–238. doi: 10.1016/j.ab.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Jarvius J, Melin J, Goransson J, Stenberg J, Fredriksson S, Gonzalez-Rey C, Bertilsson S, Nilsson M. Digital quantification using amplified single-molecule detection. Nat. Methods. 2006;3:725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Hays JB. Simple and rapid preparation of gapped plasmid DNA for incorporation of oligomers containing specific DNA lesions. Mol. Biotechnol. 2001;19:133–140. doi: 10.1385/MB:19:2:133. [DOI] [PubMed] [Google Scholar]
- 42.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl Acad. Sci. USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Arruda M, Lyamichev VI, Eis PS, Iszczyszyn W, Kwiatkowski RW, Law SM, Olson MC, Rasmussen EB. Invader technology for DNA and RNA analysis: principles and applications. Expert Rev. Mol. Diagn. 2002;2:487–496. doi: 10.1586/14737159.2.5.487. [DOI] [PubMed] [Google Scholar]
- 44.Lyamichev V, Mast AL, Hall JG, Prudent JR, Kaiser MW, Takova T, Kwiatkowski RW, Sander TJ, de Arruda M, Arco DA, et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]