Abstract
The binding of the Escherichia coli biotin holoenzyme synthetase to the two ligands, biotin and bio-5`-AMP, is coupled to disorder-to-order transitions in the protein. In the structure of the biotin complex, a "glycine-rich" loop that is disordered in the apo-enzyme is folded over the ligand. Mutations in three residues in this loop result in significant changes in the affinity of the enzyme for both biotin and bio-5`-AMP. The kinetic basis of these losses in the affinity resides primarily in changes in the unimolecular rates of dissociation of the complexes. In this work, isothermal titration calorimetry has been employed to examine the detailed thermodynamics of binding of three loop mutants to biotin and bio-5`-AMP. The energetic features of dissociation of the protein•ligand complexes also have been probed by measuring the temperature dependencies of the unimolecular dissociation rates. Analysis of the data using the Eyring formalism yielded entropic and enthalpic contributions to the energetic barrier to dissociation. The thermodynamic results coupled with the known structures of the apo-enzyme and biotin complex have been used to formulate a model for progression from the ground-state complex to the transition state in biotin dissociation. In this model, the transition-state is characterized by both partial disruption of noncovalent bonds and acquisition of some of the disorder that characterizes the glycine-rich loop in the absence of ligand.
Keywords: Ligand dissociation kinetics, disorder-to-order transition, thermodynamics of ligand binding
Protein•ligand interactions have been studied extensively at structural and functional levels to gain atomic-level understanding of the origins of the affinities and specificities exhibited by these systems. Gibbs free energies of binding as well as the enthalpic and entropic contributions to the total binding free energy for protein•ligand interactions are obtained from equilibrium thermodynamic measurements. From results of these solution studies, the major driving forces for these binding processes are delineated and attempts are then made to relate the energetics to structural features gleaned from X-ray crystallographic and/or nuclear magnetic resonance studies. One major challenge to elucidating the structure:function relationships in ligand-binding processes is the common occurrence of structural changes in the ligand and/or protein upon complexation.
Experimental strategies employed to elucidate structure-energetic relationships in noncovalent association of proteins with ligands include measurements of the consequences of structural perturbation for the functional energetics of the system. The impact of a structural perturbation on the Gibbs free energy, enthalpy, and entropy of the binding reaction are determined. Through studies of a number of, for example, single-site mutants of the protein component of a binding system, estimates of the energetic contributions of individual atomic-level interactions to the total binding free energy can be assessed. This approach also allows determination of additivity in the system.
In examining the effects of structural perturbations on protein•ligand interactions, one often observes that the kinetic basis of a change in the equilibrium constant for a binding process resides solely in a change in the dissociation rate of the complex (Miller et al. 1983). Changes in the rate of association of a protein with a ligand as a result of structural perturbation also may be observed (see Li et al. 2001 for examples). For systems that are controlled by the dissociation kinetics, a fruitful avenue for elucidating the structural basis of the stability of a protein-ligand complex is to gain a detailed understanding of the process of "dissociation" of the complex to the free protein and ligand. One approach for examining the dissociation process is application of the Eyring formalism to analysis of the temperature dependence of the dissociation rate. Results of such an analysis can be used to gain insight into the major barrier to the dissociation process by allowing partitioning of the transition-state free energy into enthalpic and entropic components. This treatment of the process of dissociation of a ligand from a protein is admittedly simplistic in that it assumes a single kinetic barrier to ligand dissociation. However, extensive studies of the streptavidin•biotin binding system underscore the utility of this approach in revealing energetic and structural features of the "rate-determining" step in dissociation of a protein•ligand complex (Stayton et al. 1999; Hyre et al. 2000). Combined ground-state and transition-state thermodynamic analysis of the protein-ligand interactions provide more complete information about the factors controlling specificity.
The Escherichia coli repressor of biotin biosynthesis/biotin holoenzyme synthetase, BirA, binds to two small ligands, biotin and biotinyl-5`-AMP, in its functional cycle (Lane et al. 1964). Biotin functions as a substrate along with ATP in the synthesis of bio-5`-AMP (Lane et al. 1964). The resulting adenylate is the activated intermediate in biotin transfer to a specific lysine residue of the Biotin Carboxyl Carrier Protein subunit of Acetyl-CoA carboxylase. Binding of both the substrate, biotin, and intermediate, bio-5`-AMP, to BirA is thermodynamically favorable and the Gibbs free energies for the interactions measured at 20°C and pH 7.5 are −10 and −14 kcal/mole, respectively (Xu and Beckett 1994). Kinetically, the difference in affinity of BirA for the two ligands is manifested primarily in differences in the magnitudes of the unimolecular dissociation rates of the complexes (Xu and Beckett 1994). In addition, binding of both ligands to BirA is characterized by biphasic kinetics with the first step reflecting formation of a collision complex and the second step corresponding to a unimolecular conformational change in the complex. Dissociation of either ligand from the protein is limited by reversal of the conformational change (Xu et al. 1995). A high-resolution structure of the BirA•biotin complex has been determined (Weaver et al. 2001). Comparison of the structure of protein in the complex to that of the apoprotein (Wilson et al. 1992) reveals that binding of biotin involves a disorder-to-order transition in a protein loop that directly contacts the ligand in the complex (Fig. 1 ▶). A segment of this loop is characterized by the sequence 115GRGRXG120 that is conserved in the sequences of all known biotin holoenzyme ligases (Tissot et al. 1997). Energetic penalties associated with binding of the two ligands to BirA variants with loop mutations at positions 115, 118, and 119 range from 1 to 5 kcal/mole in the Gibbs free energy of the binding interactions (Kwon and Beckett 2000). Moreover, the kinetic origin of the altered equilibrium dissociation constant governing binding of each mutant protein to biotin or bio-5`-AMP lies solely in a change in the unimolecular rate of dissociation of the complex (Table 1; Kwon and Beckett 2000). The positions of the three mutations in the protein structure are highlighted in Figure 1 ▶. Although all three amino acids are located in the "glycine-rich" loop that is organized over the biotin ligand in the complex, only one of the side chains is in close proximity to the ligand. The glycine at position 115, mutation of which to serine results in the largest effects on biotin and bio-5`-AMP binding, lies directly over the biotin ring system in the complex. The arginine side chain of residue 118 forms a salt bridge with the side chain of D176 and does not directly contact the biotin. Effects on ligand binding measured for the R118G mutant are intermediate between those measured for the G115S and R119W mutations. This latter mutation results in only a modest change in the affinity of BirA for biotin and, as indicated in the figure, R119 projects away from the protein in the structure. This residue is located at the monomer-monomer interface in the BirA dimer structure (Weaver et al. 2001).
Fig. 1.
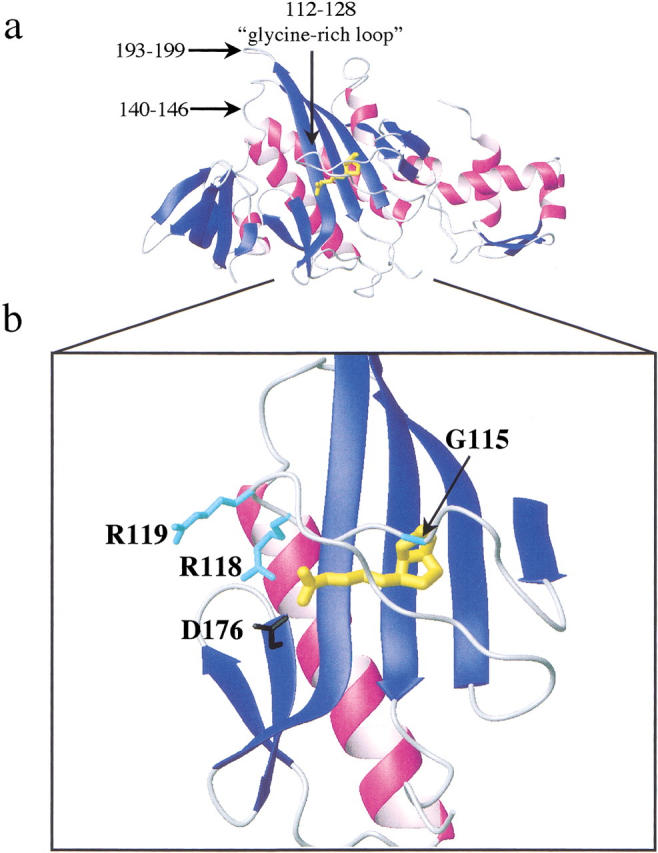
(a) Model of the three-dimensional structure of the BirA•biotin monomer isolated from the second monomer in the dimer structure (Weaver et al. 2001). The biotin ligand is shown in yellow under the glycine-rich loop (112–128). The two additional loops, 140–146 and 193–199, that were resolved in the structure of the complex but not in the apoBirA structure also are indicated in the figure. (b) Detail of the interaction of the glycine-rich loop with biotin in the complex. The side chains of the three residues that are the subject of this work are shown in cyan. The D176 side chain that forms a salt bridge with R118 is shown in black. Models were generated using MolMol (Koradi et al. 1996) with the file 1HXD as input.
Table 1.
Kinetic and equilibrium constant governing binding of BirA variants to biotin and bio-5`-AMPa
| Variant/ ligand | koff (s−1) | kon(M−1 s−1) | KD(M) |
| biotin | |||
| wt | 0.33 (±0.01) | 7.4 (±0.3) × 106 | 4.5 (±0.2) × 10−8 |
| G115S | 80 (±20) | 9 (±2) × 106 | 9 (±3) × 10−6 |
| R118G | 29 (±5) | 1.6 (±0.2) × 107 | 1.8 (±0.4) × 10−6 |
| R119W | 0.49 (±0.04) | 2.2 (±0.2) × 106 | 2.5 (±0.3) × 10−7 |
| bio-5`-AMP | |||
| wt | 0.00027 (±0.00003) | 6.0 (±0.6) × 106 | 4.5 (±0.7) × 10−11 |
| G115S | 0.84 (±0.03) | 7.0 (±0.7) × 106 | 1.2 (±0.1) × 10−7 |
| R118G | 0.119 (±0.006) | 6 (±1) × 106 | 2.0 (±0.3) × 10−8 |
| R119W | 0.00009 (±0.00001) | 4.9 (±0.2) × 106 | 1.8 (±0.2) × 10−11 |
a All parameters were previously reported in Kwon and Beckett (2000). The equilibrium dissociation constants (KD) were calculated from the measured unimolecular dissociation rate constants (koff) and bimolecular association rate constants (kon) using the equation 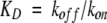 . Parameters were determined in Standard Buffer (10 mM TrisHCl, pH 7.50 ± 0.02, 200 mM KCl, 2.5 mM MgCl2 at 20.0 ± 0.1°C).
. Parameters were determined in Standard Buffer (10 mM TrisHCl, pH 7.50 ± 0.02, 200 mM KCl, 2.5 mM MgCl2 at 20.0 ± 0.1°C).
In this work, the detailed thermodynamics of interaction of the three BirA mutants, G115S, R118G, and R119W, with the two ligands biotin and bio-5`-AMP, are examined. Isothermal titration calorimetry has been used to measure the heats of binding of the proteins to the two ligands and to determine the heat capacity changes associated with the binding processes. The combined results of calorimetric measurements of binding enthalpies and kinetic measurements of equilibrium constants were used to obtain thermodynamic profiles for each binding process at 20°C. The energetic factors governing kinetic stabilities of the complexes were obtained from measurements of the temperature dependencies of dissociation of the protein•ligand complexes. Analysis of the kinetic data using the Eyring formalism yielded thermodynamic profiles for the conversion of each ground-state complex to the transition state in the dissociation process. Equilibrium thermodynamic parameters for the mutants reveal that changes in the Gibbs free energy of binding are either enthalpically or entropically based. Patterns of transition-state enthalpy and entropy changes are complex and do not directly correlate with ground-state perturbations. The results were interpreted in the context of the known disorder-to-order transition that is coupled to ligand binding by BirA. These results, in combination with previous results of kinetic studies of this system, suggest that, in addition to partial disruption of noncovalent bonds, conversion of the complex from the ground state to the transition state in dissociation is accompanied by acquisition of flexibility by the glycine-rich loop.
Results
Equilibrium thermodynamics of biotin and bio-5`-AMP binding to the BirA mutants
Results of kinetic measurements of binding of wild-type and mutant BirA proteins to biotin and bio-5`-AMP at 20°C are shown in Table 1 (Kwon and Beckett 2000). Equilibrium dissociation constants for the binding reactions, which were calculated from the kinetic parameters, are shown as well. The Gibbs free energies of interaction of the four proteins with the two ligands were calculated from the equilibrium constants and are shown in Table 2. As indicated by the magnitudes of the Gibbs free energies, the energetic penalties for the two binding processes associated with the three mutations range from 1.0 to 3.1 kcal/mole for biotin binding and −0.4 to 4.7 kcal/mole for adenylate binding at 20°C. Isothermal titration calorimetry (ITC) has been employed to further characterize the thermodynamic profiles for binding of the mutants to the two ligands. These measurements were performed under stoichiometric conditions so that in the first injection the protein is fully saturated with ligand. Moreover, in determination of the heats of binding of bio-5`-AMP to the proteins, all measurements were performed at sufficiently low protein concentration (1 μM) to avoid complication from ligand-induced dimerization of BirA (Eisenstein and Beckett 1999). Results of representative ITC traces are shown in Figure 2 ▶. In the traces, the first injection results in a large exothermic peak, while the peaks associated with the final four injections, which provide the heat of dilution of the ligand, also are exothermic but smaller. The total molar heat of biotin binding to the wild-type protein obtained from the measurement shown in Figure 2a ▶, is −9.5 kcal/mole. This binding heat differs significantly from that previously reported (Xu et al. 1996). Because all proteins used in the current study have a histidine tag fused at the C-terminus, the contribution of this modification was investigated by repeating the measurements with the unmodified protein. However, results of these measurements yielded an average value of the heat of biotin binding of −10.3±0.4 kcal/mole compared with the previously reported value of −13.0±0.2 kcal/mole (Xu et al. 1996). A similar discrepancy was observed in measurements of the heats of binding of bio-5`-AMP to wild-type BirA. While previous results indicated a ΔH° of −7.3±0.2 kcal/mole, results of the more recent measurements yielded a value of −6.3±0.1. The wild-type protein with the histidine tag yielded a heat of bio-5`-AMP binding of −5.8±0.3 kcal/mole. The deviations of the results obtained for the unmodified wild-type protein from those reported in the previous study cannot be attributed to different protein preparations because the same discrepancy was found in calorimetric measurements performed on a number of preparations of wild-type BirA. Instrumentation and the fact that all measurements were performed in-house are the major differences between the previous measurements and those reported in this work. Whereas the former measurements were performed on the MicroCal Omega 2 model at the Johns Hopkins Biocalorimetry Center, the current measurements were performed using the VP-ITC located at the University of Maryland.
Table 2.
Thermodynamic parameters for binding of small ligands to BirA variantsa
| Variant/ligand | ΔG°(kcal/mol)b | ΔH°(kcal/mol)c | -TΔS°(kcal/mol)d |
| biotin | |||
| wt | −9.9 (±0.1) | −9.4 (±0.3) | −0.5 (±0.3) |
| G115S | −6.8 (±0.2) | −5.0 (±0.3) | −1.8 (±0.4) |
| R118G | −7.7 (±0.1) | −9.4 (±0.3) | −1.7 (±0.3) |
| R119W | −8.9 (±0.1) | −9.3 (±0.4) | 0.4 (±0.4) |
| bio-5`-AMP | |||
| wt | −14.0 (±0.1) | −5.8 (±0.3) | −8.1 (±0.4) |
| G115S | −9.3 (±0.1) | −2.2 (±0.1) | −7.1 (±0.1) |
| R118G | −10.3 (±0.1) | −6.1 (±0.3) | −4.2 (±0.3) |
| R119W | −14.4 (±0.1) | −7.5 (±0.2) | −6.9 (±0.2) |
a Measurements were performed in Standard Buffer [10 mM Tris-HCl, 200 mM KCl, 2.5 mM MgCl2, pH 7.50 ± 0.01 at 20.0 ± 0.1°C] at 20.0 ± 0.1°C.
b Gibbs free energies for binding are from Kwon and Beckett (2000).
c Reported standard molar binding enthalpies represent the average of at least three independent measurements with standard errors shown in parentheses.
d Calculated using the expression 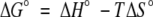 .
.
Fig. 2.

Isothermal titration calorimetry measurements of binding of (a) His-tagged wild-type BirA and (b) R118G variant to biotin. Measurements were performed in Standard Buffer at 20.0±0.1°C. In both measurements, the cell contained 1.7 mL of protein solution at a concentration of 1 μM. (a) 5–10 μL injections of a 250-μM biotin solution and (b) 5–50 μL injections of a 720-μM biotin solution.
Figure 2b ▶ provides results of measurement of the heat of biotin binding to the R118G mutant in standard conditions. This trace differs significantly from that for the similar measurement performed with wild-type BirA shown in Figure 2a ▶. The differences are because of the fact that the weak binding of biotin to the R118G mutant (Kwon and Beckett 2000) combined with the low solubility of the biotin ligand necessitated employing injections of relatively large volume to guarantee saturation of the protein with ligand in the first injection. The molar enthalpy of binding obtained from this measurement is −9.3 kcal/mole, a value similar in magnitude to the average value obtained for binding of the wild-type, histidine-tagged protein to biotin. Results of all other calorimetric measurements, like those shown in Figure 2 ▶, yielded exothermic values for binding of all mutants to the two ligands in Standard Buffer conditions (Table 2). However, in biotin binding, only the enthalpy measured for the G115S mutant differs significantly from that measured for the wild-type protein, while for bio-5`-AMP, binding of G115S is less exothermic and R119W binding is more exothermic.
Linkage of protonation to ligand binding
As indicated in Table 2, binding heats for the initial measurements were obtained in buffer containing Tris, a reagent characterized by a large heat of protonation. It is possible that the apparent binding heats obtained from these measurements include a contribution from protonation. In this scenario, each total heat of binding reports both an intrinsic binding enthalpy as well as a contribution from the heat of proton binding to, or dissociation from, the buffering agent. To determine if proton binding or release contributes significantly to biotin or bio-5`-AMP binding to the BirA variants, ITC measurements also were performed in buffer prepared using MOPS, a reagent characterized by a very small heat of protonation (Jelesarov and Bosshard 1994; Baker and Murphy 1996; Bradshaw and Waksman 1998). The corrected heats of binding of the proteins to the two ligands in Standard Buffer prepared with Tris and MOPS are shown in Table 3. The measured enthalpies were utilized to estimate the proton linkage to binding, which also are shown in Table 3. In all cases, release or uptake of protons upon binding was negligible, a result consistent with previously reported results for binding of the wild-type protein to the two ligands (Xu et al. 1996).
Table 3.
Dependence of the measured binding enthalpies on the enthalpy of buffer ionizationa
| Variant/ligand | Tris (10 mM) ΔH° (kcal/mol)b | MOPS (10 mM) ΔH° (kcal/mol)b | Δnc |
| biotin | |||
| wt | −9.4 (±0.3) | −5.7 (±0.4) | −0.62 |
| G115S | −5.0 (±0.3) | −4.4 (±0.4) | −0.09 |
| R118G | −9.4 (±0.3) | −6.4 (±0.1) | −0.51 |
| R119W | −9.3 (±0.4) | −6.3 (±0.1) | −0.35 |
| bio-5`-AMP | |||
| wt | −5.8 (±0.3) | −5.4 (±0.2) | −0.08 |
| G115S | −2.2 (±0.1) | −1.6 (±0.1) | −0.09 |
| R118G | −6.1 (±0.3) | −5.5 (±0.1) | −0.11 |
| R119W | −7.5 (±0.2) | −6.7 (±0.1) | −0.14 |
a Measurements were performed in Standard Buffer (10 mM Tris-HCl, 200 mM KCl, 2.5 mM MgCl2, pH 7.50 ± 0.01 at 20.0 ± 0.1°C) or [10 mM MOPS, 200 mM KCl, 2.5 mM MgCl2, pH 7.50 ± 0.01 at 20.0 ± 0.1°C] at 20.0 ± 0.1°C.
b Reported standard molar binding enthalpies are the average of at least three independent determinations with standard errors in parentheses.
c Estimated number of protons released upon ligand binding.
Thermodynamic profiles for the binding processes
Molar entropies of binding of the ligands to the BirA mutants at 20°C were calculated from the Gibbs free energies and the enthalpies using the equation:
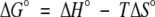 |
As indicated by the results presented in Table 2, energetic penalties associated with the mutations are complex. First, while replacing G with S at position 115 is evidenced primarily in the enthalpic cost to binding, the R to G substitution at position 118 and the R to W substitution at position 119 result in entropic penalties in biotin binding. The patterns observed for bio-5`-AMP binding to the mutants are similar with enthalpy dominating the penalty for changing the G to S at 115 and a loss in favorable binding entropy for replacement of R with G at position 118. The R119W mutation results in small energetic changes in binding of both ligands.
Heat capacities for binding of small ligands to BirA variants
Enthalpies of binding of the mutants to biotin and bio-5`-AMP were determined over the temperature range from 10°C to 30°C. These temperature-dependent binding enthalpies were used to determine the molar heat capacities for the binding processes, the results of which are shown in Figure 3 ▶. All of the binding reactions are characterized by negative heat capacity changes, the magnitudes of which are shown in Table 4. The values reported for the histidine-tag fusion of the wild-type protein are similar to those previously reported for the wild-type protein (Xu et al. 1996). For each mutant, the heat capacity change for bio-5`-AMP binding is larger than that for biotin binding. In biotin binding, the heat capacity changes associated with binding of the mutants are smaller than that measured for the wild-type protein. The magnitudes of the heat capacity changes are similar for binding of all four proteins to bio-5`-AMP.
Fig. 3.

Temperature dependencies of the molar enthalpies for binding of (a) biotin and (b) bio-5`-AMP to (□) wild-type BirA, (♦) G115S, (•) R118G, and (▵) R119W. Measurements were performed in Standard Buffer prepared at the working temperature. The molar binding enthalpy at each temperature represents the average of three independent determinations. For data points in which no error bars are apparent, the magnitude of the error is within the symbol.
Table 4.
Summary of heat capacity changes for binding of biotin and bio-5`-AMP to BirA variantsa
| ΔCp° (biotin) cal/mol/K | ΔCp° (bio-5`-AMP) cal/mol/K | |
| wt | −190 ± 30 | −200 ± 50 |
| G115S | −110 ± 20 | −220 ± 30 |
| R118G | −80 ± 30 | −330 ± 20 |
| R119W | −80 ± 30 | −250 ± 50 |
a Molar heats of binding were measured by isothermal titration calorimetry at temperatures ranging from 15 to 30°C, and molar heat capacities were obtained from the slopes of the linear dependencies of the binding heat on temperature (Fig. 3 ▶). Each standard molar binding enthalpy used in the analysis represents the average of at least three independent determinations.
Thermodynamic analysis of the kinetics of dissociation of the protein•ligand complexes
In measurements of binding of the BirA mutants to biotin and bio-5`-AMP, it was found that the kinetic basis of the decreased affinities of the proteins for the ligands is uniformly centered in the unimolecular rates of dissociation of the protein-ligand complexes (Kwon and Beckett 2000). No changes or very minor changes in the bimolecular association rates for complex formation from those measured for binding of wild-type BirA to the two ligands were observed. To further characterize the kinetic barrier to dissociation of the complexes, the temperature dependencies of the unimolecular dissociation rates were measured. The results were analyzed using the Eyring formalism to obtain activation enthalpies and entropies for the dissociation processes.
Rates of dissociation of all protein-biotin complexes were measured using stopped-flow fluorescence. Protein•biotin complexes were combined rapidly with excess bio-5`-AMP and the resulting time-dependent decrease in the intrinsic protein fluorescence was monitored. Results of measurements of dissociation of biotin from the R119W mutant at 25°C are shown in Figure 4a ▶. Nonlinear, least-squares analysis of the data using a single exponential model yielded a rate constant of 0.9 s−1. All traces obtained in measurements of biotin dissociation were well described by a single exponential equation and the resolved unimolecular rate constants increased with increasing temperature. Results of analysis of the data using the Eyring formalism are shown in Table 5. With the exception of the G115S mutant, the activation enthalpies of dissociation of the complexes are similar in magnitude.
Fig. 4.
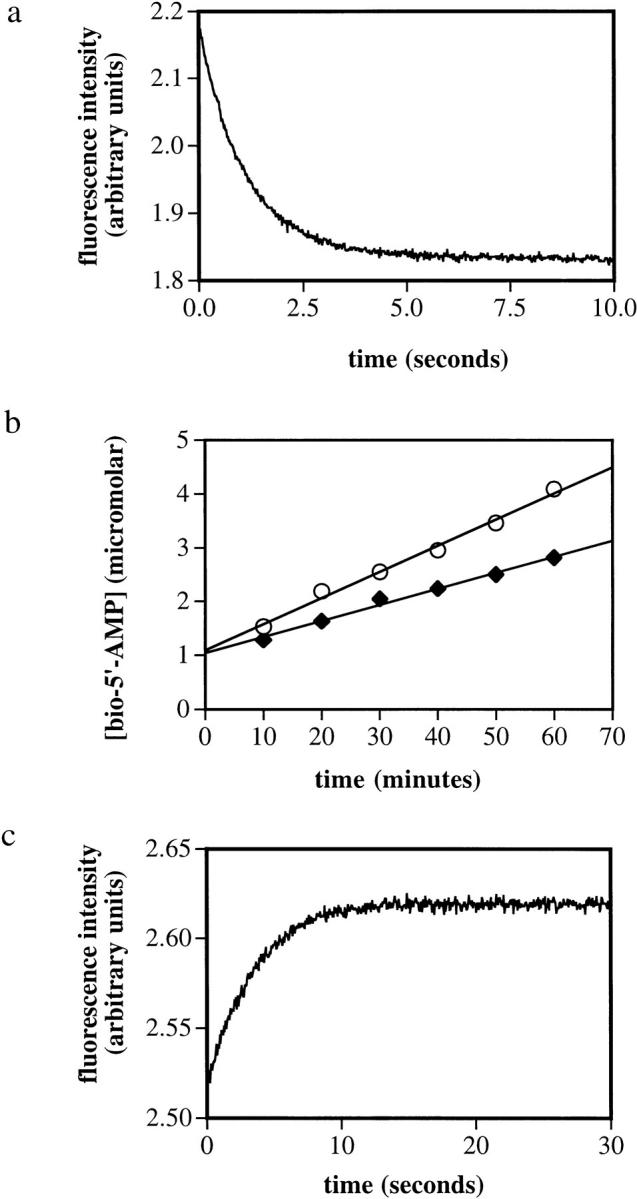
Kinetic measurements of dissociation of complexes of BirA variants bound to biotin and bio-5`-AMP. (a) Stopped-flow measurement of dissociation of the R119W•biotin complex at 25°C. A solution containing 2 μM R119W and 5 μM biotin was rapidly mixed with 100 μM bio-5`-AMP, and the resulting time-dependent decrease in intrinsic protein fluorescence was measured. (b) Measurement of dissociation of complexes of bio-5`-AMP bound to wild-type BirA (○) and R119W-BirA (♦). The rate constants governing unimolecular dissociation of the complexes are proportional to the slopes of the best-fit lines. (c) Stopped-flow fluorescence measurement of dissociation of the R118G•bio-5`-AMP complex at 25°C. A solution containing 2 μM R118G-BirA and 2.5 μM bio-5`-AMP was rapidly mixed with a 200-μM biotin solution, and the resulting time-dependent increase in intrinsic protein fluorescence was monitored.
Table 5.
Summary of activation parameters for dissociation of protein ligand complexesa
| Variant/ ligand | koff(s−1) | ΔG‡ (kcal/mole) | ΔH‡b (kcal/mole) | -TΔS‡ (kcal/mole) |
| biotin | ||||
| wt | 0.33 ± 0.01 | 17.7 ± 0.4 | 22.9 ± 0.8 | −5.1 ± 0.5 |
| G115S | 80 ± 20 | 14.5 ± 0.4 | 16 ± 2 | −2 ± 2 |
| R118G | 29 ± 5 | 15.3 ± 0.3 | 24.4 ± 0.6 | −9.1 ± 0.6 |
| R119W | 0.49 ± 0.04 | 17.7 ± 0.1 | 24.1 ± 0.8 | −6.4 ± 0.4 |
| bio-5`-AMP | ||||
| wt | 0.00027 ± .00003 | 22.0 ± 0.1 | 20 ± 4 | 4 ± 1 |
| G115S | 0.84 ± 0.03 | 16.9 ± 0.6 | 7 ± 1 | 10 ± 1 |
| R118G | 0.142 ± 0.006 | 18.4 ± 0.1 | 16 ± 3 | 2 ± 3 |
| R119W | 0.00009 ± 0.00001 | 22.3 ± 0.1 | 20 ± 2 | −0.3 ± 1 |
a Activation energies and entropies are given at 20°C.
b For the Eyring analysis, measurements were performed in Standard Buffer (10 mM Tris-HCl, 200 mM KCl, 2.5 mM MgCl2, pH 7.50 ± 0.01 at the working temperature) at temperatures ranging from 10 to 35°C. Data were obtained at 5° intervals.
Unimolecular rates of dissociation of the protein•bio-5`-AMP complexes were determined using two techniques. The complexes formed with the wild-type and R119W proteins and the adenylate are characterized by very slow dissociation rates (Xu and Beckett 1994; Kwon and Beckett 2000), which necessitated measuring the time course of enzyme-catalyzed synthesis of the adenylate to determine the kinetics of dissociation (Xu and Beckett 1994). Results of measurement of dissociation of complexes formed between the wild type and R119W•mutant with bio-5`-AMP obtained at 25°C using this technique are shown in Figure 4b ▶. The time course is characterized by a rapid initial exponential burst followed by a slow linear phase the slope of which is proportional to the unimolecular rate constant governing dissociation of the complex. The rates obtained from these measurements are 0.008 and 0.005 s−1 for the wild-type and mutant proteins, respectively. The rates of dissociation of complexes formed between the R118G and G115S mutants and bio-5`-AMP are sufficiently rapid to require measurement using rapid mixing techniques. These measurements were performed by rapidly mixing each complex in a 1:1 vol/vol ratio with a solution containing a large molar excess of biotin and monitoring the resulting time-dependent increase in the intrinsic protein fluorescence. Results of measurements of dissociation of the R118G•bio-5`-AMP complex at 25°C are shown in Figure 4c ▶. As with biotin dissociation, all stopped-flow traces could be analyzed using a single exponential equation to obtain the unimolecular rate constants for bio-5`-AMP dissociation. The resolved rate obtained from analysis of the data in Figure 4c ▶ is 0.294 s−1. Kinetic measurements of bio-5`-AMP dissociation were performed for all four proteins, wild type and 3 mutants, over a temperature range from 10°C to 35°C. Activation parameters obtained from analysis of the data using the Eyring formalism are shown in Table 5. The values of the activation enthalpies determined for dissociation of the G115S and R118G complexes differ significantly from that determined for dissociation of the complex formed between wild-type BirA and bio-5`-AMP.
Thermodynamic profiles for dissociation of the protein•ligand complexes
Using the rate constants measured at 20°C and the activation enthalpies determined from the Eyring analyses as described above, the thermodynamic profiles for dissociation of the two ligands from the four proteins were calculated at 20°C (Table 5). Similar profiles were obtained for wild-type BirA and the R119W mutant. The perturbation in the thermodynamic profile for dissociation of biotin from G115S is evidenced primarily in the decrease in the enthalpic barrier to dissociation. By contrast, dissociation of biotin from R118G is characterized by a more favorable activation entropy. For all proteins, both the enthalpic and entropic terms are barriers to dissociation of bio-5`-AMP. While the magnitudes of both terms are similar for wild-type BirA and R119W, the increased rates of dissociation of the complexes formed by the G115S and R118G mutants reflect decreases in the enthalpic barrier to dissociation. In addition, the entropic barrier to dissociation of the G115S•bio-5`-AMP complex is considerably higher than that obtained for the wild-type protein.
Discussion
Structural perturbations of the complexes formed by mutants are minimal
Elucidation of the molecular details of kinetic and thermodynamic control of ligand-binding specificity entails measurements of the functional consequences of changing the identity of a large number of amino-acid residues in a binding protein. Ideally, the changes in the amino acids are conservative. The three mutations in BirA examined in this work are not conservative. These mutants initially were obtained from genetic screens in which changes in the biotin concentration dependence of bacterial growth was used as an indication of a defect in the enzymatic function of the biotin holoenzyme ligase (Barker and Campbell 1981a,b), and they were subsequently shown to bind with altered affinity to both biotin and bio-5`-AMP (Kwon and Beckett 2000). Moreover, as discussed below, results of a number of measurements suggest that the structures of the complexes of these mutants with the ligands are similar to those formed by the wild-type protein. The changes in the energetics of biotin and bio-5`-AMP binding measured for these mutants are substantial and the kinetic basis for the lowered affinities resides primarily in dissociation of the complexes. These two observations render the mutants attractive candidates for further dissection of the equilibrium thermodynamics and kinetic barrier to dissociation.
In interpreting the detailed thermodynamics of interaction of single-site mutants of a protein with ligands, it is important to verify that the structures of the bound and free states of the protein are not significantly perturbed as a consequence of the mutations. The mutants examined in this work are located in a region of the BirA structure that is disordered in the unliganded state (Wilson et al. 1992). Therefore, structural perturbations to the unliganded state of the protein resulting from the mutations should be minor. Functional studies of the protein-ligand complexes also are consistent with minor or no differences of the structures of these complexes from that of the wild-type protein. First, complexes of the mutants bound to bio-5`-AMP are capable of catalyzing transfer of biotin to the biotin acceptor protein (BCCP) (Kwon et al. 2000; Weaver et al. 2001b) and binding to bioO. In fact, the G115S mutant, which exhibits the lowest affinity for biotin and bio-5`-AMP, binds to bioO with affinity similar to that of the wild-type repressor (Kwon et al. 2000). Second, all protein-biotin complexes are competent for binding of ATP and subsequent synthesis of bio-5`-AMP. The magnitudes of perturbations in the KM for ATP are less than twofold of and kcat values are all within fivefold of that measured for the wild-type protein (Kwon and Beckett 2000).
The results of thermodynamic measurements presented in this work also are consistent with the absence of major perturbations in the structures of the complexes of the mutants bound to the two ligands. First, the magnitudes of the heat capacity changes associated with the binding, while not identical, are similar for all proteins. Assuming that heat capacity changes reflect burial of polar and nonpolar surface area as well as folding in this system, the similar magnitudes of ΔCp suggest similar structures of the complexes. Finally, the lack of change in the contribution of protonation to either binding event observed for the mutants is consistent with the idea that the structural consequences of the amino-acid replacements are local in nature.
Energetic profiles for ligand dissociation from the mutant proteins
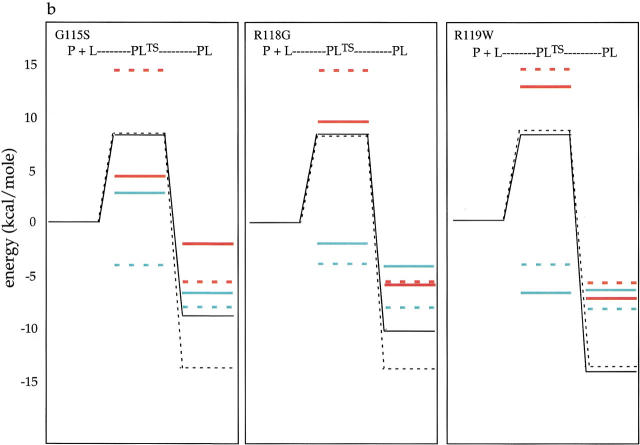
The equilibrium binding parameters were combined with the activation parameters obtained from the kinetic measurements to compose profiles for ligand dissociation from the proteins (Fig. 5a,b ▶). In these profiles, the reference state, which corresponds to the free protein and ligand, in energy, enthalpy and entropy is assumed to be at zero energy for all protein-ligand pairs. The free energy profile for each protein:ligand pair is shown in black. In addition, the enthalpic and entropic changes associated with the equilibrium-binding process, as well as the conversion of the ground-state complex (PL) to the transition state in dissociation (PLTS), are shown. The transition-state energetics in dissociation shown in each panel were calculated by adding the activation parameters obtained from the Eyring analysis of kinetic data (ΔG‡, ΔH‡, and −TΔS‡; Table 5) to the ground-state energetic parameters (ΔG°, ΔH°, and −TΔS°; Table 2). In each panel, the results obtained with the mutant protein are shown with those obtained from measurements obtained for wild-type BirA.
Fig. 5.
Energetic profiles for (a) biotin and (b) bio-5`-AMP binding to wild-type BirA and the three mutants G115S, R118G, and R119W. In each panel, the energy of the reference state corresponding to the free protein and ligand is set at zero. The Gibbs free energy changes are shown in black, enthalpic changes are in red, and entropic changes are shown in cyan. In each panel, the results obtained with the indicated mutant (solid lines) are shown with the results of measurements performed on the complex of the wild-type protein with the ligand (dashed lines). P+L, free protein and ligand (the reference state); PL, protein:ligand complex; PLTS, transition state.
Structural interpretation of the perturbations to the equilibrium thermodynamic parameters
The thermodynamic profiles for binding of biotin and bio-5`-AMP to the mutant and wild-type proteins are shown in Figure 5 ▶. In these reaction-coordinate diagrams, the free state (P + L) is assumed to be characterized by an energy of zero for all ligand/protein pairs. Inspection of the ground-state energetics for the ligand-binding reactions reveals that patterns of energetic penalties resulting from the mutations are conserved for binding of the two ligands. For example, in binding of the two ligands to the G115S mutant, the energetic penalty is primarily enthalpic. The ΔΔH° for biotin binding is 4.4 kcal/mole, while the ΔΔG° is 3.1 kcal/mole. Similarly, the ΔΔH° for bio-5`-AMP binding is 3.6 kcal/mole, while the ΔΔG° is 4.7 kcal/mole. Biotin binding by the mutant is entropically more favorable by −1.0 kcal/mole, and adenylate binding is less favorable by 1.0 kcal/mole. For the R118G mutant, it is the entropic term that is most significantly altered in the equilibrium-binding process. At 20°C, the ΔΔG° for biotin binding is 2.2 kcal/mole and the entropic penalty is 2.2 kcal/mole. In bio-5`-AMP binding ΔΔG° is 3.7 kcal/mole and the entropic penalty is 4.0 kcal/mole. For both mutants, the magnitude of the energetic penalty is significantly greater for bio-5`-AMP binding than it is for biotin binding. For the R119W mutant, there is a small entropic penalty associated with biotin binding of ∼−1.0 kcal/mole. The affinity of the R119W mutant for bio-5`-AMP is actually slightly greater than that measured for the wild-type protein and the mutant interaction is characterized by both a more favorable enthalpy and a less favorable entropy.
Consideration of the structures of the unliganded and liganded forms of BirA provides insight into the observed changes in the energetics of biotin binding. In analyzing the structural basis of the thermodynamic results, it is assumed that only the loop containing the "glycine-rich" sequence becomes ordered upon binding of the ligand to the protein monomer. The other two loops that are observed at the dimer interface in the complex are assumed to become ordered only upon the ligand-induced dimerization (Weaver et al. 2001). Justification for this assumption arises from the observation that mutations in these other two loops do not affect small-ligand binding (Kwon et al. 2000 and unpubl.). Inspection of the structure of the complex suggests an explanation for the observed enthalpic penalty for ligand binding associated with the G115S mutant (Fig. 1 ▶). In the complex, the glycine residue is in close proximity to the biotin-ring system and replacement of the residue with a serine may introduce steric bulk at the interface between the ligand and the loop that translates into a large decrease in the enthalpic term for binding. The observation of a large enthalpic penalty for bio-5`-AMP binding suggests that these same steric factors function in binding of the mutant to this ligand. However, the source of the larger magnitude for both the ΔΔG° and ΔΔH° terms is not readily apparent. The side chain of R118 does not directly contact the biotin moiety in the complex (Fig. 1b ▶.). Rather, this residue forms a salt bridge with a second residue, D176, which appears to stabilize the ordered conformation of the "glycine-rich" loop. The origin of the entropic penalties for equilibrium binding of the R118G mutant to the two ligands may reflect the greater configurational entropy available to the glycine residue (Fig. 5a ▶). The entropic cost of ordering the loop is higher for the sequence containing the glycine replacement. The structural basis for the entropic penalty associated with biotin binding to the R119W mutant is not apparent from the structural model shown in Figure 1 ▶. The side chain of this residue projects into solution from the tip of the glycine-rich loop and it is not clear why changing it to a tryptophan would result in an entropic penalty to biotin binding.
The heat capacity changes associated with binding of the two ligands to the wild-type and mutant proteins were determined (Fig. 3 ▶; Table 5). In biotin binding, all of the mutants display significantly smaller heat-capacity changes. Assuming that the heat-capacity change reflects burial of polar or nonpolar surface area, this result suggests that there may be small but significant changes in the structures of the complexes of mutant proteins bound to biotin. However, as discussed above, the functional consequence of these changes are minor. By contrast, only the R118G mutant exhibits a significantly different molar heat-capacity change in bio-5`-AMP binding. The heat-capacity change associated with this binding process is ∼1.5 times greater in magnitude than that measured for the wild-type protein. However, in the absence of a structure of the BirA:adenylate complex, it is difficult to speculate on the structural origins of this difference in binding thermodynamics.
Perturbation of the kinetic barriers to ligand dissociation
The patterns of perturbation of the transition-state barrier to dissociation of both biotin and bio-5`-AMP observed for the mutants are more complex than those measured for the ground-state energetic parameters governing ligand binding. Reaction-coordinate diagrams for ligand release are shown in Figure 5 ▶. As pointed out above, the transition-state energetics were calculated by adding the activation parameters to the ground-state equilibrium parameters. In discussion of the energetic barrier to ligand dissociation, not only the transition-state free energy but also the enthalpic and entropic components to the kinetic barrier must be considered. The increased rate of biotin release from the G115S mutant translates into ∼ a 3.2-kcal decrease in the energetic gap between the complex (PL) and the transition state (PLTS) in dissociation relative to that measured for the wild-type BirA:biotin complex. Analysis of the temperature-dependence of the rate of biotin release reveals that this perturbation to barrier height reflects a significant 6.8 kcal/mole decrease in the enthalpic barrier to dissociation accompanied by a 3.0 kcal/mole increase in the entropy change. For the R118G mutant, the drop in the energetic barrier to biotin release is evidenced primarily in a decreased transition-state entropy (−TΔSTS). Indeed, for this mutant, the activation enthalpy is slightly larger than that measured for biotin dissociation from wild-type BirA. Perturbations to the thermodynamic contributions to the transition-state barrier in bio-5`-AMP dissociation from the protein are also complex. While the enthalpic barrier to dissociation is substantially decreased by 13 kcal/mole for the G115S mutant, the entropic barrier is increased by 6 kcal/mole. In dissociation of the adenylate from the R118G mutant, both the enthalpic and entropic barriers to adenylate release are decreased.
The thermodynamic features of biotin dissociation from wild-type and mutant BirA proteins and the known structures of apoBirA and the biotin complex can be used to infer structural changes that accompany conversion of the ground-state complex to the transition state in biotin dissociation. In this discussion, an implicit assumption is that solvation contributions are equivalent in dissociation of biotin from all BirA variants. When comparing the structures of apoBirA with that of the biotin-bound protein, differences are localized to three surface loops that undergo disorder-to-order transitions (Wilson et al. 1992; Weaver et al. 2001). However, as indicated in the Introduction, several lines of evidence support the idea that in the monomeric state of the biotin complex, only the glycine-rich loop composed of residues 112–128 is ordered.
Dissociation of the wild-type BirA•biotin complex is characterized by a large enthalpic barrier that undoubtedly reflects the partial disruption of noncovalent bonds in progression from the complex to the transition state in dissociation. By contrast, the entropic term in progression to the transition state (−TΔSTS) in dissociation is negative or favorable. This favorable entropy may reflect partial disordering of the glycine loop as the reaction proceeds to the transition state. Thus, at the transition state in biotin dissociation, not only are noncovalent bonds disrupted but the glycine loop regains some of the disorder associated with it in the free or "open" state. Results obtained with the G115S and R118G mutants are consistent with this hypothesis. The enthalpic barrier to biotin dissociation from the G115S mutant is significantly lower than that determined for the wild-type protein, a result consistent with the less favorable enthalpy of the equilibrium-binding process. The steric interference introduced by the serine residue compromises the fit between the ligand and protein in the complex. As fewer bonds have to be broken/distorted in progressing to the transition state, the enthalpic barrier to dissociation is decreased. The less favorable entropic term in biotin dissociation from the G115S mutant is consistent with the notion that in the transition state, the loop containing one less glycine residue is less disordered than the same loop in the dissociation of biotin from wild-type BirA. By contrast, in the R118G mutant for which the progression to the transition state is entropically more favorable than for wild-type BirA, the additional glycine residue allows the loop of the mutant to acquire greater disorder in the transition state than it does in the wild-type protein.
At this time, structural interpretation of perturbations in the energetics of bio-5`-AMP binding are not possible because no structure of this complex is yet available. However, two pieces of evidence suggest that considerable nonadditivity exists in this system. First, for the G115S and R118G mutants the energetic consequences of mutation for bio-5`-AMP binding are much greater than those for biotin binding. Second, the complexity of the patterns and magnitudes of the energetic effects of all three mutations on the transition-state parameters governing bio-5`-AMP binding are consistent with cooperativity between the portions of the protein that interact with the adenylate and biotin moieties of bio-5`-AMP.
Relationship to biotin binding by streptavidin
Avidin and streptavidin are classical, high-affinity, biotin-binding proteins. The latter protein is a tetramer that binds four biotin molecules noncooperatively with an equilibrium dissociation constant of ∼10−13M. Like BirA, the ligand-binding site of each streptavidin monomer possesses a loop that undergoes a disorder-to-order transition in the course of biotin binding. The streptavidin:biotin binding reaction has been the subject of extensive thermodynamic, kinetic, and structural analyses, some of which are specifically directed toward understanding the function of the loop in the binding reaction (Freitag et al. 1997, 1999; Chu et al. 1998; Stayton et al. 1999). Specifically, the consequences of deletion of the streptavidin loop for biotin binding have been investigated. Unlike the extensive nine-residue segment of the 112–128 loop of BirA including amino acids 116–124, binding of biotin to streptavidin results in ordering of only a four-residue segment of each monomer. Nevertheless, deletion of the streptavidin loop results in a 107-fold increase in the equilibrium dissociation constant governing the binding reaction, and the destabilization of the interaction reflects a large enthalpic penalty to binding. Comparison of the structures of biotin complexes of wild-type streptavidin with the deletion mutant yields no information about the origin of this large enthalpic penalty, as the binding contacts in the loop deletion mutant are intact. By contrast, in the BirA:biotin complex, several of the side chain or backbone groups of residues in the 116–124 region that undergo the disorder-to-order transition form noncovalent bonds with biotin molecule in the complex. For example, W123, a residue that is conserved in all biotin-holoenzyme ligases, packs against the hydrocarbon chain of the valerate moiety of biotin. The backbone amides of G115 and R118 form hydrogen bonds with the ureido and valerate moieties, respectively, of the vitamin. Structural alterations in the BirA loop thus are anticipated to result in the enthalpic penalties to biotin binding observed in the G115S mutant. Binding of biotin to streptavidin is characterized by an unfavorable entropy, which has been proposed to reflect, in part, the penalty associated with the disorder-to-order transition of the flexible loop. In the case of the biotin holoenzyme ligase, the entropic term in equilibrium binding is negligible. Thus, while there may be an entropic penalty associated with ordering of the loop, it is compensated for by favorable contributions including solvent release.
Comparison of the kinetic profiles associated with release of biotin from BirA and streptavidin reveals similarities. Dissociation of biotin from each of these proteins is characterized by a large activation enthalpy and a favorable activation entropy. In both cases, the favorable entropy of conversion to the transition state in dissociation may reflect partial regaining of configurational entropy by the flexible loop, side chains of the loop, and/or the biotin ligand.
Implications for understanding ligand binding specificity
The biotin repressor is an example of a number of proteins that undergo disorder-to-order transitions in the course of ligand binding. Measurement of the consequences of structural perturbation in the glycine-rich loop of BirA for both equilibrium thermodynamic and kinetic features of the binding reaction has allowed development of a model for structural changes that accompany the progression of the ground-state complex to the transition state in dissociation. Protein-ligand interactions that are characterized by very favorable equilibrium binding energetics typically are characterized also by unusually high kinetic stabilities of the complexes to dissociation. Furthermore, structural perturbations to either the protein or ligand component of a complex result in changes in equilibrium-binding constant that frequently reflect only changes in unimolecular dissociation rate of the complex. Therefore, elucidation of the structural and energetic features of the dissociation process is essential to understanding specificity. Results provided in this work suggest that in ligand-binding processes that are coupled to disorder-to-order transitions, both the degree of complementarity between the protein and ligand in the complex and the relative disorder acquired in the transition state in dissociation may be utilized in tailoring ligand-binding affinity.
Materials and methods
Chemicals and biochemical
All chemicals used in preparation of buffers were at least reagent or analytical grade. The d-biotin was purchased from Sigma. Bio-5`-AMP was synthesized and purified using a modification of the procedure described in Lane et al. 1964 (Abbott and Beckett 1993).
Protein preparation
Histidine-tagged wild-type and mutant BirA proteins were prepared as described in Kwon and Beckett (2000). Protein concentrations were determined experimentally by ultraviolet absorption using extinction coefficients calculated from the amino-acid composition (Gill and von Hippel 1989).
Isothermal titration calorimetry
A MicroCal VP-ITC was used for the calorimetric measurements to determine the total heat of binding of ligands to BirA variants. All protein samples used for measurements were dialyzed exhaustively against buffer (Standard Buffer: 10 mM Tris-HCl, 200 mM KCl, 2.5 mM MgCl2 pH 7.5) the pH of which was adjusted at the experimental temperature. In each calorimetric measurement, 1.7 mL of a 1 μM protein sample were loaded in the reaction cell and the ligand, biotin, or bio-5`-AMP, was prepared as the titrant in the injection syringe. The concentrated bio-5`-AMP or biotin was diluted into the buffer used for protein dialysis. To improve baseline stability, all samples were degassed before use by stirring under vacuum at a temperature ∼4°C below the experimental temperature. A stoichiometric excess of ligand over protein was added in the first injection to saturate the protein. Four additional injections were used to determine the heat of dilution. Contributions of protonation to binding at pH 7.5 were determined by measurement of the molar heat of binding in Standard Buffer prepared using 10 mM Tris-HCl or 10 mM MOPS (3-(N-morpholino)propane sulfonate) as buffering agents. The large difference in ionization enthalpies of the two buffers allows facile determination of the contributions of protonation to the ligand-binding reactions (Jelesarov and Bosshard 1994; Baker and Murphy 1996; Bradshaw and Waksman 1998).
Data analysis
Origin version 5.0 software (MicroCal) was used for integration of peak areas, and corrected heat changes were calculated by subtraction of the average heat of dilution from the integrated area of the first injection. At least three measurements were performed for each set of conditions, and average molar binding enthalpies were calculated from the multiple determinations. The heat capacity changes for the binding processes were determined by global linear least-square analysis of the binding enthalpy versus temperature data using Microsoft Excel. At least three independent measurements of the binding enthalpy determined at each temperature were used in the analysis of the data to obtain the heat capacity changes associated with the binding processes.
Kinetic measurements
Stopped-flow fluorescence measurements
Stopped-flow fluorescence measurements were performed using a KinTek SF2001 instrument. The excitation wavelength was set at 295 nm for all measurement, and fluorescence emission was monitored above 340 nm using a cutoff filter (Corion Corporation).
Preformed protein-ligand complexes in Standard Buffer [10 mM Tris-HCl, pH 7.50 ± 0.01 at 20.0±0.1°C, 200 mM KCl, 7.5 mM MgCl2] prepared at the appropriate temperature were rapidly mixed in a 1:1 (vol:vol) ratio with solution containing a large molar excess of competing ligand, biotin, or bio-5`-AMP. The resulting time-dependent increase or decrease in fluorescence was monitored. Concentrations of competing ligands were sufficiently high so that the rate-limiting step was the release of ligand from the protein-ligand complexes.
The time course of BirA-catalyzed synthesis of bio-5`-AMP
The dissociation of bio-5`-AMP from wild-type BirA or the R119W variant was measured in Standard Buffer at 20°C using a method for monitoring the time course of BirA-catalyzed synthesis of bio-5`-AMP (Xu and Beckett 1994). Synthesis of bio-5`-AMP was quantitated by monitoring the incorporation of 32P from [α-32P]ATP into bio-5`-AMP at reaction conditions of 1μM enzyme, 500 μM ATP, 100 μM biotin, and 2 units/mL inorganic pyrophosphatase in Standard Buffer. Approximately 5,000,000 cpm of [α-32P]ATP were used for each reaction. The pyrophosphatase was used to drive the reaction equilibrium toward completion. Reactions were initiated by addition of biotin to the preequilibrated reaction mixture containing all other reaction components. Time points were obtained by the quenching of 3 μL aliquots of the reaction mixture into 1 μL of 4 mM unlabeled, chemically synthesized bio-5`-AMP. A 1-μL aliquot of each quenched reaction solution was spotted on a cellulose TLC plate (Sigma-Aldrich), and the 32P-bio-5`-AMP and α-32P-ATP were resolved using a mobile phase containing tert-amyl alcohol/formic acid/H2O (3/2/1 v/v/v). The TLC plate was exposed to a phosphor screen for at least 4 h, and quantitation was accomplished using the Molecular Dynamics phosphor-imaging system (Storm and Image Quant v 3.3). Radioactivity present in the spots corresponding to ATP and bio-5`-AMP was quantitated at each time point, and the bio-5`-AMP concentration was calculated by multiplying the initial ATP concentration by the result of division of the optical density of the bio-5`-AMP spot by the sum of the densities of the two spots.
Data analysis
All primary data from stopped-flow fluorescence measurements were analyzed using the software provided with the instrument. Time courses for dissociation were well described by a single exponential equation in all cases. At least 10 measurements of each dissociation rate were used to obtain an average dissociation rate. Time courses of BirA-catalyzed synthesis were analyzed using the following equation:
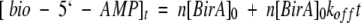 |
where [bio-5`-AMP]t is the concentration of the adenylate at time t, n is the stoichiometry of the burst complex, [BirA]0 is the total BirA monomer concentration, and koff is the unimolecular rate constant governing dissociation of the enzyme•adenylate complex. Linear regression of the adenylate concentration versus time data yielded the slope, n[BirA]0koff, which is proportional to the rate constant governing dissociation of the complex. Analysis of the rate data to obtain the activation parameters for the dissociation process was performed using the following form of the Eyring relationship:
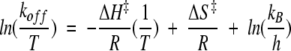 |
where koff is the unimolecular dissociation rate constant, T is the temperature in Kelvin, ΔH‡ is the activation enthalpy, ΔS‡ is the activation entropy, R is the gas constant, kB is the Boltzmann constant, and h is Planck's constant. All linear regression was performed using Microsoft Excel. Entropic contributions to the activation energy (−TΔS‡) also were calculated from the activation-free energies and enthalpies at 20°C using the following relationship:
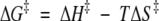 |
Acknowledgments
This work was supported by NIH Grant GM46511.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.33502.
References
- Abbott, J. and Beckett, D. 1993. Cooperative binding of the Escherichia coli repressor of biotin biosynthesis to the biotin operator. Biochemistry 33 9649–9656. [DOI] [PubMed] [Google Scholar]
- Baker, B.M. and Murphy, K.P. 1996. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. BiophysJ 71 2049–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, D.F. and Campbell, A.M. 1981a. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J. Mol. Biol. 146 451–467. [DOI] [PubMed] [Google Scholar]
- Barker, D.F. and Campbell, A.M. 1981b. Genetic and biochemical characterization of the birA gene and its product: Evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J. Mol. Biol. 146 469–492. [DOI] [PubMed] [Google Scholar]
- Bradshaw, J.M. and Waksman, G. 1998. Calorimetric investigation of proton linkage by monitoring both the enthalpy and association constant of binding: Application to the interaction of the Src SH2 domain with a high-affinity tyrosyl phosphopeptide. Biochemistry 37 15400–15407. [DOI] [PubMed] [Google Scholar]
- Chu, V., Freitag, S., Le Trong, I., Stenkamp, R.E., and Stayton, P.S. 1998. Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system. Protein Sci. 7 848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein, E. and Beckett, D. 1999. Dimerization of the Escherichia coli biotin repressor: Corepressor function in protein assembly. Biochemistry 38 13077–13084. [DOI] [PubMed] [Google Scholar]
- Freitag, S., Le Trong, I., Klumb, L., Stayton, P.S., and Stenkamp, R.E. 1997. Structural studies of the streptavidin binding loop. Protein Sci. 6 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag, S., Chu, V., Penzotti, J.E., Klumb, L.A., To, R., Hyre, D., Le Trong, I., Lybrand, T.P., Stenkamp, R.E., and Stayton, P.S. 1999. A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway. Proc. Natl. Acad. Sci. 96 8384–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Hyre, D.E., Le Trong, I., Freitag, S., Stenkamp, R.E., and Stayton, P.S. 2000. Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system. Protein Sci. 9 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelesarov, I. and Bosshard, H.R. 1994. Thermodynamics of ferredoxin binding to ferredoxin:NADP+ reductase and the role of water at the complex interface. Biochemistry 33 13321–13328. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- Kwon, K. and Beckett, D. 2000. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 9 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, K., Streaker, E.D., Ruparelia, S., and Beckett, D. 2000. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J. Mol. Biol. 304 821–833. [DOI] [PubMed] [Google Scholar]
- Lane, M.D., Rominger, K.L., Young, D.L., and Lynen, F. 1964. The enzymatic synthesis of holotranscarboxylase from apotranscarboxylase and (+) biotin: II. Investigation of the reaction mechanism. J. Biol. Chem. 239 2865–2871. [PubMed] [Google Scholar]
- Li, Y., Lipschultz, C.A., Mohan, S., and Smith-Gill, S.J. 2001. Mutations of an epitope hot-spot residue alter rate-limiting steps of antigen-antibody protein-protein associations. Biochemistry 40 2011–2022. [DOI] [PubMed] [Google Scholar]
- Miller, D.M. 3rd, Olson, J.S., Pflugrath, J.W., Quiocho, F.A. 1983. Rates of ligand binding to periplasmic proteins involved in bacterial transport and chemotaxis. J. Biol Chem. 258 13665–13672. [PubMed] [Google Scholar]
- Stayton, P.S., Freitag, S., Klumb, L.A., Chilkoti, A., Chu, V., Penzotti, J.E., To, R., Hyre, D., Le Trong, I., Lybrand, T.P., and Stenkamp, R,E. 1999. Streptavidin-biotin binding energetics. Biomol. Eng. 16 39–44. [DOI] [PubMed] [Google Scholar]
- Tissot, G., Douce, R., and Alban, C. 1997. Evidence for multiple forms of biotin holocarboxylase synthetase in pea (pisum sativum) and in Arabidopsis thaliana: Subcellular fractionation studies and isolation of a cDNA clone. Biochem. J. 323 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, L.H., Kwon, K., Beckett, D., and Matthews, B.W. 2001a. Corepressor-induced organization and assembly of the biotin repressor: A model for allosteric activation of a transcriptional regulator. Proc. Natl. Acad. Sci. 98 6045–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, L.H., Kwan, K., Beckett, D., and Matthews, B.W. 2001b. Competing protein:protein interactions are proposed to control the biological switch of the E. coli biotin repressor. Protein Sci. 10 2618–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K.S., Shewchuk, L.M., Brennan, R.G., Otsuka, A.J., and Matthews, B.W. 1992. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates biotin- and DNA-binding domains. Proc. Natl. Acad. Sci. 89 9257–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., and Beckett, D. 1994. Kinetics of biotinyl-5`-adenylate synthesis catalyzed by the Escherichia coli repressor of biotin biosynthesis and the stability of the enzyme-product complex. Biochemistry 33 7354–7360. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Nenortas, E., and Beckett, D. 1995. Evidence for distinct ligand-bound conformational states of the multifunctional Escherichia coli repressor of biotin biosynthesis. Biochemistry 34 16624–16631. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Johnson, C.R., and Beckett, D. 1996. Thermodynamic analysis of small ligand binding to the Escherichia coli repressor of biotin biosynthesis. Biochemistry 35 5509–5517. [DOI] [PubMed] [Google Scholar]