Abstract
It was recently found that the myristoyl group of CAP-23/NAP-22, a neuron-specific protein kinase C substrate, is essential for the interaction between the protein and Ca2+-bound calmodulin (Ca2+/CaM). Based on the N-terminal amino acid sequence alignment of CAP-23/NAP-22 and other myristoylated proteins, including the Nef protein from human immunodeficiency virus (HIV), we proposed a new hypothesis that the protein myristoylation plays important roles in protein–calmodulin interactions. To investigate the possibility of direct interaction between Nef and calmodulin, we performed structural studies of Ca2+/CaM in the presence of a myristoylated peptide corresponding to the N-terminal region of Nef. The dissociation constant between Ca2+/CaM and the myristoylated Nef peptide was determined to be 13.7 nM by fluorescence spectroscopy analyses. The NMR experiments indicated that the chemical shifts of some residues on and around the hydrophobic clefts of Ca2+/CaM changed markedly in the Ca2+/CaM-Nef peptide complex with the molar ratio of 1:2. Correspondingly, the radius of gyration determined by the small angle X-ray scattering measurements is 2–3 Å smaller that of Ca2+/CaM alone. These results demonstrate clearly that Nef interacts directly with Ca2+/CaM.
Keywords: Myristoylation, calmodulin, HIV-1 Nef, NMR, protein protein interaction
The nef gene from human immunodeficiency virus type 1 (HIV-1) encodes a 25–30-kD myristoylated protein produced early during the infection by translation from several singly and multiply spliced mRNA species (Kim et al. 1989; Robert-Guroff et al. 1990; Purcell and Martin 1993; Schwartz et al. 1995). In the infected cells, Nef localizes predominantly at the plasma membrane and preferentially associates itself with the cytoskeleton (Franchini et al. 1986; Kienzle et al. 1992; Niederman et al. 1993), and it has been thought that one of the roles of the myristoylation is to anchor the Nef protein to the plasma membrane. Myristoylation is an absolute requirement for biological activity (Zazopoulos and Haseltine 1992; Skowronski et al. 1993; Chowers et al. 1994; Greenway et al. 1995).
Calmodulin (CaM) is a small calcium-binding protein (16.7 kD) involved in a wide range of cellular Ca2+-dependent signaling pathways through various enzymes, including protein kinases, protein phosphatases, nitric oxide synthase, inositol triphosphate kinase, nicotinamide adenine dinucleotide kinase, and cyclic nucleotide phosphodiesterase (Crivici and Ikura 1995). We recently found a novel mechanism of target recognition of CaM. It had been demonstrated that CAP-23/NAP-22 isolated from rat brain is Nα-myristoylated, and this modification is involved in its interaction with CaM in the presence of Ca2+ (Takasaki et al. 1999). The CaM-binding site was narrowed down to the myristoyl moiety together with the N-terminal basic domain of nine amino acid residues, GGKLSKKKK. The basic Nα-myristoylated peptide corresponding to the N-terminal CaM binding site of CAP-23/NAP-22 (mC/N9) clearly has different properties from known CaM-binding peptides. The most striking feature is that the binding of mC/N9 to CaM is dependent on the presence of the myristoyl moiety, whose general function so far has been assumed to be the membrane targeting of myristoylated proteins. Additionally, the interaction between mC/N9 and Ca2+/CaM has several other unique features: (1) mC/N9 does not resemble any canonical CaM-binding domain in the amino acid sequence, (2) mC/N9 is likely to adopt a nonhelical conformation even in the complex with Ca2+/CaM, and (3) the interaction is also controlled by phosphorylation of the peptide. All of these features suggest the novelty of the interaction. Very recently, a small angle X-ray scattering (SAXS) study revealed that large structural changes of Ca2+/CaM occur by the binding of mC/N9 to CaM (Hayashi et al. 2000). Moreover, the interaction between CAP-23/NAP-22 and CaM is controlled by phosphorylation of serine residue at the N-terminus. From these observations, we identified that mC/N9 is a new CaM binding motif, differing from the sequence motifs of calmodulin recognition such as motifs termed 1-8-14 and 1-5-10 based on the position of conserved hydrophobic residues or IQ motifs seen in many proteins (Rhoads and Friedberg 1997). Moreover, from the N-terminal amino acid sequence alignment of CAP-23/NAP-22 and other myristoylated proteins, we hypothesized that some myristoylated proteins, including HIV-1 Nef, bind to CaM through the N-terminal domain (Hayashi et al. 2000).
HIV-1 Nef has a sequence of GGKWSKPR at the N-terminus, which, in addition to the myristoylation, has some functionally important features similar to those of CAP-23/NAP-22 (GGKLSKKKK), the basic residues (lysine), and the target residue of protein kinase C (serine) (Hayashi et al. 2000). Thus, we hypothesized that there is a strong possibility that Nef interacts directly with CaM, as observed in CAP-23/NAP-22. As noted, the SAXS study is a useful method to detect direct interaction between target peptides and CaM through measurable structural changes of CaM (Matsushima et al. 2000). In the present study, to investigate the interaction, we performed structural studies of Ca2+/CaM in the presence of a myristoylated N-terminal octapeptide (myr-GGKWSKPR; Nef peptide) of Nef by nuclear magnetic resonance spectroscopy (NMR) and SAXS. The NMR and SAXS studies revealed that structural changes of Ca2+/CaM occurred in the myristoylated octapeptide, indicating clearly that Ca2+/CaM bound to the myristoylated octapeptide. The present study provides the first evidence of the direct interaction between HIV-1 Nef protein and CaM.
Although participation of HIV-1 Nef protein (Biggs et al. 1999) and CaM (Della Rocca et al. 1997) in the mitogen-activated protein (MAP) kinase cascade for the regulation of the intracellular signal transduction was individually implicated, the direct interaction between HIV-1 Nef protein and CaM has thus far not been indicated. The findings of the present study urge a reconsideration of the role of the protein myristoylation and provide insights into the function of Nef itself.
Results
Determination of dissociation constants by fluorescence spectroscopy
To clarify that the N-terminal domain is indeed the calmodulin-binding domain of Nef, synthetic myristoylated and nonmyristoylated peptides with different amino acid residues were prepared and tested for their abilities to bind calmodulin. Binding of peptides to calmodulin was carried out by measuring the fluorescence change of dansyl-calmodulin upon binding of the target peptide (Matsubara et al. 1997). The addition of equivalent mole of myristoylated peptide (myr-GGKWSKPR) to dansyl-calmodulin caused an increase in the intensity and a shift of the peak maximum of the emission spectra (data not shown). The dissociation constant was determined to be 13.7 nM. However, no significant change in the fluorescence spectra was observed when the nonmyristoylated peptide (GGKWSKPR) or the acetylated peptide (acetyl-GGKWSKPR) was added to calmodulin. These results indicate that the myristoyl moiety is directly involved in the Nef-calmodulin interaction.
NMR spectroscopy
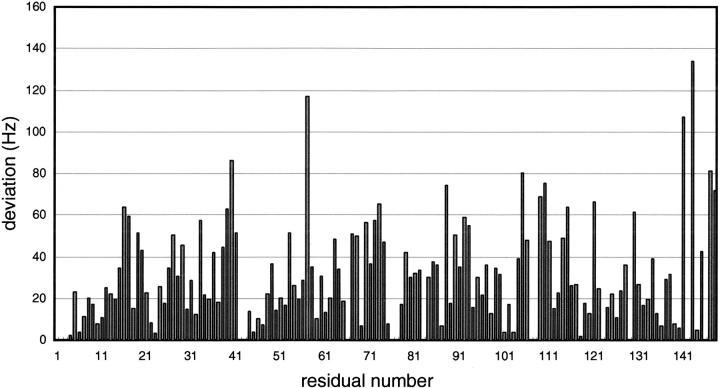
We studied the interaction between Nef peptide and Ca2+/CaM using two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectroscopy. The NMR spectra for Ca2+/CaM in the absence and presence of Nef peptide are shown in Figure 1 ▶. When 1 molar equivalent of the Nef peptide was added, some peaks were shifted slightly and no significant shift was observed in the 1H-15N HSQC NMR spectra of Ca2+/CaM. However, some marked shifts of the peak were observed with the addition of 2 molar equivalents of the Nef peptide (Fig. 1 ▶). The residues that showed larger shifts contained Gly40, Ala57, Leu105, and Phe141 (Fig. 2 ▶). As illustrated in Figure 3 ▶, their residues were primarily found in the hydrophobic clefts of the N- and C-domain of CaM.
Fig. 1.
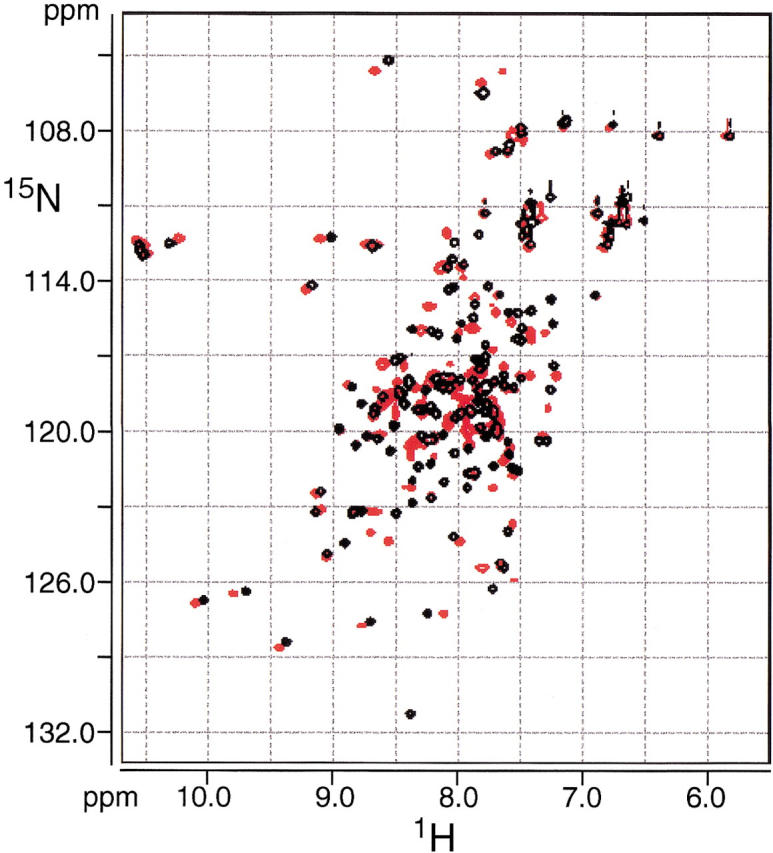
Overlay of 1H-15N HSQC spectra of Ca2+/CaM (black) and Ca2+/CaM-Nef peptide complex (red). The sample contained 0.5 mM CaM, 120 mM NaCl, 2.5 mM CaCl2, and 50 mM deuterated TrisHCl (pH7.5) in 90% H2O and 10% D2O.
Fig. 2.
The observed changes (Hz) in the 1H-15N HSQC cross-peak position upon forming Ca2+/CaM-Nef peptide complex. The changes in cross-peak positions were quantified by [(Δ15NHz)2 + (Δ1HHz)2]1/2.
Fig. 3.
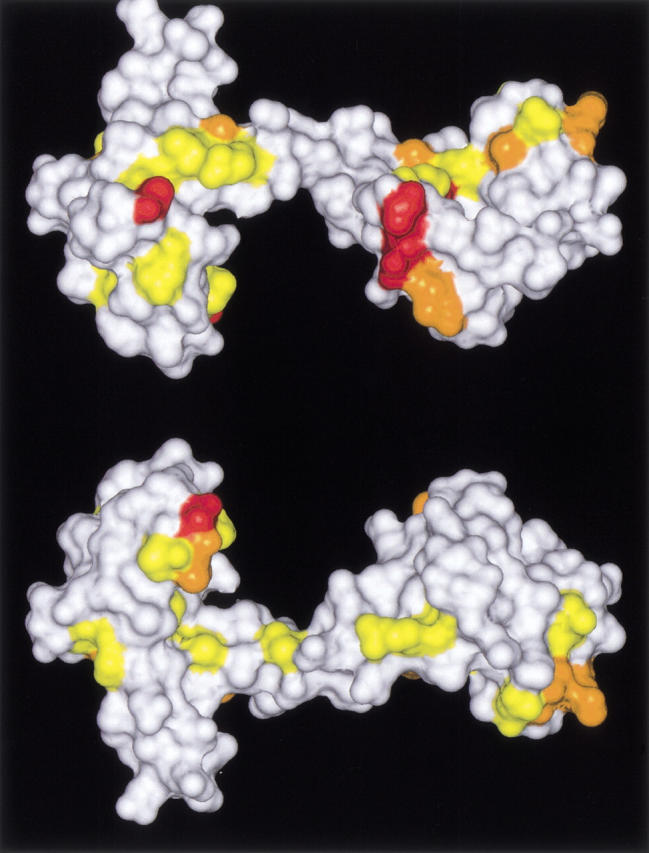
Arrangements on the Connolly surface of Ca2+/CaM (both sides) (Protein data bank accession number 1CLL) of residues whose chemical shifts were changed upon forming Ca2+/CaM-Nef peptide complex. Red, orange, and yellow indicate the locations of amino acids whose [(Δ15NHz)2 + (Δ1HHz)2]1/2 were over 80 (Gly40, Ala57, Leu105, Phe141, Gln143), between 80 and 60 (Phe16, Leu39, Ala73, Ala88, Met109, Thr110, Lys115, Val121, Ile 130, Lys148), and between 60 and 40 (Ser17, Phe19, Asp20, Ile27, Thr29, Gly33, Met36, Ser38, Gln41, Asn53, Ile63, Glu67, Phe68, Thr70, Met72, Arg74, Thr79, Arg90, Phe92, Asp93, Arg106, Asn111, Glu114, Met145), respectively. The models were constructed and rendered on an IRIS Indigo 2 workstation (SGI) using Insight II (Molecular Simulations) and SYBYL/BASE software (Tripos).
SAXS
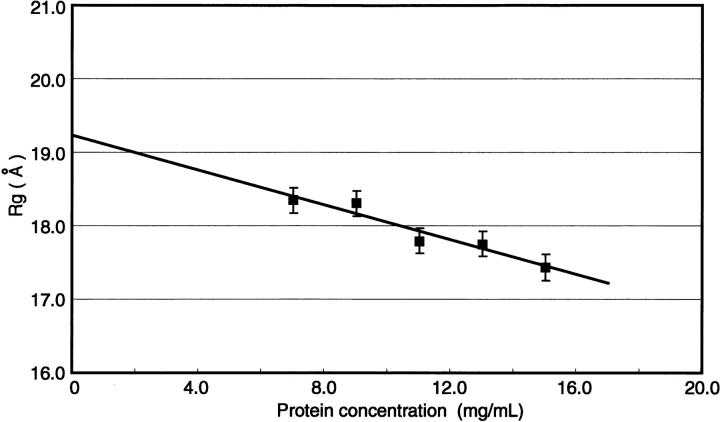
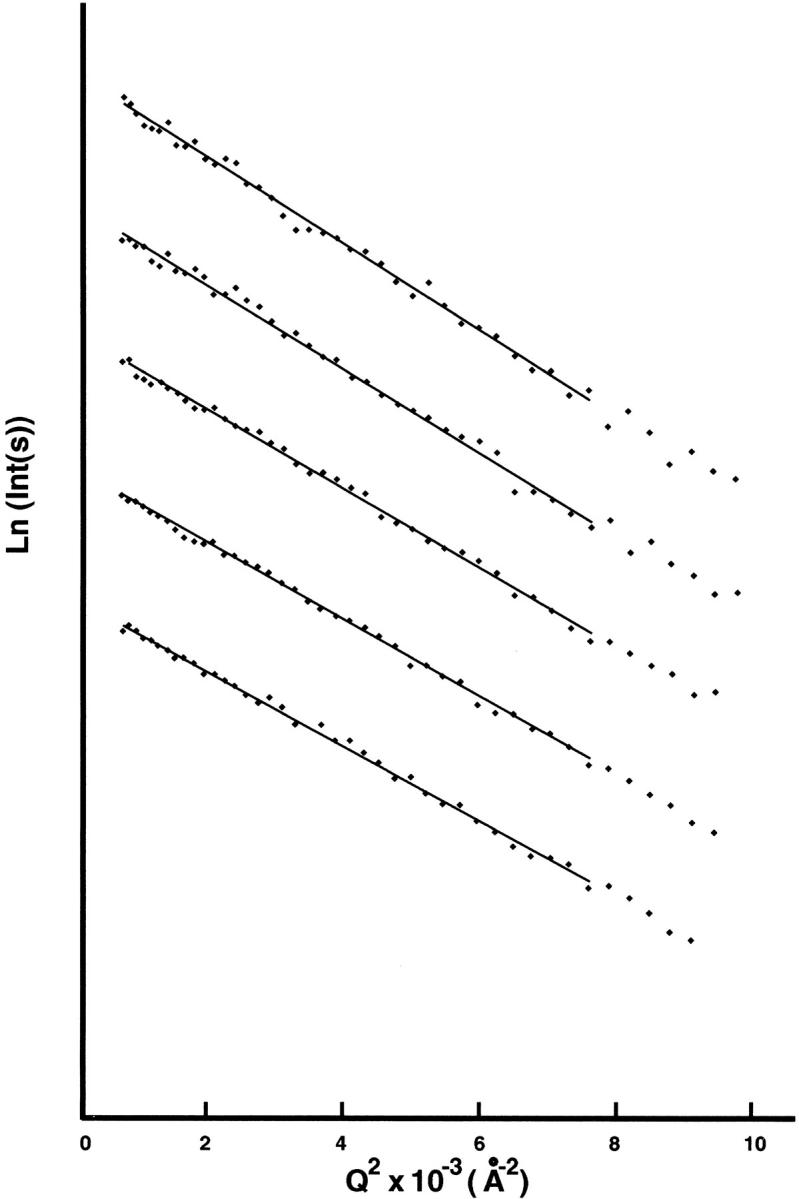
The Guinier plots for Ca2+/CaM-Nef peptide mixtures with the molar ratio of 1:2 at five protein concentrations are shown in Figure 4 ▶. Figure 5 ▶ shows the radius of gyration (Rg) as a function of protein concentration. The Rg values of the Ca2+/CaM-Nef mixtures are given in Table 1. For comparison, Table 1 also contains Rg values for other Ca2+/CaM complexes including Ca2+/CaM-mC/N9 complex (Hayashi et al. 2000). The Rg value for Ca2+/CaM-Nef complex (19.3±0.3Å) is consistent with that for Ca2+/Ca-mC/N9 complex. Both Rg values are smaller than that for Ca2+/CaM and larger than those for Ca2+/CaM complexes containing TFP, W-7, mastoparan, melittin, cyclosporin-A, substance P, and synthetic peptides corresponding to the calmodulin-binding domains of MLCK (M13), phosphorylase kinase (RhK5), and Ca2+-pump (C24W).
Fig. 4.
Guinier plots for Ca2+/CaM-Nef peptide complex (Ca2+/CaM-Nef peptide = 1:2). Protein concentrations: 7.0, 9.0, 11.0, 13.0, 15.0 mg/mL.
Fig. 5.
The radius of gyration, Rg, for Ca2+/CaM-Nef peptide complex (Ca2+/CaM-Nef peptide = 1:2).
Table 1.
Radius of gyration Rg and maximum dimension dmax for Ca2+/CaM-Nef peptide complex
| Rg[Å] | dmax[Å] | Reference | |
| Ca2+/CaM-Nef peptidea | 19.3 ± 0.3 | 58 | * |
| Ca2+/CaM-mC/N9a | 19.8 ± 0.3 | 50 | (Hayashi et al. 2000) |
| Ca2+/CaMa | 21.9 ± 0.3 | 62 | (Hayashi et al. 2000) |
| Ca2+/CaMa | 21.5 ± 0.3 | 69 | (Matsushima et al. 1989) |
| Ca2+/CaM-M13a | 16.4 ± 0.2 | 49 | (Heidorn et al. 1989) |
| Ca2+/CaM-W-7a | 17.6 ± 0.3 | 47 | (Osawa et al. 1999a) |
* This study.
a Values at zero protein concentration obtained by SAXS experiment.
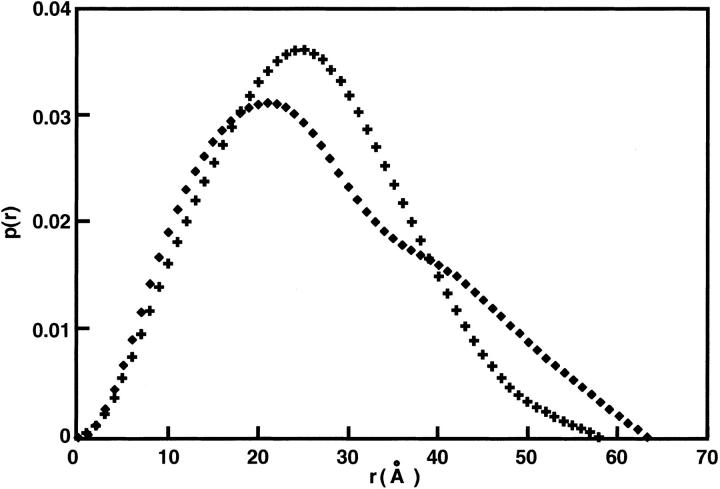
Figure 6 ▶ shows the pair distance distribution function (ITLp(r) ) for Ca2+/CaM in the presence of Nef-peptide at a molar ratio of 1:2 and Ca2+/CaM alone (Hayashi et al. 2000). The p(r) for Ca2+/CaM alone has a peak near 20Å (principally representing interatomic distances within each domain of Ca2+/CaM) and a shoulder near 40Å (mainly representing interdomain distances) (Matsushima et al. 1989). In contrast, in the Ca2+/CaM-Nef peptide complex, a peak appears not at 20Å but around 25Å. Moreover, the maximal pair distance (dmax) of the Ca2+/CaM-Nef peptide mixtures is about 4Å smaller than that of Ca2+/CaM in isolation. Additionally, the p(r) values of the Ca2+/CaM-Nef peptide mixtures are relatively symmetrical. These results of the Ca2+/CaM-Nef peptide complex correspond to those observed in other complexes including the Ca2+/CaM-mC/N9 complex. Thus, we conclude that Ca2+/CaM forms a complex with Nef peptide. The Ca2+/CaM complex with Nef peptide adopts a unique and larger globular structure that is not similar to those of the other known Ca2+/CaM complexes.
Fig. 6.
Pair distance distribution function, p(r), for Ca2+/CaM-Nef peptide complex (Ca2+/CaM-Nef peptide = 1:2). (Cross), Ca2+/CaM-Nef peptide complex (Ca2+/CaM-Nef peptide = 1:2); (Square), Ca2+-saturated CaM alone (Hayashi et al. 2000; Matsushima et al. 2000).
Discussion
Direct protein–protein interaction between Ca2+/CaM and HIV-1 Nef peptide
The addition of equivalent mole of myristoylated N-terminal octapeptide of HIV-1 Nef (myr-GGKWSKPR) to dansyl-calmodulin caused the fluorescence change as in peptides derived from other CaM-binding proteins (Matsubara et al. 1997; Takasaki et al. 1999), and the dissociation constant was determined to be 13.7 nM. Although this value suggests that the binding of the myristoylated Nef peptide to Ca2+/CaM is among the highest of CaM-binding proteins (O'Neil and DeGrado 1990), the formation of Ca2+/CaM-Nef peptide complex with the molar ratio of 1:2 observed using NMR spectroscopy and SAXS analyses was not observed using fluorescence spectroscopy. Saturation of the fluorescence change by the binding of Nef peptide to the first binding site can be considered the reason why the fluorescence change by the binding of more than two Nef peptide molecules was not seen. The other possibility is that the second binding site is far from the dansyl-group.
The present NMR and SAXS analyses demonstrated that the structure of Ca2+/CaM changed markedly in the presence of the myristoylated Nef peptide. Similarly, the binding of the myristoylated nonapeptide (mC/N9) to Ca2+/CaM induced large structural changes of CaM. Both radii of gyration (Rg) are consistent with each other (Table 1), and the pair distance distribution p(r) values showed similar behaviors (Fig. 6 ▶). The results clearly indicate that the myristoylated HIV-1 Nef peptides as well as mC/N9 can bind Ca2+/CaM. It can be concluded that HIV-1 Nef as well as CAP-23/NAP-22 interacts directly with Ca2+/CaM.
The involvement of the protein myristoylation in protein–protein interactions has been implied in various studies (Chow et al. 1987; Kawamura et al. 1994; Senin et al. 1995), but it has never been clearly demonstrated. Our study is the first direct demonstration of the involvement of the myristoylation in protein–protein interactions. Because protein myristoylation has been implicated in the regulation of various signal transduction proteins (Towler et al. 1988; Resh 1996), and because, in addition to signal transduction proteins, there are many other potential myristoylated proteins whose myristoylations can be predicted from their homologous amino acid sequences, there is a possibility that myristoylation-dependent protein–protein interaction plays important roles in some of these cases, including HIV-1 Nef protein (Hayashi et al. 2000). Therefore, a more precise examination of the molecular mechanism of its interaction is an important subject to investigate.
Myristoylated N-terminal octapeptide in HIV-1 Nef as a novel Ca2+/CaM binding motif
The present findings indicate the formation of Ca2+/CaM-Nef peptide complex with the molar ratio of 1:2. In addition, Ca2+/CaM-mC/N9 complex has been suggested to form a 1:2 complex (Hayashi et al. 2000). It is likely that mC/N9 adopts a nonhelical conformation even in the complex. As noted, since mC/N9 and Nef peptide are similar to each other, Nef peptide is thought to adopt a nonhelical conformation in the complex.
There are two well known recognition motifs for calcium-dependent CaM interactions. The two motifs of calmodulin recognition are termed 1-8-14 and 1-5-10, based on the position of conserved hydrophobic residues (Rhoads and Friedberg 1997). Of their motifs, the structures of the Ca2+/CaM-target complexes with the Ca2+/CaM binding regions of myosin light chain kinase (MLCK) and CaMKII are now available. In addition, the structure of the Ca2+/CaM complex with the Ca2+/CaM binding region of Ca2+-CaM-dependent protein kinase kinase (CaMKK) was determined (Osawa et al. 1999b). The Ca2+/CaM binding regions are characterized as a 1-16 motif. Their target peptides form 1:1 complexes with Ca2+/CaM and adopt an α-helix. A 1:2 protein-peptide complex was observed in the C-terminal domain from petunia glutamate decarboxylase (PGD), and the peptide was shown to bind to Ca2+/CaM in an α-helical conformation (Yuan and Vogel 1998).
Consequently, we conclude that Nef peptide and CAP-23/NAP-22 C/N9 are a unique Ca2+/CaM binding motif, as was our contention in a previous paper (Hayashi et al. 2000).
Structure of the Ca2+/CaM—HIV-1 Nef peptide complex
As noted, very interestingly, the (Rg) values of Ca2+/CaM-Nef peptide complex and Ca2+/CaM-mC/N9 are consistent with each other (Table 1), and the p(r) values showed similar behaviors (Fig. 6 ▶). Thus, the final overall shape conformation appears to be similar to a "relaxed" globular structure proposed in the mC/N9-Ca2+/CaM complex, presumably differing from the known compact globular structures of CaM induced by target peptides of MLCK (Ikura et al. 1992) and CaMKII (Osawa et al. 1999b).
Interaction of Nef peptide with the hydrophobic cleft in Ca2+/CaM
The CaM molecule in both Ca2+-bound and Ca2+-free states adopts an `elongated' structure that comprises two globular domains connected by a highly flexible linker (Seaton et al. 1985; Kretsinger et al. 1986; Persechini and Kretsinger 1988; Heidorn et al. 1989; Barbato et al. 1992; Finn et al. 1995; van der Spoel et al. 1996). In contrast, the structures of Ca2+-bound CaM complexed with peptides from the target enzymes adopt a compact globular shape caused by the bending of the domain linker (Ikura et al. 1992; Meador et al. 1992, 1993). Besides the traditional mechanism of target recognition of CaM described above, some novel mechanisms have been revealed. It has been shown that a single unique complex of Ca2+/CaM is formed with two peptides which correspond to the C-terminal region of PGD. The formation of a 1:2 protein–protein complex is unusual; normally, Ca2+/CaM forms 1:1 complexes with the majority of its target proteins (Yuan and Vogel 1998). It has also been shown that a peptide corresponding to the N-terminal portion of the CaM-binding domain of plasma membrane calcium pump binds only to the C-terminal half of CaM, and that in binding to the peptide, CaM does not form any collapsed structures as was observed in the previous traditional studies (Elshorst et al. 1999).
The binding of two molecules of a CaM antagonist, N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W-7), to Ca2+/CaM induces a globular structure of Ca2+/CaM (Osawa et al. 1999a), although the globular structure is similar to those observed in the 1:1 Ca2+/CaM-peptide complexes, including respective synthetic peptides corresponding to the calmodulin-binding domains of MLCK (M13), phosphorylase kinase (RhK5), and Ca2+-pump (C24W). W-7 contains a hydrophobic aromatic group with a positively charged group via aliphatic chain. In the 1:2 Ca2+/CaM-W-7 complex, the naphthalene ring of the two W-7 molecules interacts with the hydrophobic pockets of the two N- and C-terminal CaM domains. Thus, it is quite possible that Ca2+/CaM in the 1:2 Ca2+/CaM-Nef peptide complex adopts a globular structure.
The chemical shifts of some residues located in the hydrophobic clefts of the N- and C-domains of CaM were changed upon the forming of the Ca2+/CaM-Nef peptide complex, suggesting that the binding site of the Nef peptide is located in their hydrophobic clefts (Fig. 3 ▶). As well as the 1:2 Ca2+/CaM-W-7 complex, the two Nef peptides are suggested to bind to the two domains of CaM individually. The Rg for the 1:2 Ca2+/CaM-Nef peptide complex was larger than that of the Ca2+/CaM-W-7/TFP complex. It is clear that the molecule size of the myristoylated Nef peptide is larger than the W-7 molecule. This may explain the larger Rg value observed for the Ca2+/CaM-Nef peptide complex. As noted, the CaM molecule in the Ca2+/CaM Nef peptide complex assumes a "relaxed" globular conformation similar to the CaM-NAP-22-derived peptide complex (Hayashi et al. 2000).
The residues whose chemical shifts changed quite markedly contain Gly40, Ala57, Leu105, Phe141, and Gln143 (colored red in Fig. 3 ▶). Of these residues, Leu105, Phe141, and Gln143 are located on the contact surface of the Ca2+/CaM-MLCK peptide (M13) complex; the three residues of CaM make contact directly with the target (MLCK peptide) (Ikura et al. 1992). The residue of Gly40 does not make contact with the target on the Ca2+/CaM-MLCK peptide complex. However, Leu39 at the neighboring residue of Gly40 is located on the contact surface of the MLCK complex, and Gly40 itself is located near the contact surface. Besides these residues, residues whose chemical shifts were considerably changed (colored orange in Fig. 3 ▶) are primarily located on and around the contact surface. These observations strongly suggest that the two Nef peptide molecules bind to the hydrophobic clefts of CaM as seen in the M13 peptide, although, besides the binding site, the manners of binding are considered to be different from each other because other features (e.g., molar ratio and requirement of myristoylation) are distinctly different from each other. In addition to the case of these myristoylated proteins (CAP-23/NAP-22 and Nef), an example in which not one but two peptides bind to Ca2+/CaM has been reported in relation to PGD (Yuan and Vogel 1998).
The role of protein Nα-myristoylation
Protein Nα-myristoylation, one of the most common forms among protein fatty acylations, was first characterized in the catalytic subunit of cAMP-dependent protein kinase and in the regulatory subunit of calcineurin (Aitken et al. 1982; Carr et al. 1982). The target proteins are modified with myristate, a 14-carbon saturated fatty acid, and the enzymology of the myristoylation reaction is now well understood (Resh 1999). The covalent attachment of fatty acids to proteins is now a widely recognized form of protein modification, and many fatty acylated proteins have been found to play key roles in regulating cellular functions. It has thus far been generally assumed that hydrophobic acyl groups including the myristoyl group are involved in protein–membrane interactions. Thus, as for Nef, the role of the myristoylation was predicted to anchor it to the plasma membrane. In contrast, we have demonstrated that CAP-23/NAP-22 isolated from rat brain is Nα-myristoylated, and this modification is involved in its interaction with CaM in the presence of Ca2+ (Takasaki et al. 1999).
Functional implication of the interaction between HIV-1 Nef protein and CaM
The calcium-mediated regulation of Src family tyrosine kinases proceeds through a CaM-dependent mechanism (Della Rocca et al. 1997). The activation of Src family tyrosine kinases results in phosphorylation of the Shc adaptor protein, recruitment of the Grb2-Sos complex to the membrane, and activation of Ras through the guanine nucleotide exchange. Subsequent activation of Raf initiates the cascading of phosphorylation events leading to MAP kinase activation. On the other hand, Nef is required for signal transduction events involving MAPKs through induction of the activator protein-1 (AP-1) (Biggs et al. 1999). As HIV-1 Nef protein and CaM are involved in the MAP kinase cascading for the regulation of the intracellular signal transduction, the occurrence of the direct interaction between HIV-1 Nef protein and CaM is not unreasonable, although it had not been indicated to date. The present findings are the first evidence of a direct interaction. The physiological significance of such an interaction is not well understood, and further investigations are in progress.
Materials and methods
Sample preparation
Myristoylated Nef peptide was designed according to the N-terminal amino acid sequence of Nef predicted from the human immunodeficiency virus type 1 nef gene sequence (Shugars et al. 1993), and purchased from Research Genetics (Huntsville, AL). Rat CaM was expressed in Escherichia coli and purified to homogeneity as described (Hayashi et al. 1998).
Fluorescence measurements
Binding of the N-terminal Nef peptides to dansyl-calmodulin was analyzed with a JASCO FP-777 spectrofluorometer in a 1-cm quartz cuvette as described (Matsubara et al. 1997, 1998). With the excitation wavelength set at 340 nm, emission spectra of dansyl-calmodulin (200 nM) in the presence or absence of peptides were recorded in 20 mM Tris-HCl, pH 7.4, 100 mM NaCl containing 1 mM CaCl2 or 2 mM EGTA. Binding of the peptides to calmodulin was monitored by recording the fluorescence emission at 490 nm. Dissociation constants of calmodulin-peptide complexes were determined by a direct fit of the data to the mass (Matsubara et al. 1997).
NMR spectroscopy
All NMR experiments were carried out on a Bruker DMX-500 spectrometer using a 5-mm broadband, z-axis gradient-shielded probe at 298K. Two-dimensional 1H-15N HSQC spectra were obtained by pulse field gradient selection (Grzesiek and Bax 1993). The sweep width was 12 ppm in the 1H dimension and 30 ppm in the 15N dimension, with the 1H carrier set at 500.1324 MHz and the 15N carrier at 50.6814 MHz. The size of the HSQC spectra was a 1024 × 1024 real data matrix with eight scans for each experiment. Proton chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate as 0 ppm. Nitrogen-15 chemical shifts were referenced to liquid NH3.
The sequential assignments for the protein backbone nuclei of the 15N/13C-labeled Ca2+/CaM were mainly achieved by a set of experiments, CBCA(CO)NNH, CBCANNH, HNCO, and HN(CA)CO. Sidechain assignments were obtained using HBHA(CO)NNH, H(CCCO)NNH, CC(CO)NNH, and HCCH-TOCSY experiments. The details will be described elsewhere (Y. Ito, T. Shibata, and N. Hayashi, in prep.). The assignments for the protein backbone nuclei of the 15N/13C-labeled Ca2+/CaM-Nef peptide complex were achieved by the two-dimensional 1H-15N HSQC spectral changes induced by titrations of the Nef peptide to the Ca2+/CaM.
NMR spectra were processed on a Silicon Graphics Indigo2 workstation using Bruker XWIN-NMR and MSI Felix 95.0 software packages.
SAXS
The recombinant CaM was dissolved in Tris buffer (50 mM Tris-HCl, pH7.6) containing 120 mM NaCl and a sufficient amount (five molar equivalents relative to CaM) of CaCl2. The solutions for the Ca2+/CaM and Ca2+/CaM-Nef peptide complex were prepared with five equivalents of Ca2+ ion, taking into account the molar ratio of Ca2+ ions to CaM in the Ca2+/CaM crystal structure or the solution structure; CaM:Ca2+ = 1:4 (Seaton et al. 1985; Kretsinger et al. 1986; Persechini and Kretsinger 1988; Heidorn et al. 1989; Barbato et al. 1992; Finn et al. 1995). The protein concentration was determined by the quantitative amino acid analysis. The solutions for the Ca2+/CaM-Nef peptide mixtures with a molar ratio of 1:2 were prepared at CaM concentrations of 6.0, 9.0, 11.0, 13.0, and 15.0 mg/mL. For the comparison, the solutions of Ca2+/CaM were also prepared at a concentration of 9.0 mg/mL.
The measurements were performed using synchrotron orbital radiation with an instrument for SAXS installed at BL-10C of Photon Factory, Tsukuba (Ueki et al. 1985). An X-ray wavelength of 1.488Å was selected. The samples were contained in a quartz cell with a volume of 80 μL, and the temperature was maintained at 25±0.1°C by circulating water through the sample holder. The reciprocal parameter, Q, equal to 4πsinθ/λ, was calibrated by the observation of peaks from dried chicken collagen, where 2θ is the scattering angle and λ is the X-ray wavelength. Scattering data were collected for 200 sec at individual protein concentrations.
Two methods of data analysis were used. The first method was that of Guinier (Biggs et al. 1999), which gives the radius of gyration, Rg. The range of Q (Å-1) used for the Guinier plots was 0.02 to 0.09 for Ca2+/CaM-Nef peptide mixtures and 0.02 to 0.07 for the Ca2+-saturated CaM in isolation. The second method was the calculation of the pair distance distribution function, p(r), which is the frequency of the distances r within a macromolecule obtained by combining any volume element with any other volume element (Glatter 1982). The p(r) was calculated by direct Fourier transformation (Glatter 1982). Data to Q (Å−1) = 0.7 were used for the p(r) analysis. The maximal pair distance, dmax, was also estimated from the p(r) function; p(r) becomes zero at values of r equal to or greater than the maximum dmax of the particle.
Acknowledgments
This work was supported in part by grants-in-aid from the Fujita Health University (to K.T.), from the Ryoichi Naito Foundation for Medical Research (to N.H.), from the Tokai Science Foundation (to N.H.), from the Aichi Cancer Research Foundation (to N.H.), by a grant-in-aid for Scientific Research C 12208042 and 13770064 from the Ministry of Education, Science, Sports and Culture of Japan, and by a grant-in-aid for the Fujita Health University High-tech Research Center from the Ministry of Education, Science, Sports and Culture of Japan. SAXS measurements were performed with the approval of the Photon Factory Advisory Committee (proposal no. 00G147).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
HIV, human immunodeficiency virus
CaM, calmodulin
Ca2+/CaM, Ca2+-bound calmodulin
Nef peptide, myristoylated peptides corresponding to the N-terminal region of HIV-1 Nef
SAXS, small angle X-ray scattering
NMR, nuclear magnetic resonance spectroscopy
mC/N9, myristoylated peptide corresponding to the N-terminal CaM binding site of CAP-23/NAP-22
Nα, α-amino
MAP kinase, mitogen-activated protein kinase
HSQC, heteronuclear single quantum coherence
MLCK, myosin light chain kinase
M13, a peptide based on the calmodulin-binding domain of MLCK
PGD, petunia glutamate decarboxylase
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.23702.
References
- Aitken, A., Cohen, P., Santikarn, S., Williams, D.H., Calder, A.G., Smith, A., and Klee, C.B. 1982. Identification of the NH2-terminal blocking group of calcineurin B as myristic acid. FEBS Lett 150: 314–318. [DOI] [PubMed] [Google Scholar]
- Barbato, G., Ikura, M., Kay, L.E., Pastor, R.W., and Bax, A. 1992. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 31: 5269–5278. [DOI] [PubMed] [Google Scholar]
- Biggs, T.E., Cooke, S.J., Barton, C.H., Harris, M.P., Saksela, K., and Mann, D.A. 1999. Induction of activator protein 1 (AP-1) in macrophages by human immunodeficiency virus type-1 NEF is a cell-type-specific response that requires both hck and MAPK signaling events. J. Mol. Biol 290: 21–35. [DOI] [PubMed] [Google Scholar]
- Carr, S.A., Biemann, K., Shoji, S., Parmelee, D.C., and Titani, K. 1982. n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc. Natl. Acad. Sci. 79: 6128–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, M., Newman, J.F., Filman, D., Hogle, J.M., Rowlands, D.J., and Brown, F. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327: 482–486. [DOI] [PubMed] [Google Scholar]
- Chowers, M.Y., Spina, C.A., Kwoh, T.J., Fitch, N.J., Richman, D.D., and Guatelli, J.C. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68: 2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivici, A. and Ikura, M. 1995. Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24: 85–116. [DOI] [PubMed] [Google Scholar]
- Della Rocca, G.J., van Biesen, T., Daaka, Y., Luttrell, D.K., Luttrell, L.M., and Lefkowitz, R.J. 1997. Ras-dependent mitogen-activated protein kinase activation by G protein-coupled receptors. Convergence of Gi- and Gq-mediated pathways on calcium/calmodulin, Pyk2, and Src kinase. J. Biol. Chem. 272: 19125–19132. [DOI] [PubMed] [Google Scholar]
- Elshorst, B., Hennig, M., Forsterling, H., Diener, A., Maurer, M., Schulte, P., Schwalbe, H., Griesinger, C., Krebs, J., Schmid, H., Vorherr, T., and Carafoli, E. 1999. NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry 38: 12320–12332. [DOI] [PubMed] [Google Scholar]
- Finn, B.E., Evenas, J., Drakenberg, T., Waltho, J.P., Thulin, E., and Forsen, S. 1995. Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 2: 777–783. [DOI] [PubMed] [Google Scholar]
- Franchini, G., Robert-Guroff, M., Ghrayeb, J., Chang, N.T., and Wong-Staal, F. 1986. Cytoplasmic localization of the HTLV-III 3` orf protein in cultured T cells. Virology 155: 593–599. [DOI] [PubMed] [Google Scholar]
- Glatter O. 1982. Small angle X-ray scattering. In Data analysis (eds. O. Glatter and O. Kratky), pp. 119–196. Academic Press, New York.
- Greenway, A., Azad, A., and McPhee, D. 1995. Human immunodeficiency virus type 1 Nef protein inhibits activation pathways in peripheral blood mononuclear cells and T-cell lines. J. Virol. 69: 1842–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek, S. and Bax A. 1993. The importance of not saturation H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115: 12593–12594. [Google Scholar]
- Hayashi, N., Izumi, Y., Titani, K., and Matsushima, N. 2000. The binding of myristoylated N-terminal nonapeptide fromo neuron-specific protein CAP-43/NAP-22 to calmodulin does not induce the globular structure observed for the calmodulin-nonmyristoylated peptide complex. Protein Sci. 9: 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, N., Matsubara, M., Takasaki, A., Titani, K., and Taniguchi, H. 1998. An expression system of rat calmodulin using T7 phage promoter in Escherichia coli. Protein Expr. Purif. 12: 25–28. [DOI] [PubMed] [Google Scholar]
- Heidorn, D.B., Seeger, P.A., Rokop, S.E., Blumenthal, D.K., Means, A.R., Crespi, H., and Trewhella, J. 1989. Changes in the structure of calmodulin induced by a peptide based on the calmodulin-binding domain of myosin light chain kinase. Biochemistry 28: 6757–6764. [DOI] [PubMed] [Google Scholar]
- Ikura, M., Clore, G.M., Gronenborn, A.M., Zhu, G., Klee, C.B., and Bax, A. 1992. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256: 632–638. [DOI] [PubMed] [Google Scholar]
- Kawamura, S., Cox, J.A., and Nef, P. 1994. Inhibition of rhodopsin phosphorylation by non-myristoylated recombinant recoverin. Biochem. Biophys. Res. Commun. 203: 121–127. [DOI] [PubMed] [Google Scholar]
- Kienzle, N., Bachmann, M., Muller, W.E., and Muller-Lantzsch, N. 1992. Expression and cellular localization of the Nef protein from human immunodeficiency virus-1 in stably transfected B-cells. Arch. Virol. 124: 123–132 [DOI] [PubMed] [Google Scholar]
- Kim, S., Ikeuchi, K., Byrn, R., Groopman, J., and Baltimore, D. 1989. Lack of a negative influence on viral growth by the nef gene of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. 86: 9544–9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsinger, R.H., Rudnick, S.E., Weissman, L.J. 1986. Crystal structure of calmodulin. J. Inorg. Biochem. 28: 289–302. [DOI] [PubMed] [Google Scholar]
- Matsubara, M., Hayashi, N., Titani, K., and Taniguchi, H. 1997. Circular dichroism and 1H NMR studies on the structures of peptides derived from the calmodulin-binding domains of inducible and endothelial nitric-oxide synthase in solution and in complex with calmodulin. Nascent alpha-helical structures are stabilized by calmodulin both in the presence and absence of Ca2+. J. Biol. Chem. 272: 23050–23056. [DOI] [PubMed] [Google Scholar]
- Matsubara, M., Yamauchi, E., Hayashi, N., and Taniguchi, H. 1998. MARCKS, a major protein kinase C substrate, assumes non-helical conformations both in solution and in complex with Ca2+-calmodulin. FEBS Lett 421: 203–207. [DOI] [PubMed] [Google Scholar]
- Matsushima, N., Hayashi, N., Jinbo, Y., and Izumi, Y. 2000. Ca2+-bound calmodulin forms a compact globular structure on binding four trifluoperazine molecules in solution. Biochem. J. 347 Pt 1 : 211–215. [PMC free article] [PubMed] [Google Scholar]
- Matsushima, N., Izumi, Y., Matsuo, T., Yoshino, H., Ueki, T., and Miyake, Y. 1989. Binding of both Ca2+ and mastoparan to calmodulin induces a large change in the tertiary structure. J. Biochem. (Tokyo) 105: 883–887. [DOI] [PubMed] [Google Scholar]
- Meador, W.E., Means, A.R., and Quiocho, F.A. 1992. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257: 1251–1255. [DOI] [PubMed] [Google Scholar]
- Meador, W.E., Means, A.R., and Quiocho, F.A. 1993. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science 262: 1718–1721. [DOI] [PubMed] [Google Scholar]
- Niederman, T.M., Hastings, W.R., and Ratner, L. 1993. Myristoylation-enhanced binding of the HIV-1 Nef protein to T cell skeletal matrix. Virology 197: 420–425. [DOI] [PubMed] [Google Scholar]
- O'Neil, K.T., and DeGrado, W.F. 1990. How calmodulin binds its targets: Sequence independent recognition of amphiphilic alpha-helices. Trends Biochem. Sci. 15: 59–64. [DOI] [PubMed] [Google Scholar]
- Osawa, M., Kuwamoto, S., Izumi, Y., Yap, K.L., Ikura, M., Shibanuma, T., Yokokura, H., Hidaka, H., and Matsushima, N. 1999a. Evidence for calmodulin inter-domain compaction in solution induced by W-7 binding. FEBS Lett. 442: 173–177. [DOI] [PubMed] [Google Scholar]
- Osawa, M., Tokumitsu, H., Swindells, M.B., Kurihara, H., Orita, M., Shibanuma, T., Furuya, T., and Ikura, M. 1999b. A novel target recognition revealed by calmodulin in complex with Ca2+- calmodulin-dependent kinase kinase. Nat Struct. Biol. 6: 819–824. [DOI] [PubMed] [Google Scholar]
- Persechini, A. and Kretsinger, R.H. 1988. The central helix of calmodulin functions as a flexible tether. J. Biol. Chem 263: 12175–12178. [PubMed] [Google Scholar]
- Purcell, D.F. and Martin, M.A. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67: 6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh, M.D. 1996. Regulation of cellular signalling by fatty acid acylation and prenylation of signal transduction proteins. Cell Signal 8: 403–412. [DOI] [PubMed] [Google Scholar]
- Resh, M.D. 1999. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Rhoads, A.R. and Friedberg F. 1997. Sequence motifs for calmodulin recognition. FASEB J. 11: 331–340. [DOI] [PubMed] [Google Scholar]
- Robert-Guroff, M., Popovic, M., Gartner, S., Markham, P., Gallo, R.C., and Reitz, M.S. 1990. Structure and expression of tat-, rev-, and nef-specific transcripts of human immunodeficiency virus type 1 in infected lymphocytes and macrophages. J. Virol. 64: 3391–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, O., Dautry-Varsat, A., Goud, B., Marechal, V., Subtil, A., Heard, J.M., and Danos, O. 1995. Human immunodeficiency virus type 1 Nef induces accumulation of CD4 in early endosomes. J. Virol. 69: 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton, B.A., Head, J.F., Engelman, D.M., and Richards, F.M. 1985. Calcium-induced increase in the radius of gyration and maximum dimension of calmodulin measured by small-angle X-ray scattering. Biochemistry 24: 6740–6743. [DOI] [PubMed] [Google Scholar]
- Senin, I.I., Zargarov, A.A., Alekseev, A.M., Gorodovikova, E.N., Lipkin, V.M., and Philippov, P.P. 1995. N-myristoylation of recoverin enhances its efficiency as an inhibitor of rhodopsin kinase. FEBS Lett. 376: 87–90. [DOI] [PubMed] [Google Scholar]
- Shugars, D.C., Smith, M.S., Glueck, D.H., Nantermet, P.V., Seillier-Moiseiwitsch, F., and Swanstrom, R. 1993. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J. Virol. 67: 4639–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski, J., Parks, D., and Mariani, R. 1993. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J 12: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki, A., Hayashi, N., Matsubara, M., Yamauchi, E., and Taniguchi, H. 1999. Identification of the calmodulin-binding domain of neuron-specific protein kinase C substrate protein CAP-22/NAP-22. Direct involvement of protein myristoylation in calmodulin-target protein interaction. J. Biol. Chem. 274: 11848–11853 [DOI] [PubMed] [Google Scholar]
- Towler, D.A., Gordon, J.I., Adams, S.P., and Glaser, L. 1988. The biology and enzymology of eukaryotic protein acylation. Annu. Rev. Biochem. 57: 69–99 [DOI] [PubMed] [Google Scholar]
- Ueki, T., Hiragi, Y., Kataoka, M., Inoko, Y., Amemiya, Y., et al. 1985. Aggregation of bovine serum albumin upon cleavage of its disulfide bonds, studied by the time-resolved small-angle X-ray scattering technique with synchrotron radiation. Biophys. Chem. 23: 115–124. [DOI] [PubMed] [Google Scholar]
- van der Spoel, D., de Groot, B.L., Hayward, S., Berendsen, H.J., and Vogel, H.J. 1996. Bending of the calmodulin central helix: A theoretical study. Protein Sci. 5: 2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, T. and Vogel, H.J. 1998. Calcium-calmodulin-induced dimerization of the carboxyl-terminal domain from petunia glutamate decarboxylase. A novel calmodulin-peptide interaction motif. J. Biol. Chem. 273: 30328–30335. [DOI] [PubMed] [Google Scholar]
- Zazopoulos, E. and Haseltine, W.A. 1992. Mutational analysis of the human immunodeficiency virus type 1 Eli Nef function. Proc. Natl. Acad. Sci. 89: 6634–6638. [DOI] [PMC free article] [PubMed] [Google Scholar]