Abstract
Human acidic fibroblast growth factor (FGF-1) has a β-trefoil structure, one of the fundamental protein superfolds. The X-ray crystal structures of wild-type and various mutant forms of FGF-1 have been solved in five different space groups: C2, C2221, P21 (four molecules/asu), P21 (three molecules/asu), and P212121. These structures reveal two characteristically different conformations for the β8/β9 β-hairpin comprising residue positions 90–94. This region in the wild-type FGF-1 structure (P21, four molecules/asu), a his-tagged His93→Gly mutant (P21, three molecules/asu) and a his-tagged Asn106→Gly mutant (P212121) adopts a 3:5 β-hairpin known as a type I (1–4) G1 β-bulge (containing a type I turn). However, a his-tagged form of wild-type FGF-1 (C2221) and a his-tagged Leu44→Phe mutant (C2) adopt a 3:3 β-hairpin (containing a type I` turn) for this same region. A feature that distinguishes these two types of β-hairpin structures is the number and location of side chain positions with eclipsed Cβ and main-chain carbonyl oxygen groups (Ψ ≅ +60°). The effects of glycine mutations upon stability, at positions within the hairpin, have been used to identify the most likely structure in solution. Type I` turns in the structural data bank are quite rare, and a survey of these turns reveals that a large percentage exhibit crystal contacts within 3.0 Å. This suggests that many of the type I` turns in X-ray structures may be adopted due to crystal packing effects.
Keywords: β-Hairpin, crystal packing, protein dynamics, β-trefoil, fibroblast growth factor
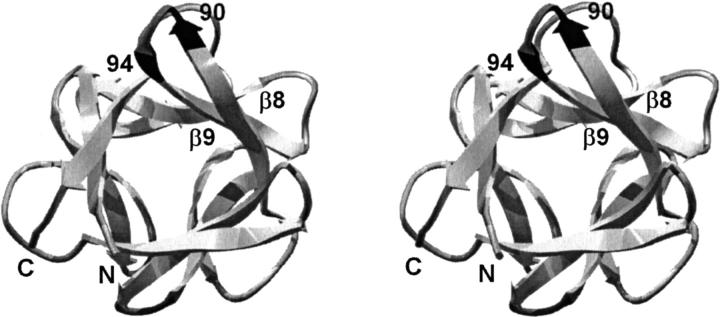
Human acidic fibroblast growth factor (FGF-1) is a member of a family of heparin-binding mitogens and hormones (Johnson et al. 1991; Jaye et al. 1992). FGF-1 exhibits a β-trefoil structure, one of the fundamental protein superfolds (Orengo et al. 1994). This fold contains six two-stranded β-hairpins (for a total of 12 β-strands), three that form an "upper" β-barrel structure and three that form a "lower" β-hairpin triplet, or triangular array, that caps the barrel at one end (Murzin et al. 1992). The β-barrel region is involved mainly in binding to FGF receptors and the triangular array contains a heparin binding region (Baird et al. 1988; Pantoliano et al. 1994; Springer et al. 1994; Blaber et al. 1996; DiGabriele et al. 1998; Pellegrini et al. 2000; Stauber et al. 2000). The arrangement of the secondary structure gives FGF-1 a pseudo threefold internal symmetry (Fig. 1 ▶).
Fig. 1.
Relaxed stereodiagram of human FGF-1 (Blaber et al. 1996) showing the location of the β8/β9 turn region and the 90–94 β-hairpin (dark gray). The view is down the pseudo threefold axis of symmetry present in the β-trefoil architecture. Region 90–94 is related to the N- and C-termini by this internal symmetry.
The turn located between the β8/β9 strands of the FGF-1, involving residue positions 90–94 (Fig. 1 ▶), has importance with regard to the dimerization of some members of the FGF family of proteins (Hecht et al. 2001). Furthermore, a glycine point mutation within this turn of FGF-1, at position 93 (a histidine in the wild-type structure) has been reported to increase the half-life of the protein (Baird et al. 1988; Arakawa et al. 1993). In the course of studying stability and structure relationships in human FGF-1 we have solved X-ray structures of wild-type and mutant FGF-1 proteins in a total of five different space groups. Two uniquely different conformations for the β8/β9 β-hairpin in FGF-1 have been identified. In some space groups a 3:3 β-hairpin containing a type I` turn is observed. In other crystal forms a 3:5 β-hairpin known as a type I (1–4) G1 β-bulge, containing a type I turn, is observed. Based on the stereochemical features unique to each type of turn, glycine mutagenesis in combination with stability studies has been employed to identify the most likely form of this β-hairpin in solution. A statistical survey of the structural databank has also been undertaken to evaluate whether the relatively rare type I` turns in crystal structures have uniquely close crystal contacts that could influence their conformation.
Results
His-tagged wild-type FGF-1 (WT*) crystals suitable for diffraction crystallized with vapor diffusion against 4.0–4.5 M formate. These crystals were relatively radiation insensitive, tolerating over 140 h of X-ray exposure with minimal decay and diffracting to better than 1.65 Å. The space group was identified as C2221 with two molecules in the asymmetric unit (Table 1). This is a different space group than the non-his-tagged wild-type protein (WT, space group P21). Molecular replacement solutions for both molecules in the asymmetric unit of this new space group were found using molecule A of the WT structure (2AFG) as a search model. Initial 2Fo–Fc maps indicated generally good agreement between the model and experimental data, with the exception of the turn involving residue positions 90–94 in both molecules. Omit maps provided unambiguous density for each of these turns, and indicated they had a type I` conformation, instead of the type I observed in the WT structure. The model refined to acceptable crystallographic residual and stereochemical parameters (Table 1). This space group is a higher symmetry form of the C2 space group observed in a previously reported his-tagged Leu44→Phe mutant (Brych et al. 2001). The higher symmetry in the C2221 space group is eliminated in the C2 space group due to an alternative side-chain rotamer of the histidine residue at position 93 in two of the four molecules in the asymmetric unit. This does not, however, affect the turn type for this region.
Table 1.
Crystallographic data collection and refinement statistics
| WT* | H93G* | N106G* | WTa | L44F*b | |
| Crystal data | |||||
| Space Group | C2221 | P21 | P212121 | P21 | C2 |
| a (Å) | 74.1 | 33.6 | 46.6 | 37.1 | 96.9 |
| b (Å) | 96.8 | 97.7 | 60.1 | 111.7 | 73.8 |
| c (Å) | 109.0 | 57.5 | 101.3 | 64.0 | 109.1 |
| β (°) | — | 104.3 | — | 90.5 | 90.0 |
| Mol/ASU | 2 | 3 | 2 | 4 | 4 |
| Matthews' constant | 2.96 | 1.85 | 2.15 | 2.10 | 2.96 |
| Max resolution (Å) | 1.65 | 2.0 | 2.1 | 2.0 | 1.70 |
| Data collection and processing | |||||
| Total/unique reflections | 254,402/ 45,684 | 102,705/ 21,128 | 245,767/ 17,287 | ||
| % compl. | 97.6 | 90.6 | 97.4 | ||
| % compl. (highest shell) | 83.5 | 75.2 | 94.6 | ||
| I/σ (overall) | 41.7 | 29.4 | 44.1 | ||
| I/σ (highest shell) | 5.5 | 3.5 | 10.9 | ||
| Wilson B (Å2) | 16.2 | 24.2 | 26.9 | ||
| R-merge (%) | 5.3 | 6.2 | 5.0 | ||
| Refinement | |||||
| Rcryst (%) | 20.0 | 22.5 | 22.6 | ||
| Rfree (%) | 24.1 | 30.0 | 30.8 | ||
| R.M.S. bond len. dev. (Å) | 0.008 | 0.006 | 0.008 | ||
| R.M.S. bond ang. dev. (°) | 1.7 | 1.6 | 1.6 | ||
| R.M.S. B factor dev (Å2) | 3.3 | 1.8 | 2.0 | ||
a Blaber et al. 1996.
b Brych et al. 2001.
* Mutations performed in the amino-terminal his-tagged background protein.
The his-tagged His93→Gly point mutant (H93G*) crystallized with vapor diffusion against solutions of PEG 4000. The space group of this crystal form was identified as P21, but was distinctly different from the P21 WT space group. In particular, the H93G* crystal form contained three molecules in the asymmetric unit (Vm = 1.85) compared to four for the WT space group (Vm = 2.1). Molecular replacement solutions for each of the three molecules in the asymmetric unit were found using molecule A of the refined WT* structure. Although the initial 2Fo–Fc maps indicated generally good agreement between the model and experimental data, it was clear that the turn involving residues 90–94 in all three molecules differed from the WT* search model. Again, omit maps provided unambiguous density with which to rebuild these turn regions. Two of these turns had reverted to type I, and the third was identified as a type IV (miscellaneous type). The model refined to acceptable crystallographic residual and stereochemical parameters (Table 1).
The N106G* mutant protein crystallized with vapor diffusion against solutions of PEG 20,000. The space group was identified as P212121 with two molecules in the asymmetric unit. Molecular replacement solutions for each of the two molecules in the asymmetric unit were found using molecule A of the refined WT* structure. As with the H93G* structure, initial 2Fo–Fc maps indicated generally good agreement between the model and experimental data, but it was clear that the turn involving residues 90–94 for both molecules in the asymmetric unit differed from the WT* search model. Similarly, omit maps provided unambiguous density with which to rebuild these turn regions. One of the turns was identified as type I, and the other as type IV (miscellaneous). The model refined to acceptable crystallographic residual and stereochemical parameters (Table 1).
Analysis of the β-hairpin involving residues 90–94 in the WT structure indicates that all four molecules in the asymmetric unit adopt a type I turn with the φ,ξ angles of positions i + 1 (residue 91) and i + 2 (residue 92) located within the AA region (preferred α-helical) of the Ramachandran plot (Table 2). Likewise, the A and C molecule of H93G* adopt a type I turn within this β-hairpin, but the B molecule is a type IV (Table 2). However, the i + 1 and i + 2 φ, ξ angles for this type IV turn are located within the Aa region (allowed α-helical) and can therefore be defined as a slightly distorted type I turn. Similarly, the A molecule in the N106G* crystal structure adopts a type I turn within this β-hairpin, while the B molecule is of type IV, but can be considered as essentially a distorted type I (Table 2). In contrast, the two molecules of the WT* structure contain a type I` turn within this β-hairpin, with the i + 1, i + 2 φ,ξ angles located within the LL region (left-handed α-helical). Likewise, all four molecules of the L44F* structure are defined as β-turn type I` with the i + 1, i + 2 φ,ξ angles located within the LL region (Table 2). All region 90–94 loop structures exhibit well-defined electron density and average main chain B factors that are equivalent, or in some cases lower, than the Wilson B factor for the respective crystal.
Table 2.
Turn classification for the β8/β9 β-hairpin in WT and mutant FGF-1 proteins
| Protein | Molecule in ASU | Turn typea | i + 1 (91) (φ/ψ) | i + 2 (92) (φ/ψ) | φψ regiona |
| WT | A | I | −54.0/−29.8 | −82.4/3.7 | AA |
| B | I | −61.5/−37.1 | −79.7/9.2 | AA | |
| C | I | −54.5/−13.0 | −93.1/19.5 | AA | |
| D | I | −74.7/−15.6 | −89.0/21.8 | Aa | |
| WT* | A | I` | 53.4/33.1 | 53.7/29.3 | LL |
| B | I` | 52.4/33.6 | 55.9/26.5 | LL | |
| H93G* | A | I | −60.5/−32.7 | −80.8/−1.6 | AA |
| B | IV | −52.8/−32.3 | −56.9/−30.4 | AA | |
| C | I | −48.3/−42.3 | −76.3/16.0 | Aa | |
| L44F* | A | I` | 54.7/30.9 | 66.6/−9.2 | LL |
| B | I` | 55.2/35.1 | 55.1/23.7 | LL | |
| C | I` | 51.2/31.4 | 66.8/−6.6 | LL | |
| D | I` | 49.9/37.0 | 54.4/23.8 | LL | |
| N106G* | A | I | −54.9/−34.4 | −62.6/−4.9 | AA |
| B | IV | −49.7/−32.8 | −51.0/−51.3 | AA |
a Protmotif v2.0 (Hutchinson and Thornton 1996).
* Mutations performed in the amino-terminal his-tagged background protein.
Differential scanning calorimetry studies of glycine point mutants at positions 91, 92, and 93 in the WT* protein show that glycine substitutions at positions 91 and 92 have virtually no effect upon stability (Table 3). However, a glycine substitution at position 93 results in a substantial increase of 8.3 kJ/mole in the stability of the protein (Table 3). In each case, the denaturation is observed to be two-state and reversible, as with the wild-type protein (Blaber et al. 1999) and within the expected error of data collection (∼0.40 kJ/molK).
Table 3.
Thermodynamic parameters for WT* and mutant proteins
| Protein | Tm (K) | ΔH (kJ/mol) | ΔΔGa (kJ/mol) | Std. errorb (kJ/molK) |
| WT* | 313.2 ± 0.7 | 261.0 ± 14.0 | 0.4 | |
| E91G* | 312.3 ± 0.1 | 248.4 ± 1.5 | 0.8 ± 0.1 | 0.2 |
| N92G* | 312.9 ± 1.4 | 249.3 ± 19.0 | 0.2 ± 1.1 | 0.4 |
| H93G* | 321.8 ± 0.1 | 351.6 ± 16.0 | −8.3 ± 0.2 | 0.5 |
| E91G/N92G* | 312.0 ± 0.5 | 247.7 ± 15.0 | 1.2 ± 0.4 | 0.4 |
a ΔΔG = ΔGWT − ΔGMUT determined at the Tm of the wild type. A negative value for ΔΔG indicates a more stable mutant.
b Standard error is for the fit to a two-state model.
* Mutations performed in the amino-terminal his-tagged background protein.
Discussion
Turn structures (that reverse the general direction of the polypeptide chain) are the most common type of nonrepetitive structure in proteins (Kabsch and Sander 1983). Venkatachalam first identified categories of turns while studying favorable conformations of short peptides (Venkatachalam 1968). Three turn conformations (types I, II, and III) were identified that form intrachain hydrogen bonds between the main chain carbonyl at position i and the main chain amide at position i + 3. Three mirror images of these turns (types I`, II`, and III`) were also identified as sterically unfavorable. Lewis and coworkers later found that 25% of turns do not contain the hydrogen bond Venkatachalam used in the definition of such turns (Lewis et al. 1973). They proposed instead that a turn be defined by a distance of 7 Å or less between the Cα at positions i and i + 3, and involve nonhelical main chain φ,ξ angles for the set of residues comprising the turn. Lewis and coworkers also calculated conformational energies for various types of turns and found that the type I` turn is 27 kJ/mole higher in energy than the type I turn (Lewis et al. 1973). Richardson subsequently defined turns on the basis of φ, ξ angles, and defined six distinct categories as well as a miscellaneous category (the type IV turn) (Richardson 1981). This definition of turn types is the most common definition in use today.
The distribution and types of turns in protein structures have been analyzed and classified by Thornton and coworkers (Wilmot and Thornton 1988). Turn definitions do not include information regarding the types of secondary structure that precede and follow the turn. However, "β-hairpins" refer to turns that connect antiparallel β-sheet secondary structures. β-Hairpins can be grouped into four general classes (A, B, C, and D) that refer to the number of residues required to complete the turn (Milner-White and Poet 1986; Sibanda et al. 1989). The "tightest" hairpins (class A) require just two residues to accomplish a reversal of the polypeptide chain direction and form the first hydrogen bond in the antiparallel β-sheet. Class B requires three residues, class C requires four, and class D requires five. Each of these general classes of β-hairpins can be further subdivided into one of two subclasses, depending upon whether one or two hydrogen bonds form in the closure of the turn. A shorthand for β-hairpin structures, which communicates the relevant structural details for the different types, has been developed by Thornton and coworkers (Sibanda et al. 1989). In this shorthand an integer prefix identifies the number of residues required for the turn. This is followed by a colon and an integer suffix that identifies the hydrogen bonding pattern in the closure of the turn (the general formula being that if two hydrogen bonds are formed at the closure, then the suffix = prefix. However, if only one hydrogen bond forms the closure, then the suffix = prefix + 2). Thus, a 2:2 β-hairpin has two residues in the turn and two hydrogen bonds in the turn closure. A 3:5 β-hairpin has three residues in the turn and a single hydrogen bond in the turn closure.
The β-hairpin nomenclature does not provide information regarding the type of turn conformation within the turn region, as this is identified using the previously described turn formalism of Richardson (e.g., type I, II, I`, II`, etc.). For example, Thornton and coworkers found that more than 50% of known 2:2 β-hairpin structures contain type I` turns (Sibanda and Thornton 1985; Sibanda et al. 1989). The tight 2:2 turn requires a twisting orientation for the main chain, which the type I` turn can readily accommodate (Chothia 1973; Sibanda et al. 1989). Amino acid positional preferences in the type I` turn have been analyzed using 205 well-defined protein structures (Hutchinson and Thornton 1994). Glycine is commonly found in the first residue of the turn, and almost exclusively at the second position of the turn. The main chain φ,ξ angles for both residues place them within the left-handed α-helical region of the Ramachandran plot. In this region of φ,ξ space ξ = +60° and results in the Cβ eclipsing the main chain carbonyl group and is an energetically unfavorable orientation. The preference for glycine at these positions in a type I` turn reflects the avoidance of this steric clash by the elimination of the Cβ group in this side chain. Although 50% of known 2:2 β-hairpin structures contain type I` turns, only 3% of all turns are type I` (Hutchinson and Thornton 1994). Thus, type I` turns represent highly specialized turns in protein structures.
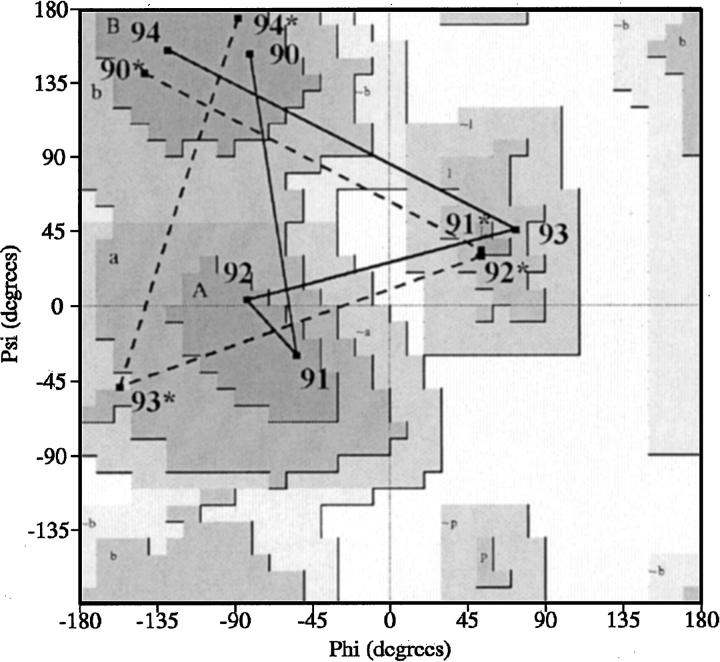
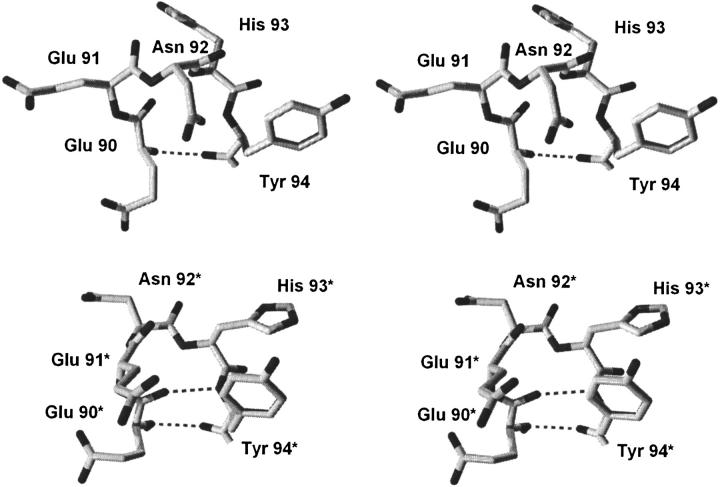
The characteristic alternative type I and type I` turn conformations for region 90–94 observed in the different space groups of FGF-1 are illustrated in a Ramachandran plot, using the representative WT (P21) and WT* (C2221) structures (Fig. 2 ▶). In the type I turn structure, residue positions Glu91 (i + 1) and Asn92 (i + 2) are located in the right-handed α-helical region, whereas position His93 (i + 3) is located in left-handed α-helical region (ξ ≅ +60°). In this conformation the Cβ of the His side chain at position 93 is eclipsed with the main chain carbonyl group, whereas the side chains of positions 91 and 92 are not (Fig. 3 ▶). However, in the type I` turn residue position 93 is now within the right-handed α-helical region, whereas residue positions 91 and 92 are in the left-handed α-helical region (ξ ≅ +60°). This conformation eliminates the eclipsed steric interaction at position 93, but introduces such interactions at positions 91 and 92 (Fig. 3 ▶). The β-hairpin containing the type I turn is formally known as a type I (1–4) G1 β-bulge (Sibanda et al. 1989). Statistical preferences of amino acids for different positions within this type of turn indicate a preference for glycine at the i + 3 position (Sibanda et al. 1989), reflecting the steric interaction between the main-chain Cβ and carbonyl oxygen groups.
Fig. 2.
Ramachandran plot showing the main-chain φ,ξ angle distributions for the β-hairpin structures defined by residue positions 90–94 in both WT (solid line) and WT* (broken line) structures.
Fig. 3.
(Upper panel) Relaxed stereodiagram of region 90–94 from the wild-type FGF-1 crystal structure (2AFG). The polypeptide chain forms a 3:5 β-hairpin known as a type I (1–4) G1 β-bulge, containing a type I turn. Residue position 93 is located in the LL region of the Ramachandran plot (Ψ ≅+60°) and the side-chain Cβ is eclipsing the main-chain carbonyl oxygen. The single hydrogen bond that defines the closure of this turn is indicated. (Lower panel) Region 90–94 from the refined his-tagged wild-type FGF-1 (WT*) structure. The polypeptide chain forms a 3:3 β-hairpin containing a type I` turn. Residue positions 91 and 92 are located in the LL region of the Ramachandran plot and the side-chain Cβ groups of these residues eclipse their main-chain carbonyl oxygens. The double hydrogen bonds that define the closure of this turn are indicated.
Which type of turn, I or I`, predominates at positions 90–94 in FGF-1 in the absence of crystal packing influences? To answer this question we evaluated the effects of Gly mutations at positions 91, 92, and 93. Gly point mutations at positions 91 or 92 had essentially no effect upon the stability of the protein (Table 3). Likewise, a double Gly mutant at positions 91 and 92 was essentially additive and, consequently, exhibited minimal effects upon stability (Table 3). However, a Gly point mutation at position 93 resulted in a dramatic increase in stability of 8.3 kJ/mole (Table 3). These results are consistent with residue position 93 being in the left-handed α-helical region of the Ramachandran plot, where removal of the side-chain Cβ group would eliminate the conformational strain associated with ξ ≅ +60°. Likewise, the results suggest that positions 91 and 92 are not experiencing conformational strain associated with ξ ≅ +60°, because removal of the side-chain Cβ group has essentially no effect upon stability. Therefore, the thermodynamic data indicates region 90–94 adopts principally the type I turn in solution.
Modeling the type I turn (from WT) into the WT* space group (C2221) results in a close contact of 0.8 Å between the His93 side chain and itself in a symmetry-related molecule. Similar analyses indicate that the type I turn cannot be accommodated in any of the space groups where the type I` turn is observed. Conversely, the type I` turn can be accommodated in all space groups where the type I turn is observed. Thus, we conclude that the type I` turn observed for region 90–94 in some spacegroups appears to be a consequence of crystal packing interactions (i.e., a high-energy conformation offset by favorable lattice energy). NMR data for the solution structure of FGF-1 has also been reported (Lozano et al. 2000). Of the 24 energetically favored structures, two exhibit a type I turn for region 90–94, while the remaining 22 structures exhibit undefined turn types, with no type I` turns present. Thus, the NMR data also suggests that the solution structure of FGF-1 favors the type I turn, rather than type I`, for positions 90–94.
The energy difference between type I and type I` turns has been calculated by Lewis and coworkers using energy minimization of N-acetyl N`-methyl ala tetrapeptide models (Lewis et al. 1973). These studies determined that the type I turn is 27 kJ/mole more stable than the type I` turn. These calculations, however, did not include potential solvent effects. Structurally, the two turns differ in the number of eclipsed configurations involving side-chain Cβ and main chain carbonyl oxygen atoms: the type I turn has one such interaction (residue 93) and the type I` turn has two such interactions (residues 91 and 92). Given that the region 90–94 adopts a type I turn in solution, the Gly mutation at position 93 eliminates the conformational strain associated with an eclipsed Cβ interaction. Thus, this conformational strain can be assigned the experimentally derived value of 8.3 kJ/mole. Therefore, to a first approximation, 8.3 kJ/mole would also represent the energetic difference between type I and type I` turns, which also differ by a single eclipsed interaction (i.e., one versus two). The experimentally derived value of 8.3 kJ/mole for the energetic difference between type I and I` turns is somewhat less than the 27 kJ/mole calculated by Lewis and coworkers(Lewis et al. 1973); however, as mentioned previously, the calculated value did not take into account solvent effects.
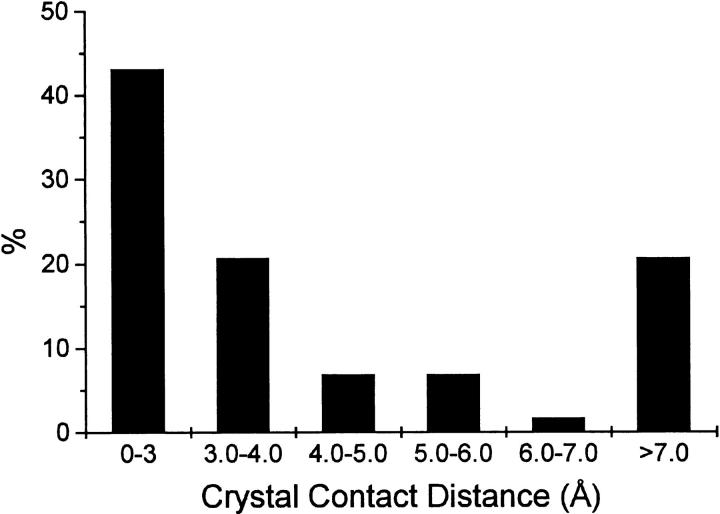
Type I` turns are a relatively rare type of turn in proteins, comprising an estimated 3% of all turn types (Hutchinson and Thornton 1994). A total of 137 examples of type I` turns were identified in the structural data bank, from 98 different proteins. The structural environment adjacent to these turns was evaluated to determine whether nearby crystal contacts exist. Of the 137 examples of type I` turns, 19 could not be analyzed further due to uncertainties in space group identification, or other considerations. Of the remaining 118 examples, 51 (43.2%) had type I` turns exhibiting crystal contacts of 3.0 Å or less (Fig. 4 ▶). These striking results suggest that adjacent crystal packing contacts exist for a large percentage of type I` turns in the structural data bank. Therefore, many type I` turns identified in the structural data bank may be stabilized by favorable lattice energy, and the actual incidence of type I` turns in proteins may be lower than the statistics derived from X-ray crystal structures.
Fig. 4.
Distribution of the distances between type I` turns and crystal contacts from 116 type I` turns in the structural databank.
Type I` turns are uncommon structural elements, and it is of interest to ask what structural or functional role might they play in proteins. FGF-9, a member of the FGF family of proteins, has been shown to dimerize in the absence of receptor binding (in contrast to other members of this family) (Hecht et al. 2001). The dimerization interface in FGF-9 actually involves the β8/β9 β-hairpin. In FGF-9 this region adopts a type I` turn, and this conformation has been proposed as the structural basis for the ability of FGF-9 to dimerize (Hecht et al. 2001). Thus, the potential dynamics of the alternative conformations of this turn identified in FGF-1 may be related to the process of dimerization and receptor binding. A different example of a type I` turn is also found in the apo-form of 2,5-diketo-d-gluconate reductase (1HW6). In the holo-form of this enzyme the type I` turn undergoes a dramatic conformational change into an extended sheet structure, resulting in up to 8 Å movements of active site residues (Khurana et al. 1998; Sanli and Blaber 2001). Within the structural data bank four examples (1LZ1, 2DRI, 2SAR, and 3DFR) were found of type I` turns directly involving active site residues in various enzymes. Thus, we propose that type I` turns may also represent useful structural markers identifying conformationally dynamic regions in protein structures.
Materials and methods
Mutagenesis and protein preparation
The WT construct comprised a synthetic gene for the 140 amino acid form of human acidic fibroblast growth factor (Gimenez-Gallego et al. 1986; Linemeyer et al. 1987). WT*, and all other his-tagged variants, included a 6-histidine amino-terminal fusion to this 140 amino acid form. All mutagenesis, expression and purification followed previously published procedures (Culajay et al. 2000; Brych et al. 2001). Purified protein was exchanged into 50 mM NaPi, 0.1 M NaCl, 10 mM (NH4)2SO4, 0.5 mM EDTA, and 2 mM DTT ("crystallization buffer") for crystallization studies or 20 mM N-(2-acetamido)iminodiacetic acid (ADA), 100 mM NaCl, pH 6.60 ("ADA buffer") for biophysical studies.
Crystallization of FGF-1
Pure FGF-1 solutions were filtered through 0.2-μm filters (Pall Life Sciences) immediately before crystallization. X-ray diffraction quality crystals of WT* grew within 1 wk at 10°C in 10-μL hanging drops by vapor diffusion against 1-mL reservoirs of 4.3–4.5 M formate in crystallization buffer. H93G* crystals were obtained within 3 wk (2 wk at 4°C and 1 wk at RT) in 10-μL hanging drops by vapor diffusion against 0.9 mL 20% PEG 4000, 10% 2-propanol, 0.1 M HEPES (pH 7.5), and 0.1 mL 30% PEG 400, 0.2 M magnesium chloride, 0.1 M HEPES (pH 7.5). N106G* crystals were obtained within 2 wk in 10-μL hanging drops by vapor diffusion against 0.9 mL 20% PEG 20,000, 0.1 M MES (pH 6.5).
Data collection, molecular replacement, and refinement
Diffraction data was collected using a Rigaku-automated X-ray Imaging System II (R-Axis II). The crystals were mounted using Hampton Research nylon mounted cryo-turns and then frozen in a cryo stream of liquid nitrogen. Diffraction data was indexed, integrated, and scaled using the DENZO software package (Otwinowski 1993; Otwinowski and Minor 1997). Molecular replacement was carried out using the MRCHK program (Zhang and Matthews 1994). Wild-type FGF-1 was used as the search model for WT*, and WT* was used as the search model for H93G* and N106G*. All structures were refined with the TNT least-squares software package (Tronrud et al. 1987; Tronrud 1992, 1996) and the O molecular graphics program (Jones et al. 1991). Analysis of turn types within the structure was accomplished using the PROTMOTIF software program (Hutchinson and Thornton 1996).
Differential scanning calorimetry
Calorimetric analyses were performed on a MicroCal model VP-DSC high-sensitivity differential scanning calorimeter (DSC) (MicroCal, Inc.). The protein sample (0.04 mM) in 0.7 M GuHCl in ADA buffer was degassed for 15 min prior to analysis. The temperature range of the thermal scan was from 278 to 353 K, with a scan rate of 15 K/h. For each experiment, buffer and protein scans were run in triplicate. Deconvolution of the calorimetric data was performed using the DSCFit software package (Grek et al. 2001).
Close contact search
Protein structures containing type I` turns were identified using published data (Hutchinson and Thornton 1994), and additional data were generously provided by Dr. E.G. Hutchinson, University of Reading. A total of 98 proteins and 139 type I` turns were analyzed. Relevant symmetry operators were applied to each coordinate file, and the distance between the type I` turn region and symmetry-related molecules was measured using the EDPDB software package (Zhang and Matthews 1995).
Acknowledgments
The authors thank Drs. T. Somasundaram and Pushparani Dhanarajan for technical assistance. We also thank Dr. E.G. Hutchinson, University of Reading, for generously supplying the compilation data of structures with type I` turns, and Ms. Deborah Kelly for helpful discussions regarding energy calculations of the turns. This work was supported by a grant GM54429-01 from the U.S.P.H.S./N.I.H. Model coordinates for all structures will be deposited in the structural data bank.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.43802.
References
- Arakawa, T., Horan, T.P., Narhi, L.O., Rees, D.C., Schiffer, S.G., Holst, P.L., Prestrelski, S.J., Tsai, L.B., and Fox, G.M. 1993. Production and characterization of an analog of acidic fibroblast growth factor with enhanced stability and biological activity. Protein Eng. 6 541–546. [DOI] [PubMed] [Google Scholar]
- Baird, A., Schubert, D., Ling, N., and Guillemin, R. 1988. Receptor- and heparin-binding domains of basic fibroblast growth factor. Proc. Natl. Acad. Sci. 85 2324–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaber, M., DiSalvo, J., and Thomas, K.A. 1996. X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry 35 2086–2094. [DOI] [PubMed] [Google Scholar]
- Blaber, S.I., Culajay, J.F., Khurana, A., and Blaber, M. 1999. Reversible thermal denaturation of human FGF-1 induced by low concentrations of guanidine hydrochloride. Biophys. J. 77 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brych, S.R., Blaber, S.I., Logan, T.M., and Blaber, M. 2001. Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core region of a β-trefoil. Protein Sci. 10 2587–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia, C. 1973. Conformation of twisted beta-pleated sheets in proteins. J. Mol. Biol. 75 295–302. [DOI] [PubMed] [Google Scholar]
- Culajay, J.F., Blaber, S.I., Khurana, A., and Blaber, M. 2000. Thermodynamic characterization of mutants of human fibroblast growth factor 1 with an increased physiological half-life. Biochemistry 39 7153–7158. [DOI] [PubMed] [Google Scholar]
- DiGabriele, A.D., Lax, I., Chen, D.I., Svahn, C.M., Jaye, M., Schlessinger, J., and Hendrickson, W.A. 1998. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature 393 812–817. [DOI] [PubMed] [Google Scholar]
- Gimenez-Gallego, G., Conn, G., Hatcher, V.B., and Thomas, K.A. 1986. The complete amino acid sequence of human brain-derived acidic fibroblast growth factor. Biochem. Biophys. Res. Commun. 128 611–617. [DOI] [PubMed] [Google Scholar]
- Grek, S.B., Davis, J.K., and Blaber, M. 2001. An efficient, flexible-model program for the analysis of differential scanning calorimetry data. Protein Pept. Lett. 6 429–436. [Google Scholar]
- Hecht, H.J., Adar, R., Hofmann, B., Bogin, O., Weich, H., and Yayon, A. 2001. Structure of fibroblast growth factor 9 shows a symmetric dimer with unique receptor- and heparin-binding interfaces. Acta Crystallogr. D Biol. Crystallogr. 57 378–384. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.G. and Thornton, J.M. 1994. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 3 2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, E.G. and Thornton, J.M. 1996. PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 5 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaye, M., Schlessinger, J., and Dionne, C.A. 1992. Fibroblast growth factor receptor tyrosine kinases: Molecular analysis and signal transduction. Biochim. Biophys. Acta 1135 185–199. [DOI] [PubMed] [Google Scholar]
- Johnson, D.E., Lu, J., Chen, H., Werner, S., and Williams, L.T. 1991. The human fibroblast growth factor receptor genes: A common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immumoglobulin domain. Mol. Cell. Biol. 11 4627–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Khurana, S., Powers, D.B., Anderson, S., and Blaber, M. 1998. Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution. Proc. Natl. Acad. Sci. 95 6768–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, P.N., Momany, F.A., and Scheraga, H.A. 1973. Chain reversals in proteins. Biochim. Biophys. Acta 303 211–229. [DOI] [PubMed] [Google Scholar]
- Linemeyer, D.L., Kelly, L.J., Menke, J.G., Gimenez-Gallego, G., DiSalvo, J., and Thomas, K.A. 1987. Expression in Escherichia coli of a chemically synthesized gene for biologically active bovine acidic fibroblast growth factor. Biotechnology 5 960–965. [Google Scholar]
- Lozano, R.M., Pineda-Lucena, A., Gonzalez, C., Angeles Jimenez, M., Cuevas, P., Redondo-Horcajo, M., Sanz, J.M., Rico, M., and Gimenez-Gallego, G. 2000. 1H NMR structural characterization of a nonmitogenic, vasodilatory, ischemia-protector and neuromodulatory acidic fibroblast growth factor. Biochemistry 39 4982–4993. [DOI] [PubMed] [Google Scholar]
- Milner-White, E.J. and Poet, R. 1986. Four classes of beta-hairpins in proteins. Biochem. J. 240 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G., Lesk, A.M., and Chothia, C. 1992. β-Trefoil fold. Patterns of structure and sequence in the kunitz inhibitors interleukins-1β and 1α and fibroblast growth factors. J. Mol. Biol. 223 531–543. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Jones, D.T., and Thornton, J.M. 1994. Protein superfamilies and domain superfolds. Nature 372 631–634. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. 1993. Oscillation data reduction program. Proceedings of the CCP4 Study Weekend: "Data collection and processing." SERC Daresbury Laboratory, England.
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pantoliano, M.W., Horlick, R.A., Springer, B.A., Van Dyk, D.E., Tobery, T., Wetmore, D.R., Lear, J.D., Nahapetian, A.T., Bradley, J.D., and Sisk, W.P. 1994. Multivalent ligand–receptor binding interactions in the fibroblast growth factor system produce a cooperative growth factor and heparin mechanism for receptor dimerization. Biochemistry 33 10229–10248. [DOI] [PubMed] [Google Scholar]
- Pellegrini, L., Burke, D.F., von Delft, F., Mulloy, B., and Blundell, T.L. 2000. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407 1029–1034. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. 1981. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34 167–339. [DOI] [PubMed] [Google Scholar]
- Sanli, G. and Blaber, M. 2001. Structural assembly of the active site in an aldo-keto reductase by NADPH cofactor. J. Mol. Biol. 309 1209–1218. [DOI] [PubMed] [Google Scholar]
- Sibanda, B.L. and Thornton, J.M. 1985. Beta-hairpin families in globular proteins. Nature 316 170–174. [DOI] [PubMed] [Google Scholar]
- Sibanda, B.L., Blundell, T.L., and Thornton, J.M. 1989. Conformation of β-hairpins in protein structures. J. Mol. Biol. 206 759–777. [DOI] [PubMed] [Google Scholar]
- Springer, B.A., Pantoliano, M.W., Barbera, F.A., Gunyuzlu, P.L., Thompson, L.D., Herblin, W.F., Rosenfeld, S.A., and Book, G.W. 1994. Identification and concerted function of two receptor binding surfaces on basic fibroblast growth factor required for mitogenesis. J. Biol. Chem. 269 26879–26884. [PubMed] [Google Scholar]
- Stauber, D.J., DiGabriele, A.D., and Hendrickson, W.A. 2000. Structural interactions of fibroblast growth factor receptor with its ligands. Proc. Natl. Acad. Sci. 97 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronrud, D.E. 1992. Conjugate-direction minimization: an improved method for the refinement of macromolecules. Acta Crystallogr. A48 912–916. [DOI] [PubMed] [Google Scholar]
- Tronrud, D.E. 1996. Knowledge-based B-factor restraints for the refinement of proteins. J. Appl. Crystallogr. 29 100–104. [Google Scholar]
- Tronrud, D.E., Ten Eyck, L.F., and Matthews, B.W. 1987. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr. A43 489–501. [Google Scholar]
- Venkatachalam, C.M. 1968. Stereochemical criteria for polypeptides and proteins. VI. Non-bonded energy of polyglycine and poly-L-alanine in the crystalline beta-form. Biochim. Biophys. Acta 168 411–416. [PubMed] [Google Scholar]
- Wilmot, C.M. and Thornton, J.M. 1988. Analysis and prediction of the different types of β-turns in proteins. J. Mol. Biol. 203 221–232. [DOI] [PubMed] [Google Scholar]
- Zhang, X.-J. and Matthews, B.W. 1994. Enhancement of the method of molecular replacement by incorporation of known structural information. Acta Crystallogr. D50 675–686. [DOI] [PubMed] [Google Scholar]
- Zhang, X.-J. and Matthews, B.W. 1995. EDPDB: A multi-functional tool for protein structure analysis. J. Appl. Crystallogr. 28 624–630. [Google Scholar]