Abstract
Vpu is an 81-residue accessory protein of HIV-1. Because it is a membrane protein, it presents substantial technical challenges for the characterization of its structure and function, which are of considerable interest because the protein enhances the release of new virus particles from cells infected with HIV-1 and induces the intracellular degradation of the CD4 receptor protein. The Vpu-mediated enhancement of the virus release rate from HIV-1-infected cells is correlated with the expression of an ion channel activity associated with the transmembrane hydrophobic helical domain. Vpu-induced CD4 degradation and, to a lesser extent, enhancement of particle release are both dependent on the phosphorylation of two highly conserved serine residues in the cytoplasmic domain of Vpu. To define the minimal folding units of Vpu and to identify their activities, we prepared three truncated forms of Vpu and compared their structural and functional properties to those of full-length Vpu (residues 2–81). Vpu2–37 encompasses the N-terminal transmembrane α-helix; Vpu2–51 spans the N-terminal transmembrane helix and the first cytoplasmic α-helix; Vpu28–81 includes the entire cytoplasmic domain containing the two C-terminal amphipathic α-helices without the transmembrane helix. Uniformly isotopically labeled samples of the polypeptides derived from Vpu were prepared by expression of fusion proteins in E. coli and were studied in the model membrane environments of lipid micelles by solution NMR spectroscopy and oriented lipid bilayers by solid-state NMR spectroscopy. The assignment of backbone resonances enabled the secondary structure of the constructs corresponding to the transmembrane and the cytoplasmic domains of Vpu to be defined in micelle samples by solution NMR spectroscopy. Solid-state NMR spectra of the polypeptides in oriented lipid bilayers demonstrated that the topology of the domains is retained in the truncated polypeptides. The biological activities of the constructs of Vpu were evaluated. The ion channel activity is confined to the transmembrane α-helix. The C-terminal α-helices modulate or promote the oligomerization of Vpu in the membrane and stabilize the conductive state of the channel, in addition to their involvement in CD4 degradation.
Keywords: HIV-1, Vpu, membrane proteins, overexpression, NMR spectroscopy, ion channel activity
Vpu is an 81-residue membrane protein from human immunodeficiency virus type 1 (HIV-1) (Cohen et al. 1988; Strebel et al. 1988). It is a typical membrane protein, with one transmembrane hydrophobic helix and two amphipathic helices in its cytoplasmic domain. Our initial NMR studies of the protein enabled the correlation of the secondary structure and topology of its helices with the ion channel activity (Marassi et al. 1999). Here we describe the expression and purification of four different constructs of Vpu, with between 36 and 80 residues; we extend the spectroscopic and ion channel comparisons to the construct of the protein containing little more than the transmembrane helix, and, importantly, we demonstrate that the sequence we use in our studies has biological activities indistinguishable from the proteins found in HIV-1-infected humans. The structural biology of Vpu is of considerable interest, since it is a key participant in the HIV-1 lifecycle. It also serves as a model system for the study of helical membrane proteins. The sequences of many small helical membrane proteins are encoded in genomes (Arkin et al. 1997), and thus the methods for expression and purification of active forms of Vpu may contribute to the preparation of samples of other membrane proteins for spectroscopic and diffraction experiments.
Vpu is unique to HIV-1, the major causative agent of the acquired immune deficiency syndrome (AIDS). Although no structural counterpart is found in other primate lentiviruses such as HIV-2 or simian immunodeficiency virus (SIV) (Terwilliger et al. 1989), there is evidence that the viral particle release enhancement activity provided by Vpu has a functional equivalent in the Env protein in some HIV-2 isolates (Bour and Strebel 1996; Bour et al. 1999b). The viral envelope protein gp41, Vpr (Piller et al. 1999), and Vpu are the only HIV-1 proteins with transmembrane hydrophobic helices. Significant amounts of Vpu are found in the internal membranes of virus-producing cells; however, the protein cannot be detected in cell-free culture fluids and therefore it is most likely not virion-associated (Maldarelli et al. 1993; Strebel et al. 1989).
Vpu has two distinct biological activities: (1) It facilitates the degradation of the CD4 receptor in the endoplasmic reticulum (ER) of infected cells (Willey et al. 1992b) by targeting it for proteolysis by the ubiquitin-proteasome pathway (Margottin et al. 1998), and (2) it enhances the release of virus particles from the plasma membrane of infected cells (Strebel et al. 1988, 1989; Terwilliger et al. 1989). Both activities contribute to increased virion production (Bour et al. 1999a) and, hence, could explain the enhanced virulence of HIV-1 infections in humans compared to HIV-2 infections. These functions appear to be associated with two different structural domains of Vpu and two different molecular activities (Bour et al. 1995; Schubert et al. 1996a; Strebel 1996). The cytoplasmic domain of Vpu, which has two highly conserved phosphorylation sites (Ser52 and Ser56), is essential for interactions with CD4 and induction of CD4 degradation in the ER (Bour et al. 1995; Schubert et al. 1996a). The N-terminal transmembrane helix, which serves as a membrane anchor, is required to regulate virus secretion, most likely by formation of an ion channel (Schubert et al. 1996b). Our initial structural characterization of Vpu by NMR spectroscopy provides strong support for this domain organization (Marassi et al. 1999).
Determining the three-dimensional structure of Vpu is the essential first step towards understanding how its molecular activities affect the biological functions essential for the viral lifecycle. Further, the design of new classes of antiinfective agents (Miller and Sarver 1997) requires knowledge about the structures of the HIV-1 encoded proteins. Because Vpu is responsible for several important biological functions and is a relatively small protein, it is an attractive candidate for structure determination. NMR spectroscopy is capable of determining the structures of membrane proteins (Opella 1997); however, it requires milligram quantities of highly purified isotopically labeled protein. Here we describe the cloning, bacterial expression, isolation, and purification of isotopically labeled Vpu and several variants of Vpu. The biological activities and initial spectroscopic results demonstrate the purity, homogeneity, and correct folding of the protein samples. We extend our earlier results (Marassi et al. 1999) to include a construct containing little more than the transmembrane helix (Vpu2–37) and the characterization of the secondary structures of the domains based on experimental NMR data.
Results and Discussion
Biological activities of Vpu
The approach we developed to obtain pure protein from E. coli relies on cyanogen bromide (CNBr) cleavage of the polypeptide corresponding to Vpu from a fusion protein. The main advantage of CNBr is its high specificity for cleavage on the C-terminus of methionine residues. Since the N-terminal residue of native Vpu is methionine, this enables the release of an 80-residue polypeptide that differs from the native protein only in that it is missing the N-terminal residue. However, this also requires the absence of other methionine residues in the sequence, and it was necessary to replace the two internal methionine residues at positions 66 and 70 with leucines. Although methionines are not conserved at these positions of Vpu, we felt that it was necessary to demonstrate that the two well characterized biological functions of Vpu, that is, facilitation of virus particle release and induction of CD4 degradation, are similar for the 80-residue double-mutant VpuML (Vpu with M66L and M70L) and the native 81-residue Vpu.
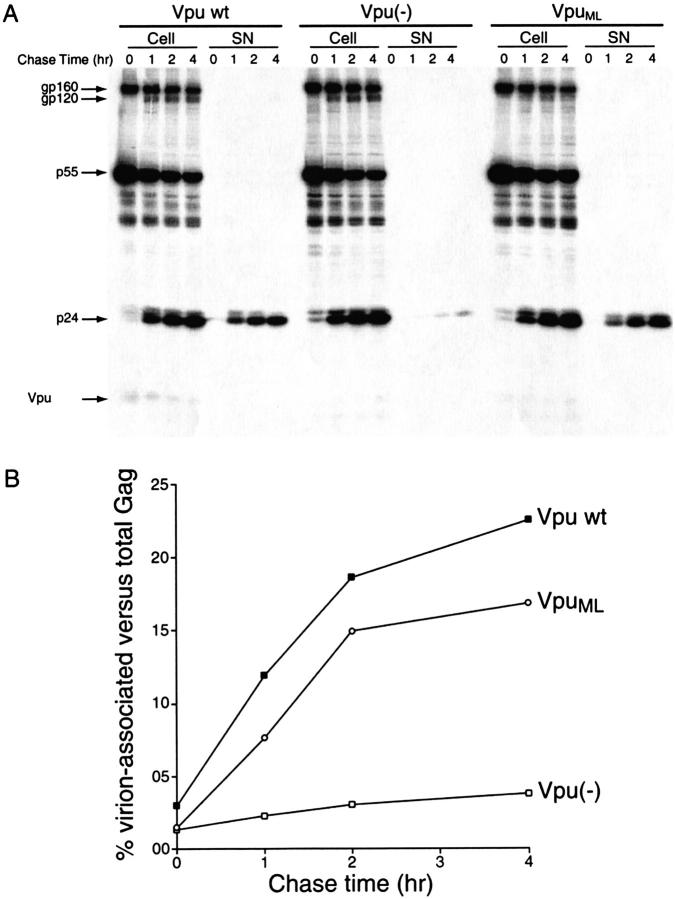
The abilities of VpuML and native Vpu to facilitate virus release were evaluated by studying the kinetics of processing and release of viral proteins from HeLa cells. Cells were transfected with plasmid DNAs encoding a Vpu-defective variant of the full-length molecular HIV-1 isolate NL4-3 (Adachi et al. 1986) together with pNL-A1 expressing wild-type Vpu (Vpu wt) or its isogenic variants pNL-A1/Udel expressing no Vpu-specific sequences (Vpu (−)), or pNL-A1/UML expressing the mutant VpuML (VpuML). Approximately 20 hours after transfection a pulse/chase analysis was performed, as described in the Materials and Methods section. Aliquots of cells were collected at the times indicated in Figure 1 ▶. Viral proteins present in the cell lysates and the viral pellet fractions were immunoprecipitated with HIV-1-positive human patient serum, separated by SDS-PAGE, and analyzed by fluorography, as shown in Figure 1A ▶. As previously reported (Willey et al. 1992 a,b), the absence of Vpu leads to a remarkable decrease in the amount of viral p24 protein secreted into the medium in the form of pelletable virions. This can be seen in Figure 1A ▶ by comparing the panels labeled "Vpu wt" and "Vpu(−)". In contrast, in cells transfected with the VpuML-expressing construct, viral particle release was efficient and virion proteins were readily detectable in the virion (SN) fraction throughout the chase period (Fig. 1A ▶, panel labeled "VpuML"). As shown in Figure 1B ▶, the efficiency of particle release was determined for each timepoint as the percentage of virus-associated p24gag and p55gag proteins compared to the total amount of gag proteins in the cell and supernatant fractions. The release of virus particles in the presence of the mutant VpuML was efficient (17% after a 4-h chase) and similar to that observed in the culture expressing wild-type Vpu (22%). The wt Vpu and VpuML cultures released 4.25 and 5.5 times, respectively, more Gag proteins over a 4-h chase period than did the Vpu-deficient culture, which had only 4% particle release efficiency. Thus, the mutations introduced in VpuML do not compromise its ability to support virus particle release. The data in Figure 1 ▶ also demonstrate that expression of VpuML was detectable by immunoprecipitation, and similar to that observed for wild-type Vpu.
Fig. 1.
VpuML supports virus particle secretion. (A) Parallel cultures of HeLa cells (2.5 × 106) were transfected with 3 μg of the Vpu-defective molecular clone pNL4-3/Udel together with the Vpu-expressing constructs pNL-A1 (+Vpu), pNL-A1/Udel (−Vpu), or pNL-A1/UML (+VpuML). Cells were labeled for 30 min with 250 μCi 35S-methionine, and a pulse/chase experiment was conducted as described in the Materials and Methods section. Viral proteins from the lysates of cells and pelleted virions were immunoprecipitated with an HIV-1-reactive human serum, separated on 12% SDS-polyacrylamide gels, and analyzed by fluorography. Fluorograms depicting cell lysates (Cell) and virion fractions (SN) are shown. HIV proteins are indicated on the left. (B) p24gag and p55gag proteins detected in the cell lysates and the virus pellet were quantified by image analysis. The efficiency of viral particle release was calculated as the percentage of Gag proteins (p24gag and p55gag) present in the virus pellet relative to the total Gag proteins detected intra- and extracellularly for each timepoint and were plotted as a function of time.
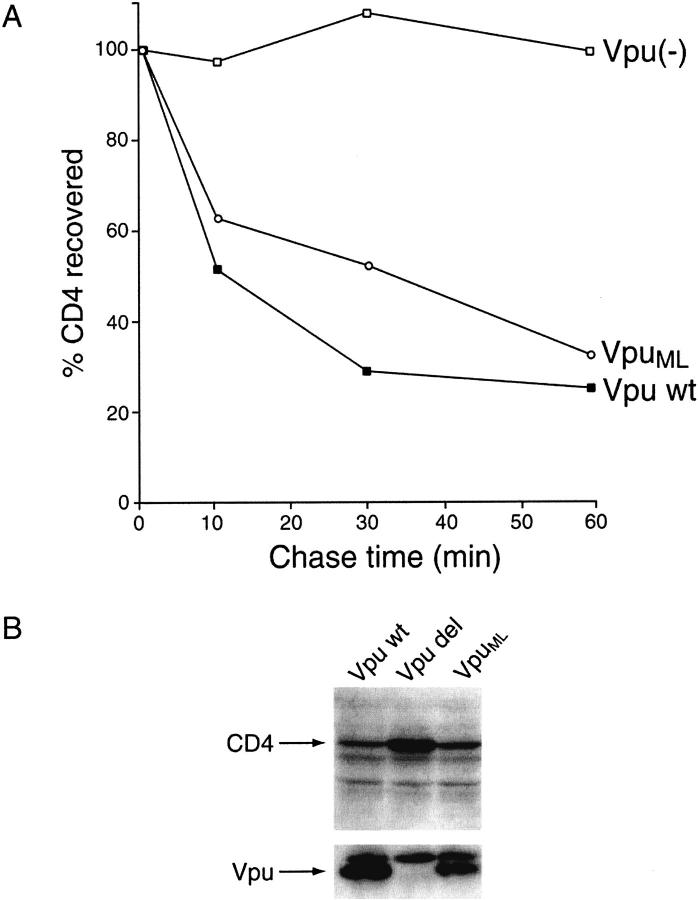
To evaluate the impact of the alterations introduced into the cytoplasmic portion of VpuML on its ability to induce degradation of CD4, the stabilities of CD4 in the presence of wild-type Vpu and of the mutant VpuML were compared using both pulse/chase (Fig. 2A ▶) and steady-state analyses (Fig. 2B ▶) of transfected HeLa cells. The Vpu-defective variant Vpudel was used as a negative control in these experiments. HeLa cells were cotransfected with pHIV-CD4 plasmid DNA expressing human CD4 and one of the following plasmids: pNL-A1, which directs the expression of wild-type Vpu (Vpu wt); pNL-A1/Udel, which lacks Vpu expression (Vpu(−)); or pNL-A1/UML, which directs the expression of the mutant VpuML (VpuML). Approximately 24 h after transfection, cells were pulse-labeled with 35S-methionine for 10 min and chased for up to 1 h in complete medium. Equal aliquots from cell lysates were immunoprecipitated with a CD4-specific polyclonal antiserum, separated by 10% SDS-PAGE, and analyzed by fluorography (data not shown). The gels were quantified using an image analyzer, and the amounts of CD4 remaining at different timepoints were calculated relative to the levels of CD4 present at the end of the pulse timepoint (0 min), which was defined as 100% (Fig. 2A ▶). Consistent with previous reports (Willey et al. 1992a,b), wild-type Vpu induced a rapid decay of CD4 within the first 10 min of the chase period; only ∼28% of the initial pulse-labeled CD4 remained at the end of the 1-h chase period. A similar rate of decay of CD4 was observed in the presence of the mutant VpuML; in the absence of Vpu, the amount of CD4 remained stable throughout the chase period.
Fig. 2.
Efficiency of VpuML to induce CD4 degradation is comparable to that of wild-type Vpu. (A) HeLa cells (5 × 106) were cotransfected with the CD4 expression plasmid pHIV-CD4 in combination with pNL-A1/UML (VpuML), pNL-A1 (wt Vpu), or pNL-A1/Udel [Vpu(−)] plasmid DNAs (24 μg) using the calcium phosphate procedure as described (Willey et al. 1992a,b). HeLa cells were pulse-labeled for 10 min with 35S-Translabel™ and chased for the indicated times. Detergent lysates of cells were immunoprecipitated with a polyclonal rabbit antiserum to CD4, separated on a 10% SDS-polyacrylamide gel, and visualized by fluorography (not shown). CD4-specific bands were quantified by image analysis, and the amounts of CD4 remaining at individual chase-times relative to the amounts of CD4 remaining at the end of the pulse (time 0) were plotted as a function of time. (B) Steady-state analysis of CD4 in Vpu-expressing cells. HeLa cells were transfected as described in the legend of Fig. 4A ▶. Cell were harvested 24 h after transfection, and cell lysates were subjected to immunoblotting using CD4- or Vpu-specific antisera. CD4- or Vpu-specific protein bands are identified on the right.
These findings were confirmed by comparing the steady-state levels of CD4 in cells expressing wild-type Vpu (Vpu wt), no Vpu (Vpu del), or the mutant VpuML (VpuML). HeLa cells were transfected as described in Figure 2A ▶. Approximately 24 h after transfection, cells were harvested and lysed as described in the Materials and Methods section. Cell lysates were separated by 12.5% SDS/PAGE, transferred to Immobilon membranes and reacted with CD4- or Vpu-specific antisera (Fig. 2B ▶). The steady-state levels of CD4 in cells expressing wild-type Vpu or VpuML were significantly lower than in cells without Vpu expression. The levels of expression of Vpu and VpuML were similar.
Taken together, these results clearly indicate that the amino acid substitutions involving the two internal methionine residues did not significantly alter the ability of Vpu to induce degradation of CD4 or facilitate virus particle release.
Vector constructions, expression, and purification
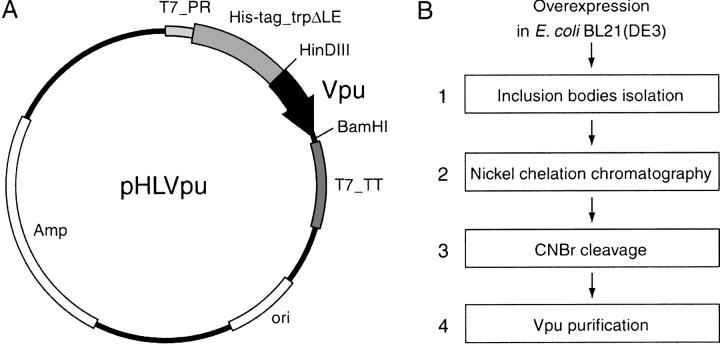
This approach to the production of Vpu in E. coli may be generally applicable to other membrane proteins, and we have used it successfully with several other small membrane proteins. For the production of Vpu, the vpu gene was cloned into the pMMHa (Staley and Kim 1994) prokaryotic expression vector. The resulting pHLVpu is illustrated in Figure 3A ▶. The His-tag_trpΔLE-polypeptide cleavage-site Vpu fusion protein produced by pHLVpu forms inclusion bodies when expressed in E. coli, and is thus protected from proteolysis. The fusion protein is not toxic to the E. coli host cell, and is expressed at levels up to 20% of total cellular protein in E. coli strain BL21(DE3). All of the variants of recombinant Vpu, which include polypeptides encompassing residues 2–81, 2–51, 2–37, or 28–81, were produced with this approach, and they are referred to as Vpu2–81, Vpu2–51, Vpu2–37, and Vpu28–81, respectively.
Fig. 3.
Schematic representation of the expression vector and purification procedure. (A) The expression vector pHLVpu. T7_PR, T7 promoter; T7_TT, T7 terminator; His-tag_trpΔLE, fusion partner; HinDIII and BamHI, restriction enzyme sites; ori, replication origin; Amp, ampicillin resistance gene. (B) Four-step purification procedures as detailed in Materials and Methods.
After expression, the separation and purification of Vpu were accomplished with the four-step protocol described in greater detail in the Materials and Methods section, and shown schematically in Figure 3B ▶: (1) The inclusion bodies containing the fusion protein were separated from the E. coli lysate by centrifugation, (2) the fusion protein was isolated by nickel affinity chromatography, (3) the Vpu polypeptide was cleaved from the fusion protein using CNBr, and (4) reverse-phase HPLC was performed to purify Vpu2–81, Vpu2–51, and Vpu2–37. The cytoplasmic construct Vpu28–8 without the hydrophobic transmembrane helix was isolated by ion-exchange and size-exclusion chromatography, followed by reverse-phase HPLC.
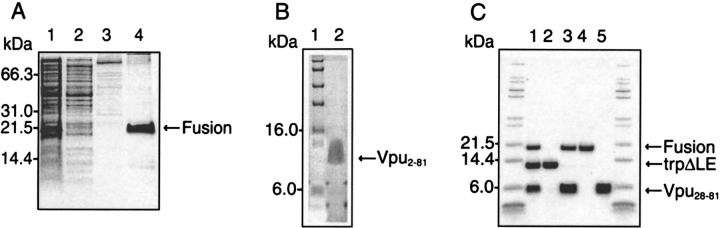
The data in Figure 4A ▶ illustrate the expression and isolation of inclusion bodies of full-length Vpu2–81. After the 4-h induction period, the cells were lysed by sonication, and the total cellular protein is shown in lane 1 of a 12% SDS-PAGE. Lanes 2 and 3 contain the soluble fractions, which were discarded after centrifugations, and lane 4 shows that the insoluble fraction (inclusion bodies) consists primarily of the fusion protein of interest. The gel of isolated Vpu2–81 after CNBr cleavage and HPLC purification is shown in Figure 4B ▶. The diffuse nature of the band is typical of a purified membrane protein under these conditions. Vpu2–51 and Vpu2–37 were purified with similar results. The less hydrophobic cytoplasmic construct Vpu28–81 was treated somewhat differently; the gel in Figure 4C ▶ demonstrates the separation of trpΔLE (lane 2) from the fusion protein plus Vpu28–81 (lane 3) by ion-exchange chromatography. The polypeptide Vpu28–81 (lane 5) was then isolated from the fusion protein (lane 4) by size-exclusion chromatography.
Fig. 4.
Examples of SDS-PAGE analyses of purification procedures as detailed in Materials and Methods. (A) Isolation of Vpu2–81 inclusion bodies. Lane 1, the total cell protein 4 h after induction for expression; lanes 2 and 3, discarded soluble fractions; lane 4, inclusion bodies fraction. (B) Purified Vpu2–81. (C) Purification of Vpu28–81. Lane 1, mixtures after CNBr cleavage reaction; lanes 2 and 3, the separation of trpΔLE from fusion and Vpu28–81; lanes 4 and 5, the separation of Vpu28–81 from the fusion protein by size-exclusion chromatography.
The compositions of the isolated proteins were confirmed by mass spectrometry. The final yields of purified Vpu2–81, Vpu2–51, Vpu2–37, and Vpu28–81 from 1L of minimal media were 1.0, 0.7, 0.7, and 8.0 mg, respectively. These purified recombinant proteins were used for solution NMR studies of the protein in phospholipid micelles, solid-state NMR studies of the protein oriented in phospholipid bilayers, and single-current measurements of the protein reconstituted in phospholipid bilayers. In separate studies, they are being used for X-ray reflectivity studies (Zheng et al. 2000) and crystallization trials.
Solution NMR and solid-state NMR studies of Vpu
The initial solution NMR and solid-state NMR studies of Vpu provided valuable information about the structure and topology of the protein (Marassi et al. 1999). The solid-state NMR spectra show that Vpu has two distinct structural domains, an N-terminal transmembrane helix and a C-terminal cytoplasmic domain with two amphipathic in-plane helices. The transmembrane helix has a tilt angle of 15° (Marassi et al. 1999) in lipid bilayers. Solution NMR spectroscopy is being used to determine the secondary structure and three-dimensional fold of Vpu in micelles. Extensive studies of sample conditions were performed to fully utilize the advantages of solution NMR spectroscopy, such as sharp lines and high resolution. Many different lipids were screened, and DHPC micelles were found to yield the best resolved solution NMR spectra. Uniform and selective labeling of all of the Vpu constructs with 2H, 15N, and 13C was performed by growing the bacteria on the appropriate media. Despite the favorable properties of the DHPC micelles, relatively high sample temperatures, and isotopic labeling, the broad lines and spectral overlap in the spectra of this highly helical protein made it very difficult to assign the backbone and side-chain resonances. The availability of the various truncated forms of the protein was essential for dealing with the limitations in the spectral data. A composite analysis of the spectra, in particular of Vpu2–37 and Vpu28–81, led to the backbone assignments of Vpu that enabled the experimental description of the secondary structure.
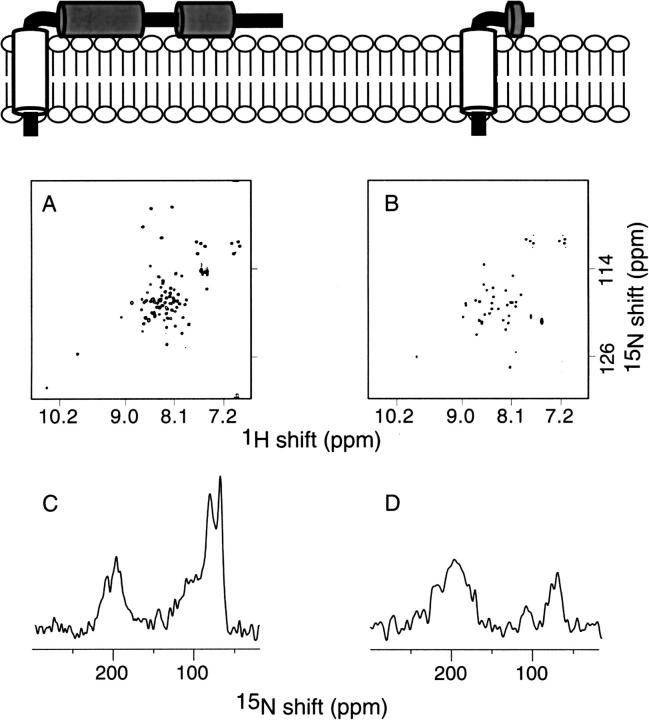
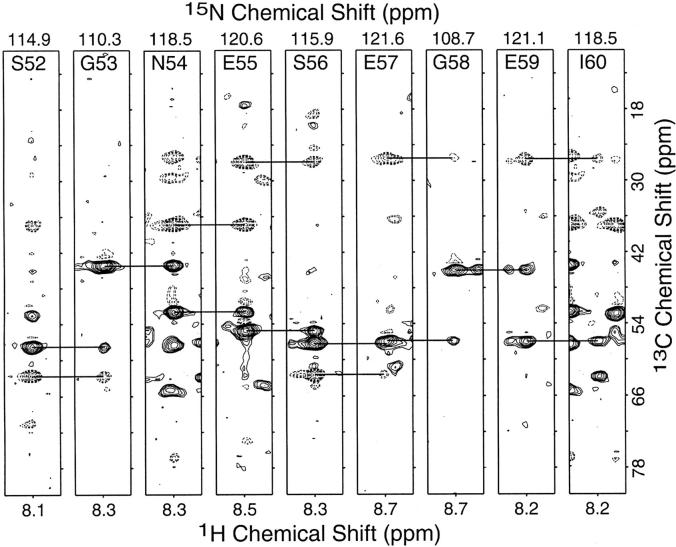
Solution NMR and solid-state NMR spectra of uniformly 15N-labeled Vpu28–81 and Vpu2–37 are compared in Figure 5 ▶. These data are complemented by those in Figure 8 ▶ and extend our structural and functional comparisons of constructs of Vpu (Marassi et al. 1999) to include Vpu2–37. Each correlation resonance in the two-dimensional solution NMR spectra (Fig. 8A,B ▶) represents a single 15N-labeled site of the polypeptides in DHPC micelles in aqueous solution. Solution NMR spectroscopy of membrane proteins in micelles is feasible because the polypeptides reorient fast enough in solution to give isotropic spectra with relatively narrow line widths. Each resonance is characterized by 1H and 15N chemical shift frequencies that reflect the local environment of the site in the protein. As expected, there are many fewer resonances in the spectrum from Vpu2–37 than that from Vpu2–81; however, many of the resonances that are in the spectrum of Vpu2–37 (Fig. 5B ▶) are superimposable on resonances found in the spectrum of Vpu2–81 (Fig. 5A ▶). This suggests that residues in the transmembrane domain of Vpu have essentially identical local environments whether or not the cytoplasmic domain is present. Membrane proteins in lipid bilayers are immobile on NMR timescales; therefore, the resonances are not motionally averaged and their frequencies reflect the orientation of the sites relative to the direction of the magnetic field. The resonance intensity near 200 ppm arises from amide sites in the transmembrane helix that have their N–H bonds approximately parallel to the field, and that near 70 ppm is from sites with N–H bonds orthogonal to the field, as found in in-plane helices. There is a substantial difference in the intensity of the resonance bands near 70 ppm in the spectra of Vpu2–37 and that from Vpu2–81. Although the smaller polypeptide has the same number of residues in its transmembrane helix as full-length Vpu, it has many fewer residues in the cytoplasmic portion, which consists of two in-plane helices in full-length Vpu. The NMR data show that the transmembrane and cytoplasmic domains fold independently and have little effect on the folding or membrane interactions of each other. Figure 6 ▶ contains spectral strips extracted at individual amide 15N chemical shift frequencies from a three-dimensional HNCACB spectrum (Wittekind and Mueller 1993) obtained from a sample of 50% 2H, U-13C, 15N cytoplasmic domain Vpu28–81 in DHPC micelles. The horizontal bars indicate the connectivities between Cα (positive contours, solid lines) as well as Cβ (negative contours, dashed lines) resonances from residues Ser52 through Ile60 of Vpu28–81.
Fig. 5.
NMR spectra of uniformly 15N-labeled Vpu2–81 and Vpu2–37 proteins in micelles (top) and oriented bilayers (bottom). A and B are two-dimensional HSQC NMR spectra. (A) Vpu2–81 (B) Vpu2–37. C and D are one-dimensional solid-state 15N NMR spectra. (C) Vpu2–81 (D) Vpu2–37.
Fig. 8.
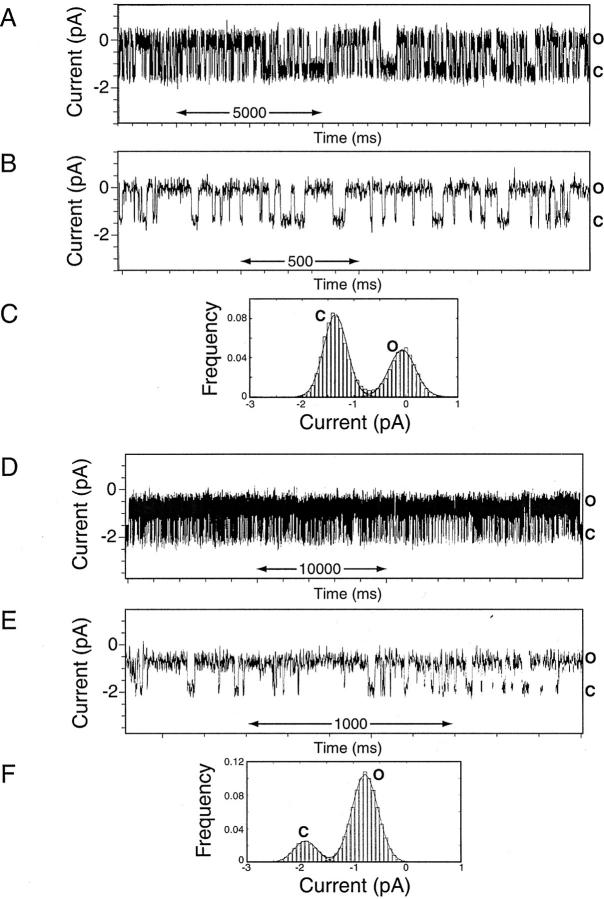
Single-channel recordings from Vpu2–37 and Vpu2–81 proteins reconstituted in lipid bilayers. (A–C) Vpu2–37. (D–F) Vpu2–81. (A and D) Single-channel recordings were obtained + 100 mV from preformed lipid bilayers in symmetric 0.5 M KCl-supplemented 10 mM Hepes, pH 7.4 following the addition of 5 μg/mL Vpu2–37 and Vpu2–81 proteins. The currents of the closed (C) and open (O) states are indicated; an upward deflection indicates channel openings. (B and E) A segment of the recording is displayed at faster time resolution. (C and F) Current histogram and Gaussian fits results obtained from continuous segments of records lasting several minutes; closed (C) and open (O) states are indicated. The records were derived from bursts of γ 12 pS channel activity occurring in the absence of 22 pS channels. For Vpu2–37, the probability of the channel being closed was Pc = 0.61 and open Po = 0.39; for Vpu2–81, Pc = 0.18 and Po = 0.82, under equivalent experimental conditions. The open channel lifetimes (τo) for the γ = 12 pS channel were well fitted by a sum of the two exponentials. For Vpu2–37, the τo1 = 1.2 ± 0.3 msec, τo2 = 22.6 ± 10.6 msec, and the closed times (τc) by τc1 = 8.1 ± 2.0, τc2 = 148.8 ± 34.4 msec (n = 7). For Vpu2–81, the τo1 = 1.8 ± 0.2 msec, τo2 = 20.8 ± 4.1 msec, and the closed times by τc1 = 1.3 ± 0.5 msec, τc2 = 23.3 ± 6.3 msec.
Fig. 6.
Strips from 3-D HNCACB experiment showing the connectivities from residues Ser52 to Ile60 from the cytoplasmic construct Vpu28–81 in DHPC micelles.
Secondary structure determination of Vpu
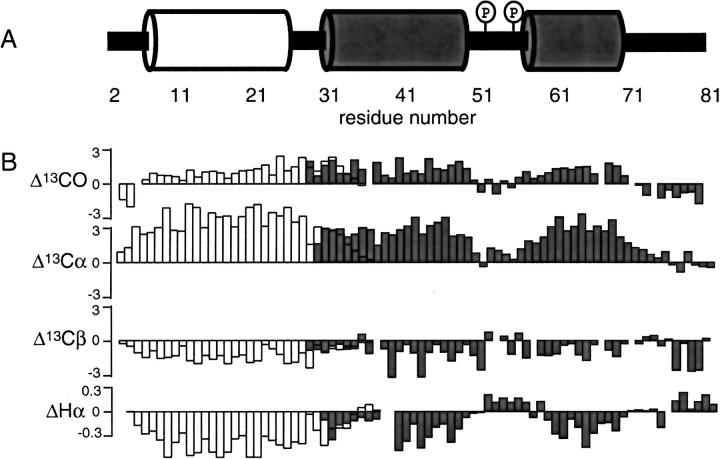
Backbone resonance assignments are necessary for determining the secondary structures of proteins by solution NMR spectroscopy. The secondary structure information is presented in the form of the chemical shift index in Figure 7B ▶ from 13C and 1Hα chemical shifts for Vpu2–37 (open bars) and Vpu28–81 (gray bars). From the analysis of the cytoplasmic construct, Vpu28–81, two regions corresponding to residues 30–49 and 58–70 show typical characteristics of α helices. The differences in the chemical shifts relative to their random coil values (downfield shifts for 13CO and 13Cα, upfield shifts for 13Cβ and 1Hα) are indicative of helical secondary structure (Wuthrich 1986; Wishart et al. 1995). On the other hand, Ser52 and Ser56, which must be phosphorylated for CD4 degradation activity, are located at the linker region (residues 51–57), which does not display spectral evidence of regular secondary structure. The resonances from the C-terminal region (residues 71–81) also do not provide evidence of regular secondary structure. Taken together, the solution NMR results show that the cytoplasmic domain consists of a 20-residue helix (residues 30–49), a linker region (residues 51–57) which contains the phosphorylation sites (Ser52 and Ser56), a 13-residue helix (residues 58–70), and the C-terminal region.
Fig. 7.
Summary of secondary structure determination of Vpu. (A) Schematic representation of secondary structure of Vpu. The numbering corresponds to the sequence of Vpu2–81, QPIQIAIAAL VVAIIIAIVV WSIVIIEYRK ILRQRKIDRL IDRLIERAED SGNESEGEIS ALVELGVELG HHAPWDVDDL. (B) Summary of secondary structure of Vpu determined based on the chemical shift differences relative to their random coil values. Data were taken from two Vpu constructs, Vpu2–37 (empty bars) and Vpu28–81 (gray bars).
The 36-residue N-terminal construct, Vpu2–37, is the smallest polypeptide containing the hydrophobic transmembrane helix that we have been able to express and purify. It is substantially shorter than the polypeptides we described previously (Marassi et al. 1999). The backbone assignments for the construct Vpu2–37 show that it contains a transmembrane hydrophobic helix. Hydrogen exchange on full-length Vpu2–81, which was monitored by the disappearance of amide proton resonances upon addition of D2O, confirms that the amide hydrogens for residues 9 to 29 have hindered exchange. This result indicates that the transmembrane helix includes residues 9 to 29. This 21-residue-long α-helix has two polar residues, Ser23 and Glu28, in the transmembrane region. For comparison, the transmembrane domain predictions from hydropathy plots (Kyte and Doolittle 1982) and the density alignment surface (DAS) method (Cserzo et al. 1997) suggest that the transmembrane helix consists of residues 6–27, and the molecular dynamic simulations suggested that residues 5–25 are the favorable transmembrane region (Fischer et al. 2000). The differences between the results of experiments and calculations may be due to the scarcity of known membrane protein structures, especially for relatively small proteins.
Ion channel activity of Vpu constructs
We previously described the ion channel activities of Vpu2–81, Vpu2–51, and Vpu28–81 (Marassi et al. 1999). The data in Figure 8 ▶ extend these studies with the comparison of the channel activities of Vpu2–37 and full-length Vpu2–81. Channel activity occurs in bursts separated by relatively long segments in which a single channel undergoes transitions between closed and open states. During the bursts of activity, the most frequent opening for Vpu2–37 has a single channel conductance, γ of 12 ± 2 pS, and a Po = 0.39 at V = 100 mV in 0.5 M KCl, pH 7.4 (Fig. 8A–C ▶). Full-length Vpu2–81 exhibits the same activity, although the two most frequent openings have γ = 22 ± 2 pS and 12 ± 2 pS; the data in Figure 8D–F ▶ show a burst in which only openings of 12 ± 2 pS occur. Segments of the recordings displayed at faster time resolution show the high signal-to-noise ratio of these records (∼5). Note, however, that the Po for Vpu2–81 is 0.82, which indicates that during the burst the channel is mainly in the open state. In contrast, for Vpu2–37, with similar γ = 12 ± 2 pS, the channel resides longer in the closed state. Simulations and modeling of Vpu suggest that the ion channel is formed by a pentamer of the polypeptides (Grice et al. 1997; Torres et al. 2001). Further, there is biochemical evidence of oligomerization (Bour et al. 1999a). Given the similar single-channel conductances of Vpu2–81 and Vpu2–37, it is reasonable to surmise that the cytoplasmic domain, consisting of the two amphipathic α helices, modulates the lifetime of the conductive oligomer and that in full-length Vpu, under these experimental conditions, the cytoplasmic domain promotes the residence of the oligomeric channel in the open state. This is reflected in a higher open channel probability and longer channel lifetimes in the open states. If this is indeed the case, it is highly significant because the oligomeric state of Vpu may be subject to regulation by cytosolic events in infected cells (Schubert et al. 1996a). Notably, the two serine residues in the linker between the two cytoplasmic helices can be phosphorylated by the protein kinase CK2. The cytoplasmic domain in the phosphorylated Vpu may, as a result of electrostatic repulsion, maintain the two helices aligned with the transmembrane domain and, therefore, preserve a patent conductive pathway of the oligomer.
Conclusions
The cloning, expression and purification of Vpu enable functional and structural studies of this membrane protein. The purified polypeptides can be obtained in quantities that are suitable for methods as demanding as NMR spectroscopy. The double mutant protein, with M66L and M70L, was shown to have the same biological activities as the wild-type protein. The backbone resonance assignments of constructs of Vpu in DHPC micelles led to the determination of the secondary structures of the domains. The transmembrane helical residues were identified. Moreover, from the comparison of channel recordings of different constructs, we identified a possible functional role for the two cytoplasmic helices of Vpu in the regulation of the oligomeric assembly and stability of the channels, in addition to their involvement in CD4 degradation.
Materials and methods
Construction of pNL-A1/UML
The double mutant VpuML, where the two internal methionine residues (M66 and M70) are substituted by leucines (M66 and 70L) was constructed by two-step PCR using pNL-A1 plasmid DNA as a template (Strebel et al. 1988). In the first step, two overlapping fragments (fragment A and fragment B) were synthesized. For fragment A, a segment corresponding to nucleotides 5740 to 6272 in the NL43 genome (Adachi et al. 1986) was amplified using oligonucleotides A5-[AATAAGAATTCTGCAAC AACTGCTG] and A3-[CCAGTTCCACCCCGAGCTCCACAA GTGCTG]. For fragment B, the sequence corresponding to nucleotides 6250 to 7258 in the NL43 genome was amplified using the B5-[GTGGAGCTCGGGGTGGAACTGGGGCACCAT GCTCCTTGGG] and B3-[ATTTGCTAGCTATCTGTTTTAAA GTGG] primers. The 3`-oligo of fragment A and the 5`-oligo of fragment B are largely complementary and contain the two nucleotide changes required to introduce the methionine to leucine conversions. In addition, an analytical SacI site was introduced into oligonucleotides A3 and B5. The two primary PCR products were gel purified and used in a 1:1 molar ratio as templates for a second round of PCR amplification using the A5 and B3 oligos as primers. The resulting 1.5 kb fragment was purified and digested with EcoRI and NheI. The resulting restriction fragment was cloned into the EcoRI/NheI sites of pNL-A1. The presence of the two desired mutations and the absence of secondary PCR-induced mutations were verified by sequence analysis of the entire PCR fragment.
The vpu-deficient plasmid pNLA1/Udel (Klimkait et al. 1990) and pNL-A1/U2/6 expressing the phosphorylation mutant Vpu2/6 (Schubert et al. 1994) were used as negative controls in some of the experiments. The plasmid pHIV-CD4, which allows the expression of wild-type CD4 under the control of the HIV-1 LTR, has been described (Willey et al. 1994).
Cells, transfection, and infection
HeLa cells (ATCC CCL2) were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). HeLa cells were grown to near confluence in 25 cm2 flasks (3 × 106 cells per flask) and transfected with a total of 6 μg of the appropriate plasmid DNA(s) using the Fugene reagent (Roche) as described by the manufacturer. For immunoblotting, cells were lysed in a buffer containing 50 mM Tris-hydrochloride (pH 8.0), 5 mM EDTA, 100 mM NaCl, 0.5% (w/v) CHAPS {3-[(3-cholamidopropyl)-dimethyl-ammonio]-1-propanesulfonate), 0.2% (w/v) deoxycholate (DOC)}. Sample buffer [2% sodium dodecyl sulfate (SDS), 1% 2-mercaptoethanol, 1% glycerol, 65 mM Tris hydrochloride pH 6.8)] was added to the cell lysates. Samples were boiled for 5 min and equal aliquots were separated by SDS-PAGE, followed by immunoblotting as described (Willey et al. 1992a).
Antisera and antibodies
Serum from an asymptomatic HIV-1 seropositive patient (TP serum) was used to detect HIV-1-specific proteins, including Vpu, by immunoprecipitation. The CD4 antigen was detected in Western blot and immunoprecipitation using the T4-4 rabbit polyclonal antibody. T4-4 was obtained from the AIDS Research and Reference Reagent Program and was originally contributed by Dr. R. Sweet (Deen et al. 1988).
Metabolic labeling, pulse chase, and immunoprecipitation
To study virus particle release, transfected HeLa cells were collected, washed once with PBS (10 mM phosphate buffer pH 7.4, 100 mM NaCl) and incubated for 15 min in methionine-free RPMI 1640 medium (Specialty Media, Lavalette, NJ) supplemented with 5% FCS to deplete the internal pool of methionine. Cells were pulse-labeled with 250 μCi 35S-methionine (ICN Biomedical, Costa Mesa, CA) for 30 min at 37°C. The medium was then removed, the cells were washed once in PBS, and equal aliquots were added to 300 μL of pre-warmed RPMI 1640/FBS for each timepoint of the chase period and incubated at 37°C. Cells were collected at the various timepoints and lysed in 400 μL of NP40-DOC buffer (20 mM Tris-HCl pH 8, 120 mM NaCl, 2 mM EDTA, 0.5% DOC, 1% NP40). The culture supernatants were filtered through 0.45 μm cellulose acetate Spin-X centrifuge tube filters (Corning Costar, Cambridge, MA) to remove remaining cells and cell debris. Virus particles were then pelleted from cell-free supernatants in a refrigerated Eppendorf microcentrifuge (4°C, 90 min, 16,000g). Pelleted virions were lysed in 400 μL of NP40-DOC buffer. Cell lysates were precleared by incubation at 4°C for 1 h with protein A-Sepharose beads (Sigma, St. Louis, MO) and immunoprecipitated with the TP patient serum. Immunoprecipitates were solubilized by boiling in sample buffer containing 2% SDS, 1% β-mercaptoethanol, 1% glycerol, 65 mM Tris-hydrochloride (pH 6.8) and separated by SDS-PAGE using 12% polyacrylamide gels. Gels were fixed, incubated for 20 min in Enlightning (NEN Research Products, Boston, MA) and dried. Radioactive bands were visualized by fluorography using Bio-Max MR films (Eastman Kodak, Rochester, NY). Quantitation of the relevant bands was performed using a Fujix BAS 2000 Bio-Image Analyzer.
CD4 degradation assay
HeLa cells were harvested 24 h posttransfection, rinsed once in PBS (10 mM phosphate buffer pH 7.4, 100 mM NaCl) and starved for 30 min in methionine- and cysteine-free RPMI 1640 medium. Cells were pulse-labeled for 10 min with 2 mCi/ml Trans 35S-Label and subjected to a chase for up to 1 h at 37°C in 1 mL of prewarmed RPMI/FBS for the indicated chase periods. Cells were collected and lysed in 400 μL of NP40-DOC buffer (20 mM Tris-HCl pH 8, 120 mM NaCl, 2 mM EDTA, 0.5% DOC, 1% NP40). Immunoprecipitation was performed as described above using the anti-CD4 polyclonal serum T4-4.
Construction of the expression vectors pHLVpu2–81, pHLVpu2–51, Vpu2–37, and pHLVpu28–81
The vpu gene of the HIV-1 isolate HTLVIIIB (Ratner et al. 1985) was amplified by PCR using the primers f_vpu-[TTCACAAGCT TAATGGTATGCAACCTATACAA] and r_vpu-[ATAACGGAT CCTTATTAGAGATCATCAACATC]. The amplified 277 bp fragment was digested with the restriction enzymes HindIII and BamHI (New England Biolabs, Beverly, MA), purified by agarose gel electrophoresis and ligated with the HindIII-BamHI-cleaved expression vector pMMHa. After transformation into E. coli strain DH5α (Novagen, Madison, WI) the 3794 bp vector pHLVpu was identified by restriction analysis, and the sequence of the cloned vpu fragment was confirmed by DNA sequencing. The plasmid pHLVpu was finally transformed into the E. coli strain BL21(DE3) (Novagen) (Studier and Moffat 1986). A stable transformed clone BL21(DE3):pHLVpu was screened for high-level expression of the fusion protein. Aliquots of the clone were stored as glycerol stocks (15% of glycerol) at −80°C.
For PCR site-directed mutagenesis of the expression vector, the procedures consist of two PCR amplification. First, two separate PCRs were performed by using (a) the primers f_vpu-[TTCACAAGCTTAATGGTATGCAACCTATACAA] and r_ml[GTGCCCCAGCTCCACCCCCAGCTCCAC], and separately (b) the primers f_ml-[GTGGAGCTGGGGGTGGAGCTGGGGCAC] and r_vpu-[ATAACGGATCCTTATTAGAGATCATCAACATC]. The amplified fragments were purified by agarose gel electrophoresis and mixed together to serve as the template for the second PCR amplification. Second, the primers f_vpu and r_vpu and the mixture of the two fragments generated in the first (a) and (b) reactions, which serves as the template, were utilized for the second PCR amplification using the same procedures. The amplified 277bp fragments were again digested with the restriction enzymes HindIII and BamHI, purified by agarose gel electrophoresis and ligated with the HindIII-BamHI-cleaved expression vector pMMHa. The consequent expression vector pHLVpu2–81 was identical to the vector pHLVpu except that the codon for Vpu Met66 and Met70, ATG, was mutated into CTG, the codon for leucine. The plasmid pHLVpu2–81 was also transformed into the E. coli strain BL21(DE3). A stable transformed clone BL21(DE3): pHLVpu2–81 was screened for high-level expression of the fusion protein. Aliquots of the clone were stored as glycerol stocks (15% of glycerol) at −80°C. A schematic representation of the expression vector pHLVpu2–81 is given in Figure 3 ▶.
In a similar approach, the expression vectors which carry the truncated constructs of Vpu (residues 2–51, residues 2–37 and residues 28–81) were also prepared and transformed into BL21(DE3), and resulted as BL21(DE3):pHLVpu2–51, BL21(DE3): pHLVpu2–37, and BL21(DE3): pHLVpu28–81.
Expression of recombinant Vpu2–81, Vpu2–51, Vpu2–37, and Vpu28–81
Five mL of LB media or minimal media (7.0 g/L Na2HPO4, 3.0 g/L KH2PO4, 0.5g/L NaCl, 0.1 mM CaCl2, 1 mM MgSO4, 50 mg/L thiamin, 1% LB (v/v), 10 g/L d-glucose, and 1 g/L (NH4)2SO4) containing 100 μg/mL of ampicillin was inoculated with 10 μL of the glycerol stock of BL21(DE3):pHLVpu2–81, BL21(DE3):pHLVpu2–51, or BL21(DE3):pHLVpu28–81. For expression of isotopically labeled proteins, 15N-(NH4)2SO4 and 13C-glucose (Cambridge Isotope Laboratories, Andover, MA) were either used as the sole nitrogen and carbon source, respectively, or used in combination for 13C/15N-labeled samples. For 2H/13C/15N-labeled samples, an appropriate percentage of D2O (50%–70%) was used together with the 13C/15N-supplemented media. After 5 h at 37°C, 1 mL of the culture was used to inoculate 100 mL of the media with ampicillin. The culture was incubated at 300 rpm at 37°C overnight. The 100mL culture was centrifuged at 6,000 rpm for 10 min at 4°C and the pellet resuspended into 1 L of media. The cells were cultivated by shaking at 37°C to obtain a cell density corresponding to an absorbance at 600 nm (A600) of 0.7. Expression of the His-tag_trpΔLE_Vpu fusion protein was induced by the addition of isopropyl-β-D-thiogalactoside (IPTG) to a final concentration of 0.4 mM. Shaking was continued for another four h at 37°C until the A600 reached 0.9. The cells were subsequently harvested by centrifugation at 6,000 rpm for 10 min at 4°C, and then stored at −80°C overnight.
Cell lysis and purification of the fusion protein
The cell pellets were resuspended in buffer I [50mM Tris pH 8.0, 15% glycerol (v/v), 1mM NaN3, and 50μg/mL lysozyme (Boehringer Mannheim)]. Cell lysis was accomplished by incubating the pellets with buffer I at room temperature for 10 min. The cell lysate was sonicated for 4 min on ice twice (duty cycle 30%, output control 5, Branson Sonifier 450, microtip) and then centrifuged at 17,000rpm for 30 min in an SS34 rotor (43,000g) at 4°C. The resulting supernatant was discarded and the pellet was resuspended and sonicated for 4 min on ice in Buffer II [50mM Tris pH 8.0, 1% deoxycholic acid (w/v), 1% IGEPAL CA-630 (Sigma) (v/v), 1mM NaN3] twice, and then centrifuged at 19,000rpm for 30 min in an SS34 rotor at 4°C. The resulting supernatant was discarded, and the pellet was sonicated for 4 min on ice in guanidine hydrochloride binding (GHB) buffer (6M GdnHCl, 5mM imidazole, 0.5M NaCl, 20mM Tris pH 8.0) twice. The resulting homogenous mixtures were transferred to GSA bottles and diluted 10-fold with Milli-Q H2O. Precipitate (inclusion bodies) was centrifuged at 13,000rpm (27,500g) for 1 h at 4°C. The resulting pellets (inclusion body fractions) were dissolved in GHB buffer and stored at 4°C overnight.
Nickel affinity chromatography (His.Bind Resin, Novagen) was used to separate the His-tag fusion protein from the inclusion body fractions. After the column was loaded with the inclusion body fractions, it was washed with 3 bed-volumes of GHB buffer. The His-tag fusion protein was eluted from the column with 2 bed-volumes of guanidine hydrochloride elute buffer (6M GdnHCl, 0.5M imidazole, 0.5M NaCl, 20mM Tris, pH 8.0).
The fractions containing the fusion protein were concentrated in an AMICON stir cell concentrator with a YM10 membrane (molecular weight cutoff (MWCO) 10 kD). The protein solution was dialyzed (MWCO 10 kD) against Milli-Q H2O and subsequently lyophilized and stored at −20°C before the cleavage reaction. Typically, 30–50 mg of fusion protein were obtained from 1 L of cell culture.
Cleavage of the fusion protein
Cyanogen bromide (CNBr) (Gross and Witkop 1961) was used to cleave Vpu from the His-tag_trpΔLE_Vpu fusion protein. The fusion protein powder was dissolved in 70% formic acid to a concentration of 10–20 mg/mL, and then a 10-fold molar excess of CNBr was added to the solution. The reaction was kept in the dark for 2 h at room temperature. The reaction mixture was dialyzed (MWCO 1 kD) immediately against Milli-Q H2O and subsequently lyophilized and stored at −20°C before the final purification steps.
Purification of recombinant Vpu2–81, Vpu2–51, Vpu2–37, and Vpu28–81
Recombinant Vpu2–81 and Vpu2–51 were purified by means of preparative reverse-phase high-pressure liquid chromatography (RP-HPLC). The lyophilized cleavage mixture was dissolved in triflouroethanol (TFE) with bath sonication for 10 min and then an equal volume of buffer A for HPLC (see below) was added to the solution. The purification of Vpu2–81 and Vpu2–51 was achieved using a Delta-Pak C4 column (15μ, 300Å, 7.8 × 300mm, Waters, Milford, MA) with a Waters Delta Prep 3000 Preparative Chromatography System. Protein elution was monitored at 220nm. Elution involved a 10-min wash with 80% buffer A, 20% buffer B [buffer A: 10% ACN (acetonitrile), 90% H2O, 0.1% TFA (trifluoroacetic acid); buffer B: 90% ACN, 10% H2O, 0.1% TFA], followed by a linear gradient to 100% buffer B over 60 min, at a flow rate of 3mL/min; 1-min fractions were collected. The fractions containing pure Vpu2–81 or Vpu2–51 were collected and the protein concentration was determined by measuring the UV absorbance of this solution at 280 nm. Typically, a yield of 1 mg of Vpu2–81 and 0.7 mg of Vpu2–51 was obtained from 1 L of cell culture. The fractions containing pure Vpu2–81 or Vpu2–51 were pooled, and a Rotovap was used to remove ACN and TFA. The pure recombinant proteins were lyophilized and stored at −20°C.
Vpu2–37 was purified in a manner analogous to the other transmembrane-containing constructs mentioned above. However, the increased hydrophobicity resulted in a noticeable decrease in solubility and a stronger tendency to irreversibly adsorb to the C4 reverse phase column; this necessitated the use of alternative gradient and injection conditions. The method was modified from one originally developed by Kukol and Arkin (1999) to purify a synthetic peptide corresponding to a similar region of Vpu. In the original protocol, the peptides were dissolved in TFA for injection onto a C4 column and eluted using a linear gradient from 100% buffer C to 100% buffer D (see below). Because of concern about the potential damage to the column resulting from injecting strong acid and the relatively poor yields, an alternative injection condition was utilized (Bollhagen et al. 1995). Briefly, 10 mg of cleavage product was dissolved in 1 mL of hexafluoro-2-propanol (HFIP) and sonicated until clear, approximately 10–15 min. Then, 4 mL of dichloromethane was added, and the resulting mixture was injected onto a Waters C4 column equilibrated with 90% buffer C, 10% buffer D (buffer C: 95% H2O, 3% 2-propanol, 2% ACN, 0.1% TFA; buffer D: 47% 2-propanol, 28% ACN, 20% TFE, 5% H2O, 0.1% TFA). Elution was achieved by a 120-min gradient to 100% buffer D at a flow rate of 3 mL/min. Fractions containing pure Vpu2–37 were pooled, and treated as described above. The identity of the resulting polypeptide was confirmed by mass spectrometry. The yield of Vpu2–37 was typically 0.7 mg/L culture.
For the isolation of Vpu28–81, the lyophilized cleavage mixture was dissolved in 8 M urea, 20 mM Tris pH 8.0, 50 mM NaCl and 1 mM NaN3 buffer, and then loaded onto a Q Sepharose Fast Flow (Pharmacia Biotech, Uppsala, Sweden) column. The Vpu28–81-containing fractions were collected while eluting with the same buffer with 500 mM NaCl. After the fractions were concentrated to 5 mg of protein per mL, they were applied onto a Superdex 75 HR 10/30 column (Pharmacia Biotech) running with 8 M urea, 20 mM Tris pH 8.0, 200 mM NaCl and 1 mM NaN3 with a flow rate of 0.7 mL/min on an FPLC system. The fractions corresponding to Vpu28–81 were further purified on a Delta-Pak C18 column (15μ, 300Å, 7.8 × 300mm, Waters) using the buffers and the same gradient described above for RP-HPLC. The ACN and TFA were removed and the protein was lyophilized to powder and stored at −20°C. The yield is 8 mg from 1 L of cell culture for Vpu28–81.
Solution NMR experiments
Solution NMR samples were prepared by mixing an appropriate amount of labeled protein with 270 μL of micelle solution containing 200 mM d40-DHPC (Cambridge Isotope Laboratories), 10% D2O, and 5 mM NaN3 at pH 4.0. The sample was spun in a microcentrifuge for 5 min, and the supernatant was transferred into a 5-mm Shigemi NMR tube (Shigemi, Tokyo, Japan) and placed in the magnet. All of the experiments were performed on either a Bruker DMX 600 or DMX 750 spectrometer equipped with pulsed field gradient, four rf channels and a 5-mm triple resonance probe. The HSQC, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH experiments were performed as described (Grzesiek and Bax 1992; Wittekind and Mueller 1993; Yamazaki et al. 1994; Mori et al. 1995) or with minor modifications for deuterated samples.
Solid-state NMR experiments
Two mg of protein was dissolved in 0.5 mL of 500mM lauryl sulfate (SDS) in water. Unilamellar vesicles were prepared by sonication of 100 mg of 4:1 dioleoyl phosphatidylcholine: dioleoyl phosphatidylglycerol in water. The protein solution was mixed with the vesicles, and 30 mL of water was added. The mixture was quickly frozen in liquid nitrogen and thawed at 22°C, and SDS was removed by dialysis. Planar bilayers oriented on glass slides were prepared from the reconstituted vesicles as described (Marassi et al. 1997). Solid-state NMR spectra were obtained on a home-built spectrometer with a Magnex 700/64 magnet, and on a Chemagnetics-Otsuka Electronics spectrometer with an Oxford 400/89 magnet, at 0°C, using single-contact cross-polarization as described (Marassi et al. 1997). In all NMR spectra, 15N and 1H chemical shifts were referenced to 0 ppm for liquid ammonia and tetramethylsilane, respectively.
Single-channel recordings in planar lipid bilayers
Lipid bilayers were assembled by apposition of two monolayers spread from a lipid solution in hexane as described (Schubert et al. 1996b; Opella et al. 1999). The lipids were diphytanoyl phosphatidylethanolamine and diphytanoyl phos-phatidylcholine (Avanti Biochemicals) at a 4:1 ratio in hexane (5 mg/mL). The aqueous subphase was composed of 0.5 M NaCl and 5 mM Hepes (pH 7.4). Purified recombinant polypeptides were dissolved in trifluoroethanol at 0.01 mg/mL and added to the aqueous subphase after bilayer formation. Single-channel currents were recorded at an applied voltage of 100 mV. Acquisition and analysis of single-channel currents were performed as described (Schubert et al. 1996b; Opella et al. 1999). Records were filtered at 1 kHz with an 8-pole Bessel filter (Frequency Devices, Haverhill, MA) and digitized at 0.1 msec per point using an Axon TL-1 interface (Axon Instruments, Foster City, CA). Data processing was performed with a pClamp 5.5 (Axon Instruments). The illustrated channel recordings are representative of the most frequently observed conductances under the specified experimental conditions. Single-channel conductance was calculated from Gaussian fits to current histograms, and the channel open and closed lifetimes were calculated from exponential fits to probability density functions using data from segments of continuous recordings lasting longer than 30 sec and with 300 events (means ± SEM). Openings shorter than 0.3 msec were ignored. Bilayer reconstitution experiments were performed at 24 ± 2°C.
Acknowledgments
This research was supported by grant PO1GM56538 from the National Institute of General Medical Sciences to S.J.O. and M.M. D.H.J. was supported by postdoctoral fellowships from the Medical Research Council of Canada. The research utilized the Resource for Solid-State NMR of Proteins supported by grant P41RR09731 from the Biomedical Research Technology Program, National Center for Research Resources, National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
HIV-1, human immunodeficiency virus type 1
AIDS, acquired immune deficiency syndrome
NMR, nuclear magnetic resonance
CNBr, cyanogen bromide
DHPC, dihexanoyl phosphatidylcholine
TROSY, transverse relaxation-optimized spectroscopy
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.37302.
References
- Adachi, A., Gendelman, H.E., Koenig, S., Folks, T., Willey, R., Rabson, A., and Martin, M.A. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin, I.T., Brunger, A.T., and Engelman, D.M. 1997. Are there dominant membrane protein families with a given number of helices? Proteins 28 465–466. [DOI] [PubMed] [Google Scholar]
- Bollhagen, R., Schmiedberger, M., and Grell, E. 1995. High-performance liquid-chromatographic purification of extremely hydrophobic peptides–transmembrane segments. J. Chromatogr. A. 711 181–186. [DOI] [PubMed] [Google Scholar]
- Bour, S., Perrin, C., and Strebel, K. 1999a. Cell surface CD4 inhibits HTV-1 particle release by interfering with Vpu activity. J. Biol. Chem. 274 33800–33806. [DOI] [PubMed] [Google Scholar]
- Bour, S., Schubert, U., and Strebel, K. 1995. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: Implications for the mechanism of degradation. J. Virol. 69 1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour, S. and Strebel, K. 1996. Human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 70 8285–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour, S.P., Aberham, C., Perrin, C., and Strebel, K. 1999b. Lack of effect of cytoplasmic tail truncations on human immunodeficiency virus type 2 ROD Env particle release activity. J. Virol. 73 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, E.A., Terwilliger, E.F., Sodroski, J.G., and Haseltine W.A. 1988. Identification of a protein encoded by the vpu gene of HIV-1. Nature 334 532–534. [DOI] [PubMed] [Google Scholar]
- Cserzo, E., Wallin, E., Simon, I., von Heijne, G., and Elofsson, A. 1997. Prediction of transmembrane alpha-helices in procariotic membrane proteins: The Dense Alignment Surface method. Protein Eng. 10 673–676. [DOI] [PubMed] [Google Scholar]
- Deen, K.C., McDougal J.S., Inscker, R., Folena-Wasserman, G., Arthos, J., Rosenberg, J., Maddon, P., Axel, J.R., and Sweet, R.W. 1988. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331 82–84. [DOI] [PubMed] [Google Scholar]
- Fischer, W.B., Forrest, L.R., Smith, G.R., and Samson, M.S. 2000. Transmembrane domains of viral ion channel proteins: A molecular dynamics simulation study. Biopolymers 53 529–538. [DOI] [PubMed] [Google Scholar]
- Grice, A.L., Kerr, I.D., and Sansom, M. 1997. Ion channels formed by HIV-1 Vpu: A modelling and simulation study. FEBS Lett. 405 299–304. [DOI] [PubMed] [Google Scholar]
- Gross E. and Witkop, B. 1961. Selective cleavage of methionyl peptide bonds in ribonuclease with cyanogen bromide. J. Am. Chem. Soc. 83 1510–1511. [Google Scholar]
- Grzesiek, S. and Bax, A. 1992. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114 6291–6293. [Google Scholar]
- Klimkait, T., Strebel, K., Hoggan, M.D., Martin, M.A., and Orenstein, J.M. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukol, A. and Arkin, I.T. 1999. Vpu transmembrane peptide structure obtained by site-specific Fourier transform infrared dichroism and global molecular dynamics searching. Biophys. J. 77 1594–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J. and Doolittle, R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157 105–132. [DOI] [PubMed] [Google Scholar]
- Maldarelli, F., Chen, M.Y., Willey, R.L., and Strebel, K. 1993. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol. 67 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi, F.M., Ma, C., Gratkowski, H., Straus, S.K., Strebel, K., Oblatt-Montal, M., Montal, M., and Opella, S.J. 1999. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc. Natl. Acad. Sci. 96 14336–14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi, F.M., Ramamoorthy, A., and Opella, S.J. 1997. Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers. Proc. Natl. Acad. Sci. 94 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin, F., Bour, S.P., Durand, H., Selig, L., Benichou, S., Richard, V., Thomas, D., Strebel, K., and Benarous, R. 1998. A novel human WD protein, h-bTrCP, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1 565–574. [DOI] [PubMed] [Google Scholar]
- Miller, R.H. and Sarver, N. 1997. HIV accessory proteins as therapeutic targets. Nat. Med. 3 389–394. [DOI] [PubMed] [Google Scholar]
- Mori, S., Abeygunaward, C., Johnson, M.O., and van Zijl, P.C.M. 1995. Improved sensitivity of HSQC spectra of exchange protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoid water saturation. J. Magn. Reson. 108 94–98. [DOI] [PubMed] [Google Scholar]
- Opella, S.J. 1997. NMR and membrane proteins. Nat. Struct. Biol. 4 845–848. [PubMed] [Google Scholar]
- Opella, S.J., Marassi, F.M., Gesell, J.J., Valente, A.P., Kim, Y., Oblatt-Montal M., and Montal M. 1999. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat. Struct. Biol. 6 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piller, S.C., Ewart, G.D., Jans, D.A., Gage, P.W., and Cox, G.B. 1999. The amino-terminal region of Vpr from human immunodeficiency virus type 1 forms ion channels and kills neurons. J. Virol. 73 4230–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner, L., Haseltine, W., Patarca, R., Livak, K.J., Starcich, B., Josephs, S.F., Doran, E.R., Rafalski, J.A., Whitehorn, E.A., and Baumeister, K. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313 277–284. [DOI] [PubMed] [Google Scholar]
- Schubert, U., Bour, S., Ferrer-Montiel, A.V., Montal, M., Maldarelli, F., and Strebel, K. 1996a. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, U., Ferrer-Montiel, A.V., Oblatt-Montal, M., Henklein, P., Strebel, K., and Montal, M. 1996b. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 398 12–18. [DOI] [PubMed] [Google Scholar]
- Schubert, U., Henklein, P., Boldyreff, B., Wingender, E., Strebel, K., and Porstmann, T. 1994. The human immunodeficiency virus type 1 encoded Vpu protein is phosphorylated by casein kinase-2 (CK-2) at positions Ser52 and Ser56 within a predicted a-helix-turn-a-helix-motif. J. Mol. Biol. 236 16–25. [DOI] [PubMed] [Google Scholar]
- Staley, J.P. and Kim, P.S. 1994. Formation of a native like subdomain in a partially folded intermediate of bovine pancreatic trypsin inhibitor. Protein Sci. 3 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel, K. 1996. Structure and function of HIV-1 Vpu. In Human Retroviruses and AIDS 1996: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences (eds. G. Myers, B.T. Korber, B.T. Foley, K-T Jeang, J.W. Mellors, S. Wain-Hobson), pp. III19–III27. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos.
- Strebel, K., Klimkait, T., Maldarelli, F., and Martin, M.A. 1989. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J. Virol. 63 3784–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel, K., Klimkait, T., and Martin, M.A. 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241 1221–1223. [DOI] [PubMed] [Google Scholar]
- Studier, F.W. and Moffat, B.A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned gene. J. Mol. Biol. 189 113–130. [DOI] [PubMed] [Google Scholar]
- Terwilliger, E.F., Cohen, E.A., Lu, Y., Sodroski, J.G., and Haseltine, W.A. 1989. Functional role of human immunodeficiency virus type 1 vpu. Proc. Natl. Acad. Sci. 86 5163–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, J., Kuko, A., and Arkin, I.T. 2001. Mapping the energy surface of transmembrane helix–helix interactions. Biophys. J. 81 2681–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey, R.L., Buckler-White, A., and Strebel, K. 1994. Sequences in the cytoplasmic domain of CD4 are necessary and sufficient to confer sensitivity to the human immunodeficiency virus type 1 Vpu protein. J. Virol. 68 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey, R.L., Maldarelli, F., Martin, M.A., and Strebel, K. 1992a. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 66 7193–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey, R.L., Maldarelli, F., Martin, M.A., and Strebel, K. 1992b. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J. Virol. 66 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D.S., Bigam, C.G., Holm, A., Hodges, R.S., and Sykes, B.D. 1995. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. J. Biomol. NMR 5 67–81. [DOI] [PubMed] [Google Scholar]
- Wittekind, M. and Mueller, L. 1993. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide protons and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. 101 201–205. [Google Scholar]
- Wuthrich, K. 1986. NMR of proteins and nucleic acids, J. Wiley, New York.
- Yamazaki, K., Lee, W., Arrowsmith, C., Muhandiram, D., and Kay L.E. 1994. A suite of triple resonance NMR experiments for the backbone assignments of 15N, 13C, 2H labeled proteins with high sensitivity. J. Am. Chem. Soc. 116 11655–11666. [Google Scholar]
- Zheng, S.Y., Strzalka, J., Ma C., Opella, S.J., Ocko, B.M., Blasie, J.K. 2000. Structural study of the HIV-1 accessory protein Vpu via synchrotron X-ray reflectivity from langmuir monolayers. Biophys. J. 78 945Pos. [DOI] [PMC free article] [PubMed]