Abstract
The solution structure of human β2-microglobulin (β2-m), the nonpolymorphic component of class I major histocompatibility complex (MHC-I), was determined by 1H NMR spectroscopy and restrained modeling calculations. Compared to previous structural data obtained from the NMR secondary structure of the isolated protein and the crystal structure of MHC-I, in which the protein is associated to the heavy-chain component, several differences are observed. The most important rearrangements were observed for (1) strands V and VI (loss of the C-terminal and N-terminal end, respectively), (2) interstrand loop V-VI, and (3) strand I, including the N-terminal segment (displacement outward of the molecular core). These modifications can be considered as the prodromes of the amyloid transition. Solvation of the protected regions in MHC-I decreases the tertiary packing by breaking the contiguity of the surface hydrophobic patches at the interface with heavy chain and the nearby region at the surface charge cluster of the C-terminal segment. As a result, the molecule is placed in a state in which even minor charge and solvation changes in response to pH or ionic-strength variations can easily compromise the hydrophobic/hydrophilic balance and trigger the transition into a partially unfolded intermediate that starts with unpairing of strand I and leads to polymerization and precipitation into fibrils or amorphous aggregates. The same mechanism accounts for the partial unfolding and fiber formation subsequent to Cu2+ binding, which is shown to occur primarily at His 31 and involve partially also His 13, the next available His residue along the partial unfolding pathway.
Keywords: β2-microglobulin, amyloid, dialysis-related amyloidosis, protein structure, NMR of proteins, unfolding
β2-microglobulin (β2-m) is the nonpolymorphic light chain of the class I major histocompatibility complex (MHC-I). It consists of 99 residues with a single disulfide bridge between the two Cys residues of the sequence at positions 25 and 80 and folds into the classical β-sandwich motif of the immunoglobulin superfamily, as shown by the crystal structure of MHC-I (Bjorkman et al. 1987). The systemic deposition of β2-m fibrils is associated to dialysis-related amyloidosis (DRA; Gejyo et al. 1985), a disease which arises in individuals with chronic renal failure as the inescapable complication of long-term hemodialysis. More than 90% of patients undergoing dialysis for ∼10 y develop symptoms such as destructive arthropathy, bone fractures, and carpal tunnel. Because of the ensuing physical disabilities, against which treatment (including surgery) is only palliative, and the widespread occurrence of renal failure (∼200,000 individuals are steadily under dialysis in Europe alone), this type of amyloidosis can be regarded as a high-cost social disease.
Amyloidoses have been recognized as conformational diseases that arise from the conversion of globular proteins to insoluble fibrillar aggregates. In spite of the diversity of the involved proteins, their amyloid fibrils share a common structure known as cross-β structure, in which partially unfolded conformations aggregate through intermolecular β-strand pairing that are arranged perpendicular to the fiber polymerization axis (Sunde et al. 1997).
Analyses of the material extracted from DRA patients have shown that only full-length wild-type β2-m and proteolysis products thereof occur in fibrils (Linke et al. 1987, 1989; Bellotti et al. 1998), along with some glycation (Miyata et al. 1993) and oxidation derivatives (Capeiller-Blandin et al. 1991) and auxiliary proteins (apoE, serum amyloid P component; Miyata et al. 1993). The most abundant species among the truncation products of β2-m (30% of β2-m in ex vivo fibrils) is the form devoid of the six N-terminal residues (ΔN6β2-m). This species was shown to have a higher tendency to self-aggregate than the full-length protein and did not form a fully folded native state at the end of the refolding procedure (Bellotti et al. 1998). A comparative investigation of full-length β2-m and ΔN6β2-m by 1H NMR, electrospray mass spectrometry, and limited proteolysis enabled us to establish the extent and the location of the structural modifications undergone by β2-m upon removal of the N-terminal hexapeptide (Esposito et al. 2000). By analyzing the changes and the persistences of the NMR pattern of both products, some features of the amyloidogenic conformation of β2-m could be suggested, namely, the identity of the β-strands involved in the intermolecular pairing and a model to explain partial unfolding and aggregation of the truncated and full-length species (Esposito et al. 2000). In a subsequent study on the fibrils of β2-m and ΔN6β2-m, overlapping patterns of limited proteolysis were observed with an additional cleavage for the full-length protein fibrils leading to the truncated species (M. Monti et al. 2001, pers. comm.). These findings, while suggesting a mechanism for the occurrence of ΔN6β2-m in vivo, which implies that the only fibrillogenic species is the full-length protein, strongly support the idea that the truncated species features the amyloidogenic conformation of β2-m, as previously anticipated (Esposito et al. 2000; Chiti et al. 2001). Going further along this rationale, we wondered which could be the structural characteristics that drive the transition of β2-m into an amyloidogenic conformation. The protein occurs naturally in the MHC-I complex and is lost from the cell surface into the plasma to reach the kidneys and eventually to be degraded. When the renal function is compromised and dialysis becomes necessary, only a considerable increase of its plasma concentration is steadily detected (25- to 35-fold; Gejyo et al. 1985) before deposition of amyloid fibrils. Although the isolated protein in solution has been studied by NMR (Okon et al. 1992), only a qualitative analysis of its secondary structure was reported that showed, however, a very close analogy with the crystal structure of the protein in the context of the MHC-I complex (Bjorkman et al. 1987; Saper et al., 1991). Because even minor deviations from the crystal structure may affect significantly the stability of a protein that naturally binds to the polymorphic component of human leukocyte antigen, a determination of the tertiary structure of isolated β2-m in solution is useful, provided the proper level of accuracy is reached to perform a meaningful comparison with the crystal structure. Here we present the 3D solution structure of isolated β2-m obtained from 1H NMR data (Protein Data Bank ID 1JNJ) and show that differences with respect to the crystal structure are indeed observed.
Results and Discussion
NMR assignments and secondary-structure identification
The 1H NMR assignment of the β2-m spectrum and its secondary-structure determination have been reported (Okon et al. 1992). In a previous work (Esposito et al. 2000), the 2D NMR spectra of β2-m, obtained in aqueous ammonium acetate (50 mM) at pH 6.6 and T = 310 K, were compared with the reported assignments. Spin systems were first identified in the TOCSY spectrum and then confirmed by checking the sequential connectivity pattern in the NOESY maps (Wüthrich 1986). For β-sheet regions, the occurrence of a number of interstrand contacts were ascertained. Overall, the identification of the resonance patterns for 92 residues (plus Met 0) out of 99 were achieved. No consistent resonance pattern were observed for the remaining 7 residues due to exchange broadening and/or inherent line-width problems. All these gaps were filled in the present study, performed under similar conditions except for the saline phosphate buffer (Table A1 of Supplemental Material). The TOCSY fingerprint data (Fig. 1) were used also for estimating JHN-Hα coupling constants (Table A2 of Supplemental Material) to extract ϕ-torsion-angle restraints (Wang and Bax 1996). In this study also the analysis of NOESY data was extended to assign and quantify systematically as many correlations as possible to provide the internuclear distance set required for restrained modeling. The qualitative consideration of the NOESY pattern enabled confirming the occurrence of the eight strands of β structure along with their connecting loops reported by Okon et al. (1992). The secondary-structure diagnostic plot from the sequential NOE-connectivity data is depicted in Figure 2 ▶. In addition to intraresidue and sequential connectivities, most of the effort was devoted to the assessment of medium- and long-range NOEs. From the NOESY spectra obtained in D2O all the interstrand connectivities were assigned and measured, including those that had escaped our former qualitative classification (Esposito et al. 2000), which enabled recognizing the limits and pairing pattern of the β strands. The latter was consistent with the expected β-sandwich motif (Saper et al. 1991) formed by two pleated sheets, respectively of 4 (sheet 1) and 3 (sheet 2) antiparallel β strands, plus a short connection strandlike stretch between the two sheets. From the NOESY spectra in D2O also the cis configuration of Pro 32 could be established, in agreement with the previous NMR and crystal-structure evidence.
Fig. 2.
Summary of the secondary-structure diagnostic NOEs observed from 500 MHz 1H 2D NOESY spectra of β2-m. Sequential and short-range NOE connectivities (dNN[i, j+1], dαN[i, j+1], dβN[i, j+1], dαN[i, j+2]) are indicated by bars of proportionally different height indicating the intensity of the various NOEs. Grossly speaking, an extensive β structure could be inferred from this figure, although the pattern appears somewhat discontinuous and hence different with respect to the analogous plot reported by Okon et al. (1992) because only the NOE connectivities that could be actually used for modeling are shown (i.e., only those with an overlap <40%).
Structure determination
The collected restraints were analyzed to remove redundant information. The refinement process led to 1541 meaningful interatomic distances, which together with 67 dihedral angle restraints and 22 stereospecific assignments formed the experimental database for the subsequent restrained modeling. These data are resumed in Table 1, which lists the statistics of restraint violations and structural parameters that establish the definition of the final 20 best structures obtained from DYANA and DISCOVER refinement. It is readily seen that these quality parameters worsen on submitting the DYANA best-conformation family to restrained molecular mechanics. The average violation numbers, for instance, increase nearly by one order of magnitude in the DISCOVER-refined ensemble, although similar figures as those obtained from DYANA are reached by shifting the violation limits to looser but still meaningful values. The degradation of the structure-quality parameters, systematically experienced upon refinement of NMR-restrained structures, is preeminently due to basic geometry and force-field-constant differences among the various modeling software (Doreleijers et al. 1998), in particular for bond lengths, angles, and group planarity constraints. Although such an outcome has to be considered as the unavoidable consequence of current methodology limits, a number of indicators have been recognized to provide a suitable basis for validation of NMR-based structures (Laskowski et al. 1996; Doreleijers et al. 1998). Besides checking the deviations of the number of restraints per residue and the root-mean-square (rms) NOE violation from their statistical mean values, very stringent parameters are the compliance of ϕ–Ψ distribution with the most favorable region of the Ramachandran plot, referred to as Ramachandran quality, and the reliability of the backbone and side-chain torsion-angle distribution expressed as G-factor (Laskowski et al. 1996). The latter is the log-odds score probability, computed from high-resolution X-ray data, of a specific angle distribution within the final structure family expressed as circular variance (Allen and Johnson 1991) or order parameter (Hyberts et al. 1992). These data are listed in Table 2 and allow one to conclude that the presented structure matches the currently accepted validation standards. A more extended documentation of the validation parameters is available in the Supplemental Material. The structure coordinates can be retrieved from Brookhaven Protein Data Bank (PDB ID 1JNJ).
Table 1.
Structure quality parameters for human β2-m from restrained dynamics (DYANA) and mechanics (DISCOVER) calculations
| Parameter | DYANA | DISCOVER |
| Number of structures | 20 | 20 |
| Average target function (/10−2 nm2) | 1.64 ± 0.20 [1.24–1.86] | n.a. |
| Average number of violations | ||
| Distance upper limits > 0.02 nm | 2.5 ± 1.4 [0–5] | 30.5 ± 2.9 [27–38] |
| Distance lower limits > 0.02 nm | 0.5 ±0.7 [0–3] | 6.1 ± 1.9 [3–9] |
| van der Waals > 0.02 nm | 0.7 ± 0.8 [0–3] | n.d. |
| Dihedral angles > 5 deg | 0 | 8.0 ± 1.1 [6–10] |
| Average violation (fall restraints) | ||
| Distance upper limits (/10−1 nm) | 0.009 ± 0.001 | 0.026 ± 0.001 |
| [0.011–0.010] | [0.024–0.028] | |
| Distance lower limits (/10−1 nm) | 0.002 ± 0.000 | 0.006 ± 0.001 |
| [0.001–0.003] | [0.004–0.008] | |
| Average pairwise RMSD (residues 0–99) | ||
| Backbone (/10−1 nm) | 1.24 ± 0.21 | 1.27 ± 0.21 |
| Heavy atoms (/10−1 nm) | 1.99 ± 0.21 | 2.17 ± 0.24 |
| Average pairwise RMSD (residues 1–97) | ||
| Backbone (/10−1 nm) | 0.83 ± 0.10 | 0.87 ± 0.11 |
| Heavy atoms (/10−1 nm) | 1.66 ± 0.13 | 1.74 ± 0.13 |
| Average energy (/kJ mol−1) | +7290 ± 518 | −1697 ± 84 |
Minimum and maximum values of the target function, violation numbers, and extents are given in brackets.
n.a., not applicable.
n.d., not determined.
Overall 1541 meaningful distance restraints were employed (679 intraresidue, 281 sequential, 102 medium-range, and 479 long-range, along with 22 stereospecific assignments (13%), 66 ϕ, and 1 χ1 dihedral restraints, giving a total of 1608 restraints. Only in one case the analysis enabled defining a range for χ1 torsion angle that was used as restraint within bounds of ± 60°; whereas, when the stereospecific assignment was defined, no χ1 torsion restraint was additionally employed. Rather than using the same experimental evidence also for imposing χ1 restraints, we preferred to check the agreement between the expected value range and the final model to verify the correctness of the diastereotopic identifications without further restraint bias.
Table 2.
Structure validation parameters for human β2-m conformers from NMR data refinement (DISCOVER)
| Parameter | This work | Consensus standarda |
| Total residues/selected residuesb | 100/83 | 74 ± 39/62 ± 36 |
| Number of conformers | 20 | 20 ± 15 |
| Restraints per residuec,d | 10.3 | 11.3 ± 4.5 |
| NOE rms violation (/10−1 nm)e | 0.079 | 0.061 ± 0.043 |
| Ramachandran qualityb | ||
| Residues in most-favored regions | 70.5% | 73.6 ± 15.5% |
| Residues in additional allowed regions | 28.2% | n.a. |
| Residues in generously allowed regions | 1.0% | n.a. |
| Residues in disallowed regions | 0.3% | n.a. |
| Average G-factorsb,f | ||
| ϕ − Ψ | −1.13 | n.a. |
| χ1 −χ2 | −1.03 | n.a. |
| χ1 only | −0.14 | n.a. |
| overall | −0.98 | n.a. |
n.a., not available.
a From Doreleijers et al. 1998.
b According to Doreleijers et al. (1998) the listed parameters, except for the NOE rms violation, were calculated on well-defined segments of the protein, namely only those residues for which the average of the circular variance of the backbone angles φ and Ψ was <0.2 were included in the selected segments.
c To avoid the redundancy of considering upper and lower bounds, only a single distance bound per internuclear separation was included in the count. Thus, only 789 NOE and 67 dihedral angle restraints were included in the count.
d Referred to the reduced NOE-restraint set above considered from well-defined segments.
e Referred to all residues.
f Low G-factors indicate low-probability conformations (Laskowski et al. 1996). As a rule of thumb, acceptable overall values range around −1.
Solution conformation of β2-m
The best-fit superposition of the refined-structure family obtained from the present NMR study is shown in Figure 3 ▶. According to standard secondary structure classification criteria (Kabsch and Sander 1983), eight strands of β structure are identified, namely, within fragments 6–11 (strand I), 23–28 (strand II), 35–36 and 39–40 (strand III), 45–46 (strand IV), 49–51 (strand V), 63–68 (strand VI), 79–84 (strand VII), and 91–94 (strand VIII). It is worth noting, at this point, that the β-strand numbering adopted by Okon et al. (1992) does not coincide with the one given for the low- (Bjorkman et al. 1987) and high-resolution (Saper et al. 1991) crystal structures. According to the latter structures, β2-m contains only seven β strands. An additional 2-residue β-strand segment, running approximately perpendicular to strand III and beginning the crossover back to the first sheet of the β-sandwich, was named strand III' in the crystal structure but became strand IV in the NMR description. As a consequence, the NMR designation of the subsequent strands carried on the numbering shift, that is, strand IV of the crystal structure was referred to as strand V and so on. Here we adopt the NMR notation.
Fig. 3.
Stereoview of the best-fit superposition of the 20 final conformers of β2-m after DISCOVER refinement (MSI) of the NMR-restrained ensemble obtained from DYANA (Günter et al. 1997). Only backbone atoms are drawn. The backbone global rms deviations (nm) along specific segments (in parentheses) of the structure ensemble in solution are 0.147 (1–5), 0.053 (6–11), 0.096 (12–22), 0.036 (23–28), 0.091 (56–62), 0.062 (41–44), 0.085 (45–46), and 0.121 (47–48).
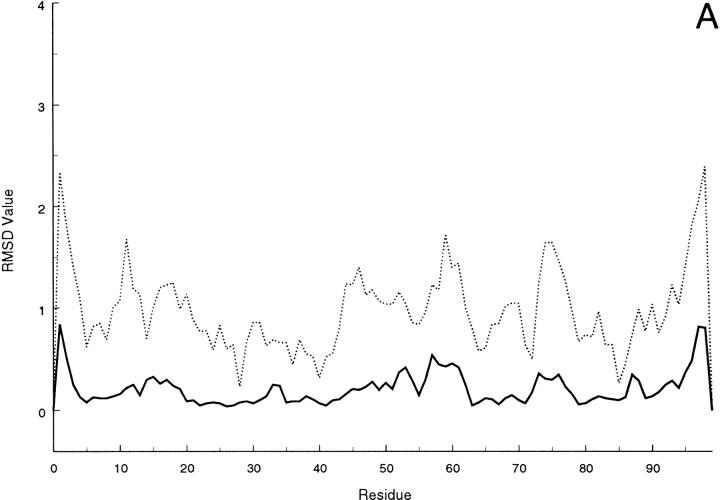
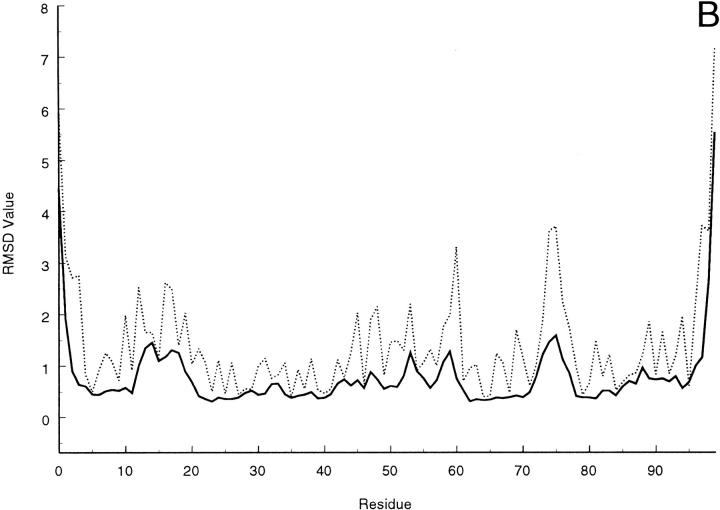
Slight variations of the strand limits are observed within the refined conformational ensemble, and the results indicate that strand IV is definitely missing in several members of the family (11 out 20); whereas, in most of them strand V does not extend beyond residue 51. As a matter of fact, only 3 structures show residue 55 involved in either a β bridge or a short β strand comprising the flanking residues. The pairing pattern involves strands I, II, VI, and V on the one hand and strands III, VII and VIII on the other in the β-sandwich motif of the molecule, with the two sheets held together by a disulfide bridge between Cys 25 and Cys 80. The strand limits found in the present NMR-structure determination are summarized in Table 3. An increased extent of conformational spreading, reflecting a reduced number of NOEs, is observed at segments 0–2 (N terminus), 13–18 (loop between strands I and II), 47–48 (loop IV-V), 53–54 (strand V), 57–61 (loop V-VI) and 96–99 (C terminus). This can be seen in Figure 4 ▶, in which the residue local and global rms deviation (Güntert et al. 1991) over the 20 final DYANA structures are depicted (similar features can be observed for the DISCOVER-refined structures).
Table 3.
Comparison of the secondary structure elements of human β2-m from present work and previous NMR and X-ray data
| Residue limits | |||
| NMR | |||
| Strand (sheet) | X-raya,b | Previous datac | Present workb,d |
| I (1) | 6–11 | 3, 6–12 | 6–11 |
| II (1) | 2–30 | 20–28, 31 | 23–28 |
| III (2) | 35–41 | 36–41 | 35–36, 39–40 |
| IV (2) | 44–45 | 44–45 | 45–46 |
| V (1) | 50–51, 55–56 | 50–52, 55–57 | 49–51 |
| VI (1) | 62–70 | 61–71 | 63–68 |
| VII (2) | 78–84 | 77–83 | 79–84 |
| VIII (2) | 91–94 | 90–95 | 91–94 |
| Turns | |||
| 41–44 | |||
| 57–61 | |||
| 84–87 | |||
a Protein Data Bank record (1HLA) from X-ray structure determination of HLA-A2 (Bjorkman et al. 1987; Saper et al. 1991).
b Secondary structure were defined according to Kabsch and Sander algorithm (1983).
c NMR-based secondary structure identification (Okon et al. 1992).
d NMR-restrained solution structure after DISCOVER refinement. Secondary structure classification is reported by collecting the information from PROCHECK-NMR analysis (Laskowski et al. 1996) over the 20-membered NMR-structure ensemble. Strands I, II, III, VII, and VII were univocally defined within the family (occurrence >50%). Strand IV was present in 9 out of 20 structures; whereas, strand V was identified in the majority of them (13), though only 7 structures included also residue 49. Likewise, strand VI encompassed segment 64–68 in 13 models, out of which the limits extended to residue 63 in 6 cases.
Fig. 4.
(A) Local and (B) global average rms deviations calculated over the 20 best DYANA conformers according to Günter et al. (1991) for backbone (solid line) or heavy atoms (dashed line).
Comparison with previous structural data
Some potentially relevant differences are recognized between the qualitative picture of β2-m secondary structure in solution (Okon et al. 1992), its crystal structure, and the solution conformational family presented here. By considering the above listed regions of conformational dispersion and the data of Table 3, it is seen that our results differ from Okon and coworkers' scheme of secondary structure that, in turn, is close to the X-ray classification. The largest difference occurs at the C-terminal three-residue moiety of strand V that is largely missing in the conformation family of Figure 3 ▶. This latter feature, together with the shortening at strand VI and the structural dispersion at segments 53–54 and 57–61, appears consistent with the proposal of a slow conformational equilibrium in the region 56–62 of β2-m (Okon et al. 1992). Also our evidence, while suggesting a fairly increased mobility at segment 58–59, confirms the occurrence of anomalously broad lines for the flanking residues (56, 57, 60, and 61) and even for distant but closely spaced ones (e.g., Pro 32). The implied conformational dynamics do not support a stable β-sheet pairing at the end of strand V and the start of strand VI.
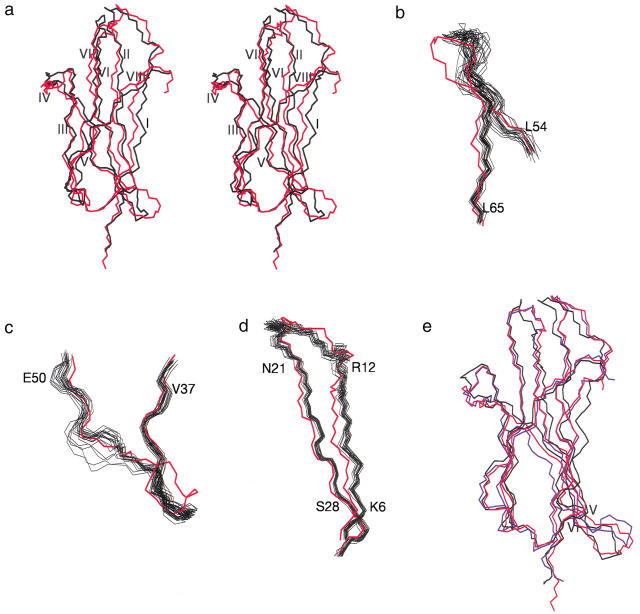
The differences between the tertiary structure of β2-m in solution and the crystal structure of the protein in the MHC-I complex are highlighted in Figure 5 ▶, in which separate panels show the overall best-fit superposition of the average solution model over the crystal structure (Fig. 5a ▶) and the details of the overlap in significantly displaced regions using the whole collection of the solution ensemble (Fig. 5b–d ▶). The overall backbone rms deviation between the crystal and mean solution structure is 0.16 nm. Any comparison is drawn with the X-ray data of MHC-I complex (Saper et al. 1991) because the crystal structure of isolated bovine β2-m (75% sequence identity; Becker and Reeke 1985) does not exhibit significant differences with respect to the structure in the complex except for a displacement at the loop between strands V and VI. Figure 5e ▶ displays the superposition of the two-crystal structure and the average-solution model. Concerning the mentioned loop, the MHCI-bound structure is somehow halfway between the isolated protein structure in solid and liquid phase (see below). It is readily seen that, in addition to the fraying pattern at both termini of the solution structural family, more pronounced at the C terminus (Fig. 3 ▶), the largest changes and, consequently, rms deviations from crystal structure occur at segments 56–62 (Fig. 5b ▶) and 41–48 (Fig. 5c ▶) and for the sheet formed by strands I and II (Fig. 5d ▶). Obviously, overlap to crystal structure is poor also for any other region of the structural family in solution with significant conformational spreading. This is, for instance, the case within segment 41–48 (Fig. 5c ▶), where the stretch 47–48 is quite dispersed within the NMR ensemble; whereas, the preceding moiety (fragment 41–46) appears much more conformationally homogeneous and, in fact, just displaced with respect to crystal structure. Net displacements, rather than conformational dispersion, are evident also for strand I and the subsequent loop with strand II (Fig. 5d ▶) and for strand VI (Fig. 5a ▶). These deviations may bear relevance to rationalize the amyloidogenic behavior of β2-m. In particular, the displacement and/or dispersion of nearly all strands and loops of the first sheet, coupled to similar features within segment 41–48 and the splitting of strand III, should lead to loosening of the packing observed in the crystal structure. The overall interaction between the two sheets of the β-sandwich should be weakened, though the disulfide bridge still holds strands II and VII in their relative positions. The conspicuous departure from the crystal-structure geometry of the segment 56–62 (the largest observed segmental displacement) is certainly linked to the loss of the β-sheet arrangement at the C-terminal moiety of strand V and the N-terminal residues of strand VI, though establishing whether or not the former induces the latter may be misleading. The whole region extends out along the plane defined by strands V and VI rather than kinking toward the core, as observed in the structure of the complex (Fig. 5a,b ▶). This back-kink is even more pronounced in the isolated bovine β2-m structure (Fig. 5e ▶; Becker and Reeke 1985). All these structural changes are induced by interaction with solvent of specific β2-m surface regions, normally in contact with the heavy chain of the histocompatibility antigen complex. The rearrangements should entail separation of contiguous hydrophobic regions (Cheng and Rossky 1998), that is, decrease of tertiary packing. Inspection of MHC-I structure (Saper et al. 1991) shows that strands I and II and the loop between them are effectively solvent-protected by the α3 domain of the heavy chain that engages 10 out of its 13 contacts with β2-m in this region (the remaining 3 involving the C-terminal dipeptide and a residue side chain of strand VI). Besides the displacement of strand I and the following loop, the solvent accessibility difference between HLA-bound and isolated β2-m on removal of the α3 domain screening explains also the loss of the salt bridge between the side chains of Glu 16 and Lys 19 that can be inferred from the structure based on NMR restraints. Conversely, the other 3 salt bridges observed in the HLA-bound β2-m (involving the side chains of pairs Asp 38-Arg 45, Asp 38-Arg 81, and Glu 77-Lys 94; Saper et al. 1991) appear conserved in the NMR structure because, already in the complex, the corresponding locations are unscreened by the heavy chain (detailed information about the assessment of H-bond and salt-bridge occurrences in NMR-based solution structures is available in Table A4 of Supplemental Material). On the other hand, the interaction between α1 and α2 domains of the heavy chain and β2-m shields from solvent exposure the following regions of the latter: the interstrand loops II-III and V-VI, the final part of strand V and the preceding bulge, and the N-terminal end of strand VI. Of the 15 contacts of β2-m with α1 and α2 domains, 13 occur at the mentioned locations; whereas, the remaining 2 are observed at the N-terminal tripeptide segment of β2-m (Saper et al. 1991). Upon exposing this β2-m interface region, the N-terminal fragment and the loop between strands V and VI diverge out of the plane of loop II-III to accommodate local solvation at the expense of the reciprocal contacts that tighten up the folding of the protein in MHC-I complex (Fig. 5 ▶). This rearrangement literally opens one of the apical regions of β2-m and breaks down the local tertiary packing. In particular, the hydrophobic contacts of the crystal structure between the side chains of Ile 1 and Arg 3 on one hand and Phe 30 and His 31 on the other are totally lost in solution, as shown by the absence of any long-range NOE among those residues. At the same time, the spatial proximity between these directly interacting residues and the loop V-VI through the contacts with the same segment of α2 domain is abolished. In the region between strands V and VI, including the loop and the flanking residues, solvation induces the mentioned conformational rearrangement that breaks the local packing and exposes not only Asp 53 and Lys 58 (already at the interface in the complex), but also Asp 59 (partially buried in the complex). Charge solvation also occurs at Arg 3 and Lys 6 (strand I) and Lys 91 (strand VIII), all of which were exposed in the complex except in one case (Lys 6) with the screen of a salt bridge to α3 domain. Here the packing is lost in isolated β2-m because solvation must not be as effective as the salt bridge required in the complex to screen a charge embedded onto a hydrophobic surface. A similar loss of packing arising from substitution of solvation for salt bridges is observed at the C-terminal stretch of β2-m. The local charge cluster flanking the hydrophobic surface at the interface with the heavy chain is held with the contribution of two salt bridges connecting the α3 domain to residues 98 and 99 of β2-m. The resulting geometry compacts the C-terminal segment on the surface with a back-turning arrangement that allows the formation of a H-bond between the side chains of Arg 97 and Asp 17. None of these features survives in isolated β2-m, showing that solvation breaks the surface contiguity not only in hydrophobic districts (Cheng and Rossky 1998) but also in hydrophilic ones.
Fig. 5.
Comparison between X-ray and solution structures of β2-m. (A) Overall backbone best-fit superposition of crystal (red) and mean solution structure (black) over the refined ensemble. The overall backbone rms deviation between the crystal and mean solution structure is 0.16 nm. The local backbone rms deviations (nm) between X-ray and mean solution structure are 0.212 (1–5), 0.156 (6–11), 0.154 (12–22), 0.092 (23–28), 0.277 (56–62), 0.196 (41–44), 0.180 (45–46), and 0.134 (47–48). Details of the best-fit superposition of the X-ray (red) and the 20 best-refined (black) structures showing (B) loop between strands V and VI, (C) strand IV, and (D) strand I and II. (E) Best-fit superposition of the mean solution structure (black) and two available crystal structures, respectively MHC-I-bound human β2-m (red) and isolated bovine β2-m (blue). In addition to the displacement of loop V-VI mentioned in text, strand V appears shifted in bovine β2-m with respect to human protein structures. However, the bovine sequence exhibits a deletion at position 48, that is, at the start of strand V, which should particularly contribute to the shift of the subsequent fragment.
Implications for β2-m amyloidogenesis
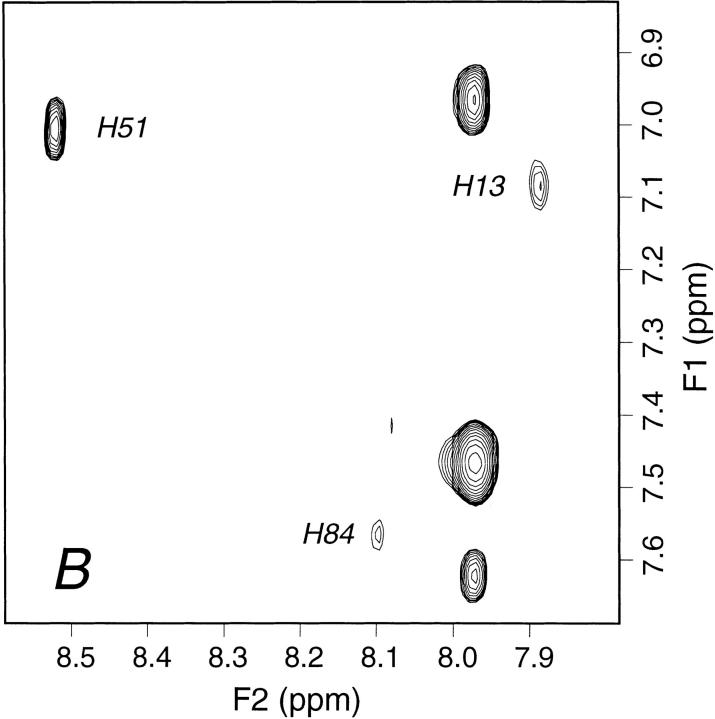
From our examination of the deviations of isolated β2-m from X-ray geometry, it follows that the loss-of-solvent protection from α1 and α2 domains brings about the most remarkable consequences on the structure of the protein. The deviations from crystal geometry of the N-terminal fragment and the loop V-VI (Figs. 3, 5a ▶ ▶) are, in fact, crucial modifications toward the amyloidogenic transition. Although the loss of screening due to α3 domain is responsible for propagating further the deviation from X-ray structure, displacements of both regions on the opposite sides of the loop II-III puts the protein in a state in which the balance between hydrophobic and hydrophilic forces is very close to the edge. As shown previously (Bellotti et al. 1998; Esposito et al. 2000), the removal of the N-terminal hexapeptide, implying the suppression of the charged Arg 3 and Lys 6, distinctly facilitates protein aggregation and fibril formation. The same effects are observed with the full-length protein upon decreasing the pH, even just below 6, especially in the presence of chaotropes worse than sodium phosphate, such as ammonium acetate (Esposito et al. 2000). The role of pH and ionic strength in β2-m fibrillogenesis has been addressed at length by a recent study (McParland et al. 2000). Based on evidence from different spectroscopic techniques, the investigators conclude that a partially unfolded state of the protein, reached by titration of one or more residues with an apparent overall pKa of 4.7, is a key intermediate in fibrillogenesis. The observed pKa may reflect the acid-base equilibrium of one or more of the 8 Glu or 7 Asp residues of the molecule, although other residues with unusually low pKa values, such as the buried His 84, cannot be ruled out. Among the other possibilities, the same investigators also mention Asp 59 and Glu 74, which, according to their calculations, should form salt bridges with Arg 3 and Arg 97, respectively (McParland et al. 2000). Besides the fact that none of these salt bridges is found in either the crystal structure and the solution conformational family presented here, salt-bridged carboxylates of Asp or Glu residues would be expected to shift their pKa well below 4.7, so the proposal appears obscure if not conflicting. Moreover fibril formation with very low yields has been reported also at physiological pH, although under conditions of very high protein concentration and very low ionic strength (Connors et al. 1985; Stoppini et al. 1997). Taken all together these results suggest that an interpretation aiming at attributing to a single-residue titration a key role for the amyloidogenic transition of β2-m may be misleading. Rather, the whole body of evidence seems to confirm our previous proposal (Esposito et al. 2000). In the isolated protein, the hydrophobic versus hydrophilic balance of the region contacting the α1 and α2 domains of HLA heavy chain is so delicate that altering the overall local charge, even by titration of a single-residue side chain, induces the amyloidogenic transition, which starts with the detachment of strand I from the first sheet of the molecule. By the end of this work a communication appeared (Morgan et al. 2001) that strongly supports our interpretation. Based on electrospray mass spectrometry evidence, the formation of a specific complex between Cu2+ and β2-m is reported that unusually destabilizes the protein folding against thermal and urea denaturation despite a nativelike conformation appears to be retained by β2-m upon Cu2+ binding. After showing that denatured β2-m binds four metal ions through the 4 His residues of the sequence (Fig. 6 ▶), the authors argue that Cu2+ binding must probably occur at one or more of the conserved His residues (His 31 and His 84) that should be essential for the stability of the protein because Cu2+ binding may affect the stability in a manner similar to His protonation. The authors further show that Cu2+ binding and destabilization uniquely promote de novo fiber formation at 37°C and neutral pH. The effects of the specific metal binding on the protein stability are totally consistent with the structural picture and the unfolding pathway outlined by this study. Even before any control, we could anticipate that Cu2+ binding must have occurred at His 31, which in solution becomes exposed due to the loss of the interactions with Ile 1 and Arg 3 side chains (Fig. 6 ▶) and which is located just at the top of the loop II-III, in a region where any minimal change of the charge status is likely to be fatal for the folding. Surprisingly, the communication by Morgan et al. (2001) also reports that the 1D NMR spectrum of β2-m is not significantly perturbed by the presence of Cu2+. At variance, we find that the 1D NMR spectrum is indeed affected, even at a Cu2+/β2-m ratio of 1:10, just for the aromatic resonances of His 31. By continuing to titrate, it is seen that already at a ratio 3:10 the His 31 Hδ2–Hɛ1 TOCSY connectivity is totally lost (Fig. 7 ▶) due to the fast-exchange averaging of the metal paramagnetic perturbation; whereas, an attenuation due to a less specific interaction starts to arise also for His 13, that is, the next available His residue along strand I, which has the displacement that starts the transition to the partially unfolded intermediate. At the same time, no effects at all are observed for either His 51 and His 84, also at metal protein ratios ∼1:1 (data not shown). Rather, a slow increase of the sample turbidity is seen, which becomes clearly visible ∼1 wk after copper addition. Because β2-m precipitation is the hallmark of its partial unfolding and aggregation into insoluble oligomers and Cu2+ promotes β2-m fiber formation and amorphous aggregate after a 6-d incubation (Morgan et al. 2001), our data show that Cu2+ reversible binding to His 31 induces a slow transition that begins with the unpairing of strand I and leads to the amyloidogenic partially unfolded intermediate of β2-m under conditions (temperature, protein concentration, pH, and buffer) that otherwise support steady sample stability for several months.
Fig. 6.
Orientation of the histidine side chains in human β2-m crystal structure (MHC-I-bound protein). The insets show the same side chains in the first 10 members of the solution structure family (black) with the corresponding crystal structure (red), all drawn in frame with the backbone best-fit superposition.
Fig. 7.
Aromatic region from 1H 2D TOCSY spectrum of 0.71 mM β2-m in 70 mM phosphate buffer, 100 mM NaCl at pH 6.6, T = 310 K, (A) in the absence and (B) in the presence of Cu2+ at a metal/protein concentration ratio of 3:10. Cu2+ titration experiments were performed by successive addition of 1 μL aliquots from a 35 mM CuCl2 solution. The histidine Hδ2–Hɛ1 cross-peak assignments are indicated. The complete bleaching of the connectivity arising from His 31 in the presence of Cu2+ is consistent with a specific reversible binding of the ion to the imidazole ring of the residue. The extent of resonance attenuation reflects an efficient averaging of the paramagnetic perturbation due to fast Cu2+ exchange rate, that is, faster than the transverse relaxation rates of His 31 aromatic hydrogens. Partial attenuation is also observed for the connectivity attributed to His 13, the next available binding site along the partial unfolding pathway; whereas, no effect is detectable for the other two histidine cross-peaks (His 51 and His 84) and also on increasing the metal ion/protein ratio up to 1:1. In addition to the imidazole ring connectivities, only the Hβ resonances of His 31 and His 13 are detectably attenuated by further Cu2+ addition (i.e., beyond ion/protein ratio of 0.3), due to spatial proximity of the paramagnetic species binding site.
Conclusions
The present investigation of the solution 3D structure of human β2-m by 1H NMR spectroscopy was performed under similar conditions as those used for the determination of the secondary structure (Okon et al. 1992) and was not affected by protein precipitation problems.
A family of β2-m conformers was obtained from restrained molecular modeling of the NMR data that was submitted to refinement. The resulting structures were shown to comply with the currently accepted standards for NMR structure validation. By comparing this solution ensemble with the β2-m crystal structure in the class I histocompatibility antigen complex (Saper et al. 1991), a number of deviations were observed. The most important rearrangements were found for (1) strands V and VI (loss of the C-terminal and N-terminal end, respectively), (2) interstrand loop V-VI, and (3) strand I, including the N-terminal segment (both items diverging from the molecular core in opposite directions). These last modifications can be hypothesized as the prodromes of the amyloid transition. The close correspondence between regions of β2-m solution structure that deviate from the crystal structure and contacts with the heavy chain in MHC-I structure highlights the role of solvation in undermining the stability of the isolated protein. The unfolding ΔG of β2-m, measured from GdnHCl titration at 293 K, 20 mM phosphate at pH 7.3, is 6.6 ± 0.2 Kcal/mole (De Lorenzi et al. 2001), that is, equal to typical values observed for amyloidogenic proteins overcoming the folding energy barrier (Hurle et al. 1994). Water exposure washes out folding free-energy contributions by breaking the contiguity of the surface hydrophobic patches at the interface with HLA heavy chain and the nearby region at the surface charge cluster of the C-terminal segment. This partial unpacking also weakens or destroys a number of hydrogen bonds and intramolecular salt bridges of β2-m. Without the contacts with the heavy chain that must stabilize the fold, even minor additional solvation injuries, subsequent to net charge variations in response to pH or ionic strength changes or also Cu2+ binding, can easily compromise the precarious hydrophobic/hydrophilic balance and trigger the transition into a suitable, partially unfolded intermediate, the very start of the amyloidogenic transformation. Based on both the present analysis and previous evidence (Esposito et al. 2000; Monti et al. 2001, pers. comm.), this transformation should begin with the unpairing of the N-terminal strand I and evolve into a conformation in which also strands III, IV, and V are lost or rearranged through intermolecular pairing involving most of the former strand V and possibly strand III prior to precipitation. The results presented here suggest that further information can be gained through the structural analysis of suitable β2-m mutants and work is already in progress along this line. However, already at this stage, an interesting hot-spot region of the protein has been identified that may be addressed for the design of proper ligands to stabilize the folding and prevent the amyloidogenic transition.
Materials and methods
NMR spectroscopy
Human β2-m with an additional Met at the N-terminus (Met 0) was obtained as previously reported (Esposito et al. 2000). 1H NMR spectra were collected at 500.13 MHz with a Bruker Avance 500 NMR spectrometer from 0.4 to 0.9 mM aqueous solutions (H2O/D2O (90/10) or 100% D2O) with 70 mM phosphate buffer, 100 mM NaCl at pH 6.6, T = 310 K. A number of 2D TOCSY (Braunschweiler and Ernst 1983), DQF-COSY (Piantini et al. 1982), and NOESY (Jeener et al. 1979) spectra were acquired with solvent suppression (Piotto et al. 1992; Hwang and Shaka 1995), 1- to 1.5-sec steady-state recovery time, mixing times (tm) of 20–50 ms for TOCSY and 50–150 ms for NOESY, t1 quadrature detection by TPPI (Marion and Wüthrich 1983), or States method (States et al. 1982), or gradient-assisted coherence selection (echo-antiecho; Keeler et al. 1994). The spin-lock mixing of the TOCSY experiment was obtained with MLEV17 (Bax and Davis 1985) or DIPSI-2 (Shaka et al. 1988) pulse trains at γB2/2π = 7–10 kHz. The acquisitions were performed over a spectral width of 7002.801 Hz in both dimensions (or sometimes 8012.820 Hz) with matrix array size of 2048–3072 points in t2 and 400–600 points in t1 and 32–128 scans/t1 FID. Reduced acquisition times were selected to observe fast-relaxing spin systems (e.g., some prolines). Data processing and analysis were performed using Felix (Molecular Simulation, MSI) software with shifted (60–90 degrees) square sinebell apodization and 9th-order polynomial baseline correction for NOESY data. All spectra were referenced on the L23 CδH3 resonance at −0.58 ppm as previously reported (Esposito et al. 2000).
Collection of the experimental restraints from NMR data
Internuclear distances were quantified from the cross-peak volumes of several NOESY spectra with tm = 100 ms using some cross-peaks from Trp 95 (Hɛ1–Hζ2, Hɛ3–Hζ3, Hη2–Hζ2) as calibrant. Because of the compactness of the structure (judged from the spread and number of NOEs), only calibrant connectivities from a rigid location of the protein were employed. The effects of local mobility (side chains of exposed residues) were assumed to fall within the restraint allowance (±20%). Whenever recognizable cross-peak overlap exceeded 40%, the corresponding connectivities were considered only as a qualitative proximity indication (≤0.50 nm). Standard pseudoatom corrections were applied (Wüthrich et al. 1983). In addition to internuclear distances, NMR data were also employed to extract dihedral angle restraints and infer hydrogen-bond occurrence. Most of the HN–Hα coupling constants for ϕ torsion angle restraints were evaluated from the measurement of the corresponding TOCSY cross-peak line width at half height (Δv½) as proposed by Wang et al. (1997). This method appears robust only if the backbone motional regime of the protein under investigation is homogeneous, as indeed was the case with β2-m. For about 10% the HN–Hα cross-peaks, with peak separations well above the upper limit of the amide-coupling constant, the theoretical maximum of 10.1 Hz was imposed, corresponding to ϕ = −120 (Wang and Bax 1996). An allowance of ±30 was introduced for each ϕ angle included in the restraint list. Additional qualitative J-coupling determinations from DQF-COSY pattern were performed for a number of Hα–Hβ pairs to achieve stereospecific assignments (Esposito et al. 1987). An additional dihedral angle restraint was imposed to force the peptide bond between His 31 and Pro 32 in a cis configuration (Ψ31 = 0°), according to the observed NOE pattern.
Explicit hydrogen bonds were not added in the restraint list despite that their locations could be inferred from observation of the expected NOE pattern for regular secondary-structure elements (Wüthrich 1986). We preferred to check the consistency between expectation and final model and compare the results with the reported exchange pattern (Okon et al. 1992; Esposito et al. 2000).
Structure generation procedures
All the available experimental information was used as input for restrained molecular dynamics (MD) simulations, performed by using the program DYANA 1.5 (Güntert et al. 1997).
DYANA runs were performed according to the default protocol for simulated annealing (6000 steps Torsion Angle Dynamics, 2000 conjugate gradients minimization steps) with 380 randomly generated starting conformations. The 20 best structures of the DYANA ensemble were submitted to restrained minimization using DISCOVER (MSI) with the AMBER force field (Weiner et al. 1984). Overall, the same calculation procedure and parameters were employed as described elsewhere (Esposito et al. 1996). In both DYANA and DISCOVER treatments, the restraint on Ψ31 was handled indirectly, that is, through the violations of the intervening distances because it is in-built in the prolyl configuration (Pro 32). For this reason, it is excluded from the restraint count and any parameters thereof (e.g., statistical quality indicators; see below). The quality of the final structures was assayed by means of the software AQUA and PROCHECK-NMR (Laskowski et al. 1996). All structures were visualized within the INSIGHTII framework (MSI; Dayringer et al. 1986 and MOLMOL (Koradi et al. 1996).
Supplemental material in the electronic appendix
1H NMR chemical shift (Table A1), JHN-Hα coupling constants (Table A2), NOE count distribution histograms (Fig. A1) from β2-m NMR data are given, along with additional structure quality parameters for the final DYANA conformers and their DISCOVER refinement from restrained modeling (Table A3) and from validation assessment (Figs. A2, A3) and the list of H-bonds and salt bridges (Table A4).
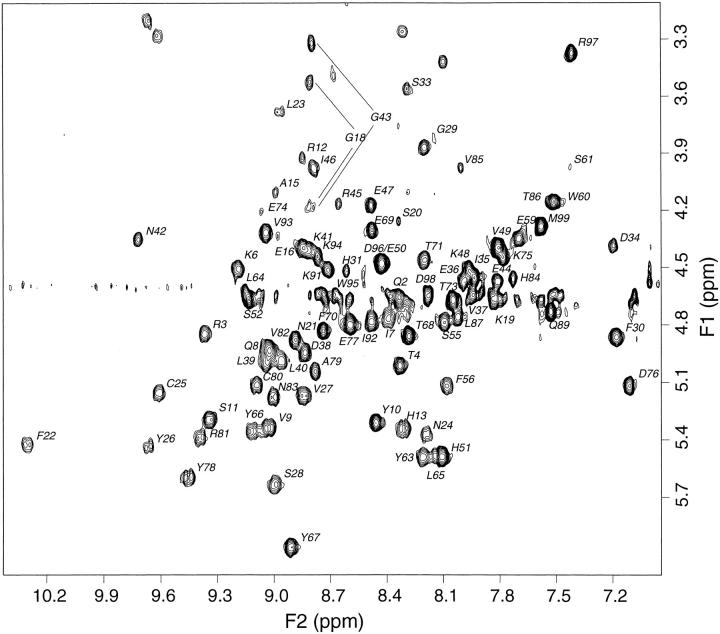
Fig. 1.
Fingerprint region from 500 MHz 1H 2D TOCSY spectrum of 0.9 mM β2-m in 70 mM phosphate buffer, 100 mM NaCl at pH 6.6, T = 310 K. The Hα–HN connectivity assignments are indicated by amino acid single-letter code. The relevant assignment information is given in Table A1 of Supplemental Material. Compared to our previous assignment (Esposito et al. 2000) the missing resonance patterns were identified especially by means of low-resolution spectra, which enabled observing the pattern of P32, P72, P14, N17, D53, and S61, particularly elusive due to broad line width. A number of novel chemical shift attributions (mainly concerning diastereotopic pairs) were also obtained.
Acknowledgments
This work was supported by MIUR (PRIN "Folding e misfolding delle proteine" and COFIN 2000 MM05221899) and Ministero della Sanità (Ricerca finalizzata sulla malattia di Alzheimer, 020ALZ00/01). The help of Dr. A. Makek is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
β2-m, β2-microglobulin
ΔN6β2-m, β2-microglobulin 7–99 fragment
1D, 2D, 3D, one, two, three dimension
DQF COSY, double quantum filtered correlation spectroscopy
DRA, dialysis related amyloidosis
GdnHCl, guanidinium chloride
HLA, human lukocyte antigen
MHC-I, class I major histocompatibility complex
NOE, nuclear Overhauser effect
NOESY, nuclear Overhauser spectroscopy
rms, root mean square
T, absolute temperature expressed in Kelvin (K)
TOCSY, total correlation spectroscopy
TPPI, time proportional phase incrementation
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.29002.
References
- Allen, F.H. and Johnson, O. 1991. Automated conformational analysis from crystallographic data. IV. Statistical descriptors for a distribution of torsion angles Acta Crystallogr. B47 62–67. [Google Scholar]
- Bax, A. and Davis, D.G. 1985. MLEV-17 based two dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65 355–360. [Google Scholar]
- Becker, J.W. and Reeke Jr., G.N. 1985. Three-dimensional structure of β2-microglobulin. Proc. Nat. Acad. Sci. 82 4225–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellotti, V., Stoppini, M., Mangione, P., Sunde, M., Robinson, C.V., Asti, L., Brancaccio, D., and Ferri, G. 1998. β2-microglobulin can be refolded into a native state from ex vivo amyloid fibrils. Eur. J. Biochem. 258 61–67. [DOI] [PubMed] [Google Scholar]
- Bjorkman, P.J., Saper, M.A., Samaroui, B., Bennet, W.S., Strominger, J.L., and Wiley, D.C. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329 506–512. [DOI] [PubMed] [Google Scholar]
- Braunschweiler, L. and Ernst, R.R. 1983. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reson. 53 521–528. [Google Scholar]
- Capeiller-Blandin, C., Delaveau, T., and Descamps-Latscha, B. 1991. Structural modifications of human β2-microglobulin treated with oxygen-derived radicals Biochem. J. 277 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.K. and Rossky, P.J. 1998. Surface topography dependence of biomolecular hydrophobic hydration Nature 392 696–699. [DOI] [PubMed] [Google Scholar]
- Chiti, F., Mangione, P., Andreola, A., Giorgetti, S., Stefani, M., Dobson, C.M., Bellotti, V., and Taddei, N. 2001. Detection of two partially structured species in the folding process of the amyloidogenic protein β2-microglobulin. J. Mol. Biol. 307 379–391 [DOI] [PubMed] [Google Scholar]
- Connors, L.H., Shirahama, T., Skinner, M., Fenves, A., and Cohen, A.S. 1985 In vitro formation of amyloid fibrils from intact β2-microglobulin Biochem. Biophys. Res. Commun. 131 1063–1068. [DOI] [PubMed]
- Dayringer, H.E., Tramontano, A., Sprang, S.R., and Fletterick, R.J. 1986. Interactive program for visualization and modelling of proteins, nucleic acids and small molecules. J. Mol. Graphics 6 82–87. [Google Scholar]
- De Lorenzi, E., Grossi, S., Massolini, G., Giorgetti, S., Mangione, P., Andreola, A., Chiti, F., Bellotti, V., and Caccialanza, G. 2001. Capillary electrophoresis investigation of a partially unfolded conformation of β2-microglobulin. Electophoresis (in press). [DOI] [PubMed]
- Doreleijers, J.F., Rullmann, J.A.C., and Kaptein, R. 1998. Quality assessment of NMR structures: A statistical survey. J. Mol. Biol. 281 149–164. [DOI] [PubMed] [Google Scholar]
- Esposito, G., Carver, J.A., Boyd, J., and Campbell, I.D. 1987. High resolution 1H NMR study of the solution structure of alamethicin. Biochemistry 26 1043–1050. [DOI] [PubMed] [Google Scholar]
- Esposito, G., Fogolari, F., Damante, G., Formisano, S., Tell, G., Leonardi, A., Di Lauro, R., and Viglino, P. 1996. Analysis of the solution structure of the homeodomain of rat thyroid transcription factor 1 by 1H-NMR spectroscopy and restrained molecular mechanics. Eur. J. Biochem. 241 101–113. [DOI] [PubMed] [Google Scholar]
- Esposito, G., Michelutti, R., Verdone, G., Viglino, P., Hernández, H., Robonson, C.V., Amoresano, A., Dal Piaz, F., Monti, M., Pucci, P., Mangione, P., Stoppini, M., Merlini, G., Ferri, G., and Bellotti, V. 2000. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 9 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gejyo, F., Yamada, T., Odani, S., Nakagawa, Y., Arakawa, M., Kunitomo, T., Kataoka, H., Suzuki, M., Hirasawa, Y., Shirahama, T., Cohen, A.S., and Schmid, K. 1985. A new form of amyloid protein associated with hemodialysis was identified as β2-microglobulin. Biochem. Biophys. Res. Commun. 129 701–706. [DOI] [PubMed] [Google Scholar]
- Güntert, P., Braun, W., and Wüthrich, K. 1991. Efficient computation of three-dimensional protein structure in solution from nuclear magnetic resonance data using the program DIANA and supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 217 517–530. [DOI] [PubMed] [Google Scholar]
- Güntert, P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Hurle, M.R., Helms, L.R., Li, L., Chan, W., and Wetzel, R. 1994. A role for destabilizing amino acid replacement in light-chain amyloidosis. Proc. Natl. Acad. Sci. 91 5446–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, T.L. and Shaka, A.J. 1995. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Mag. Reson. 112 275–279. [Google Scholar]
- Hyberts, S.G., Goldberg, M.S., Havel, T.F., and Wagner, G. 1992. The solution structure of eglin c based on measurements of many NOE and coupling constants. Protein Sci. 1 736–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeener, J., Meier, B.H., Bachmann, P., and Ernst, R.R. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71 286–292. [Google Scholar]
- Kabsh, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Keeler, J., Clowes, R.T., Davis, A.L., and Laue, E.D. 1994. Pulsed field gradients: Theory and practice. Meth. Enzymol. 239 145–207. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structure. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Linke, R.P., Hampl, H., Bartel-Schwarze, S., and Eulitz, M. 1987. β2-microglobulin, different fragments and polymers thereof in synovial amyloid in long-term hemodialysis. Biol. Chem. Hoppe Seyler 368137–144. [DOI] [PubMed] [Google Scholar]
- Linke, R.P., Hampl, H., Lobeck, H., Ritz, E., Bommer, J., Waldherr, R., and Eulitz, M. 1989. Lysine-specific cleavage of β2-microglobulin in amyloid deposits associated with haemodialysis. Kidney Int. 36 675–681. [DOI] [PubMed] [Google Scholar]
- Marion, D. and Wüthrich, K. 1983. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of proton–proton spin–spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113 967–974. [DOI] [PubMed] [Google Scholar]
- McParland, V.J., Kad, N.M., Kalverda, A.P., Brown, A., Kirwin-Jones, P., Hunter, M.G., Sunde, M., and Radford, S.E. 2000 Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry 39 8735–8746. [DOI] [PubMed]
- Miyata, T., Oda, O., Iinagi, R., Iida, Y., Araki, N., Yamada, N., Horiuchi, S., Taniguchi, N., Maeda, K., and Kinoshita, T. 1993. β2-microglobulin modified with advanced glycation end products is a major component of haemodialysis-associated amyloidosis. J. Clin. Invest. 92 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, C.J., Gelfand, M., Atreya, C., and Miranker, A.D. 2001 Kidney dialysis-associated amyloidosis: A molecular role for copper in fiber formation J. Mol. Biol. 309 339–345. [DOI] [PubMed]
- Okon, M., Bray, P., and Vucelic, D. 1992. 1H NMR assignments and secondary structure of human β2-microglobulin in solution. Biochemistry 31 8906–8915. [DOI] [PubMed] [Google Scholar]
- Piantini, U., Sørensen, O.W., and Ernst, R.R. 1982. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104 6800–6801. [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2 661–665. [DOI] [PubMed] [Google Scholar]
- Saper, M.A., Bjorkman, P.J., and Wiley, D.C. 1991. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J. Mol. Biol. 219 277–319. [DOI] [PubMed] [Google Scholar]
- Shaka, A.J., Lee, C.J., and Pines, A. 1988. Iterative schemes for bilinear operators: Application to spin decoupling. J. Magn. Reson. 77 274–293. [Google Scholar]
- States, D.J., Haberkorn, R.A., and Ruben, D.J. 1982. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Reson. 48 286–292. [Google Scholar]
- Stoppini, M., Bellotti, V., Mangione, P., Merlini, G., and Ferri, G. 1997 Use of anti-β2 microglobulin mAb to study formation of amyloid fibrils. Eur. J. Biochem. 249 21–26. [DOI] [PubMed]
- Sunde, M., Serpell, L.C., Bartlam, M., Fraser, P.E., Pepys, M.B., and Blake, C.C.F. 1997. Common core structure of amyloid fibrils by sinchrotron X-ray diffraction. J. Mol. Biol. 273 729–739. [DOI] [PubMed] [Google Scholar]
- Wang, A.C. and Bax, A. 1996, Determination of the backbone dihedral angles ϕ in human ubiquitin from reparametrized empirical Karplus equations. J. Amer. Chem. Soc. 118 2483–2494.
- Wang, Y., Nip, A.M., and Wishart, D.S. 1997. A simple method to quantitatively measure polypeptide JHNHα coupling constants from TOCSY or NOESY spectra. J. Biomol. NMR 10 373–382. [DOI] [PubMed] [Google Scholar]
- Weiner, S.J., Kollman, P.A., Case, D.A., Singh, U.C., Ghio, C., Alagona, G., Profeta Jr., S., and Weiner, P. 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106 765–784. [Google Scholar]
- Wüthrich, K. 1986. NMR spectroscopy of proteins and nucleic acids. Wiley & Sons, NY.
- Wüthrich, K., Billeter, M., and Braun, W. 1983. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton–proton distance constraints with nuclear magnetic resonance J. Mol. Biol. 169 949–961. [DOI] [PubMed] [Google Scholar]