Abstract
Centruroides sculpturatus Ewing variant 2 toxin (CsE-v2) is a neurotoxin isolated from the venom of a scorpion native to the Arizona desert. The structure of CsE-v2 was solved in two different crystal forms using a combination of molecular replacement and multiple isomorphous replacement techniques. Crystals of CsE-v2 display a temperature-dependent, reversible-phase transition near room temperature. At lower temperature the space group changes from P3221 to P3121 with an approximate doubling of the C-axis. The small-cell structure, which has one molecule per asymmetric unit, has an R factor of 0.229 at 2.8 Å resolution. The large-cell structure has two molecules per asymmetric unit and was refined at 2.2 Å resolution to an R factor of 0.255. CsE-v2 is a rigid, compact structure with four intrachain disulfide bonds. The structure is similar to other long-chain β neurotoxins, and the largest differences occur in the last six residues. The high-resolution structure of CsE-v2 corrects an error in the reported C-terminal sequence; the terminal tripeptide sequence is Ser 64-Cys 65-Ser 66 rather than Ser 64-Ser 65-Cys 66. Comparison of CsE-v2 with long-chain α toxins reveals four insertions and one deletion, as well as additional residues at the N and C termini. Structural alignment of α and β toxins suggests that the primary distinguishing feature between the two classes is the length of the loop between the second and third strands in a three-strand β sheet. The shorter loop in α toxins exposes a critical lysine side chain, whereas the longer loop in β toxins buries the corresponding basic residue (either arginine or lysine).
Keywords: Crystal structure, scorpion toxin, noncrystallographic symmetry
Scorpion venom contains a number of proteins that are responsible for the neurotoxic activity of the venom. These proteins are divided into two large classes, depending upon their length. The short neurotoxins (<40 amino acid residues) are active on various potassium channels (Possani et al. 1982; Gimenez-Gallego et al. 1988; Galvez et al. 1990; Crest et al. 1992). On the other hand, the long neurotoxins (60–70 amino acid residues) appear to function by affecting the activation or inactivation of the sodium channel (Narahashi et al. 1972; Romey et al. 1975; Catterall 1979; Couraud et al. 1980). This property makes them useful probes for investigating the topology of sodium channels. The long-chain scorpion neurotoxins can be further subdivided into two classes, α and β toxins, on the basis of their effects on the sodium channel (Jover et al. 1980). In general, α toxins are found in the venom of old-world scorpions, whereas β toxins are more common in new-world species.
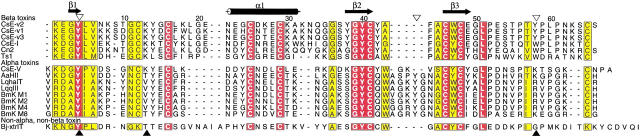
All long-chain neurotoxins have similar tertiary structures, consisting of an α helix of 2.5 turns, a three-strand antiparallel β sheet, four disulfide bonds, and a conserved hydrophobic surface (Zhao et al. 1992; Housset et al. 1994; Jablonsky et al. 1995; Landon et al. 1997). However, they show considerable variation in amino acid sequence (Fig. 1 ▶). In this regard, toxicological studies indicate that individual scorpion toxins differ significantly with respect to neurotoxicity and species specificity. Comparison of amino acid sequences of long-chain scorpion toxins shows that α and β neurotoxins differ primarily in the lengths of two loops: the loop just prior to the α helix and the loop between the second and third β strands. Compared to β scorpion toxins, α toxins have a longer loop between the β strands and a shorter loop preceding the α helix. One notably different structure of a scorpion toxin from Buthotus judaicus has been reported (Oren et al. 1998). Although it has the same core structure as other neurotoxins, it contains an additional α helix at the C terminus.
Fig. 1.
Sequence alignment of CsE-v2 with scorpion toxins whose coordinates have been reported. The α helix and β strands in CsE-v2 are indicated. AaH2, Androctonus australis Hector 2; LqhaIT, Leiurus quinquestriatus hebraeus α; LqqIII, Leirus quinquestriatus quinquestriatus 3; BmKM1, Buthus martensii Karsch M1; BmKM2, B. martensii Karsch M2; BmKM4, B. martensii Karsch M4; BmKM8, B. martensii Karsch M8; CsE-V, Centruroides sculpturatus Ewing 5; Ts1, Tityus serrulatus 1; CsE-I, C. sculpturatus Ewing 1; CsE-v1, C. sculpturatus Ewing variant 1; CsE-v2, C. sculpturatus Ewing variant 2; CsE-v3, C. sculpturatus Ewing variant 3; Cn2, Centruroides noxius Hoffman; Bj-xtrIT, Buthotus judaicus. (Prepared with ALSCRIPT [Barton 1993].)
Variant 2 scorpion toxin (CsE-v2) is a 66–amino acid protein isolated from the venom of Centruroides sculpturatus Ewing, a scorpion native to the southwestern United States (Babin et al. 1974). CsE-v2 is one of three toxins produced in the venom that are called variants because they are much less potent than other toxins originally described in this species. Otherwise, their physical and chemical properties are quite similar. Crystallographic studies of CsE-v2 were initiated to compare molecular structures and possibly identify features that affect toxicity. The structures of the other two variant toxins in Centruroides, CsE-v1 (Lee et al. 1994) and CsE-v3 (Zhao et al. 1992), as well as two potent toxins from this species, CsE-I (Jablonsky et al. 1999) and CsE-V (Jablonsky et al. 1995), have been determined (Table 1). CsE-v2 differs from CsE-v3 by only four amino acid residues; there are three conservative substitutions and one additional residue at the C terminus. CsE-v2 has nine differences from CsE-v1 in the first 27 amino acids, as well as an additional residue at the C terminus. Compared to CsV-I, the three variant toxins have one extra residue in the first β turn, but all three differ considerably from CsE-V, which has more sequence and structural homology with old-world α toxins.
Table 1.
Long chain scorpion toxin structures determined by X-ray crystallography and nuclear magnetic resonance (NMR) (only structures with published coordinates are included)
| Toxin | Abbreviation | Method | Protein Data Bank code | Reference |
| Beta toxins | ||||
| Centruroides sculpturatus | CsE-v1 | NMR | 1VNB | Lee et al. 1994 |
| Ewing variant 1 | ||||
| C. sculpturatus | CsE-v2 | X-ray | 1JZA, | This paper |
| Ewing variant 2 | 1JZB | |||
| C. sculpturatus | CsE-v3 | X-ray | 2SN3 | Zhao et al. 1992 |
| Ewing variant 3 | ||||
| C. sculpturatus Ewing 1 | CsE-I | NMR | 2B3C | Jablonsky et al. 1999 |
| Centruroides noxius Hoffmann | Cn2 | NMR | ICN2 | Pintar et al. 1999 |
| Tityus serrulatus 1 | Ts1 | X-ray | 1B7D | Polikarpov et al. 1999 |
| Alpha toxins | ||||
| C. sculpturatus Ewing 5 | CsE-V | NMR | 1NRA | Jablonsky et al. 1995 |
| Androctonus australis Hector 2 | AaH2 | X-ray | 1PTX | Housset et al. 1994 |
| Leiurus quinquestriatus hebraeus α | LqhaIT | NMR | 1LQH | Tugarinov et al. 1997 |
| Leiurus quinquestriatus quinquestriatus 3 | LqqIII | NMR | 1LQQ | Landon et al. 1997 |
| Buthus martensii Karsch M1 | BmKM1 | X-ray | 1SN1 | He et al. 1999 |
| B. martensii Karsch M2 | BmKM2 | X-ray | 1CHZ | He et al. 2000 |
| B. martensii Karsch M4 | BmKM4 | X-ray | 1SN4 | He et al. 1999 |
| B. martensii Karsch M8 | BmKM8 | X-ray | 1SNB | Li et al. 1996 |
| Non-alpha, non-beta | ||||
| Buthotus judaicus | Bj-xtrIT | X-ray | 1BCG | Oren et al. 1998 |
Another interesting aspect of the CsE-v2 structure is unusual noncrystallographic symmetry. Preliminary studies of CsE-v2 indicated that the protein crystals undergo a temperature-dependent reversible transition near room temperature (Ealick et al. 1984). At room temperature, the space group is P3221 with one molecule in the asymmetric unit, but at 4° the crystals change to space group P3121 with two molecules per asymmetric unit. There is also an approximate doubling of the C-axis, which suggests that pairs of molecules are related by noncrystallographic translational symmetry parallel to the C-axis. Here we describe the three-dimensional structure of CsE-v2 in both the large-cell and small-cell crystal forms and discuss its differences from other long-chain neurotoxins.
Results
Description of the structure
Table 2 is a summary of refinement parameters for the two structures. Superposition of the R-factor curve with theoretical curves for different mean positional errors gives estimated errors of 0.33 and 0.31 for the small- and large-cell structures, respectively (Luzzati 1952). For the small-cell structure, a Ramachandran plot shows that 90.2% of the nonglycine residues are in most-favored regions and 9.8% are in additional-allowed regions. For the large-cell structures, the corresponding values are 89.2% and 10.8%, respectively. The mean B values for molecules A and B in the large cell are 17.9 Å2 and 20.6 Å2, respectively, reflecting the poorer electron density for molecule B. Because the two molecules in the large-cell asymmetric unit were refined with noncrystallographic symmetry restraints, the structures are virtually identical. The root-mean-square difference between C-α carbons for the two molecules is only 0.09 Å. As expected, the structures of CsE-v2 in the large and small cells are quite similar. Superposition of the C-α carbons for either CsE-v2 molecule in the large cell with CsE-v2 from the small cell shows root-mean-square differences of only 0.24 Å. The largest differences (0.4–0.6 Å) occur in the first two residues at the N terminus (Lys 1-Glu 2).
Table 2.
Refinement statistics for CsE-v2
| Small cell | Large cell | |
| Space Group | P3221 | P3121 |
| Resolution range (Å) | 50.0–2.80 | 50.0–2.20 |
| Number of reflections (total) | 1609 | 6460 |
| Working set | 1528 | 6098 |
| Test set | 81 | 362 |
| Completeness (%) | 99.2 | 99.5 |
| R value | 0.229 | 0.255 |
| Free R value | 0.281 | 0.287 |
| Highest resolution shell (Å) | 2.98–2.80 | 2.34–2.20 |
| Number of reflections (total) | 260 | 1061 |
| Working set | 252 | 1007 |
| Test set | 8 | 54 |
| Completeness (%) | 96.7 | 99.6 |
| R value | 0.286 | 0.319 |
| Free R value | 0.283 | 0.395 |
| Number of protein atoms | 498 | 1000 |
| Number of water molecules | 0 | 27 |
| Estimated coordinate error (Å) | 0.33 | 0.31 |
| Deviations from ideality | ||
| Bond lengths (Å) | 0.007 | 0.008 |
| Bond angles (°) | 1.4 | 1.4 |
| Dihedral angles (°) | 24.5 | 22.5 |
| Improper angles (°) | 1.3 | 1.3 |
| Temperature factor refinement | ||
| Mean B value (Å2) | 20.1 | 19.2 |
| Rmsd main-chain bond (Å2) | 1.69 | 2.07 |
| Rmsd main-chain angle (Å2) | 2.77 | 3.00 |
| Rmsd side-chain bond (Å2) | 2.70 | 3.49 |
| Rmsd side-chain angle (Å2) | 3.98 | 4.73 |
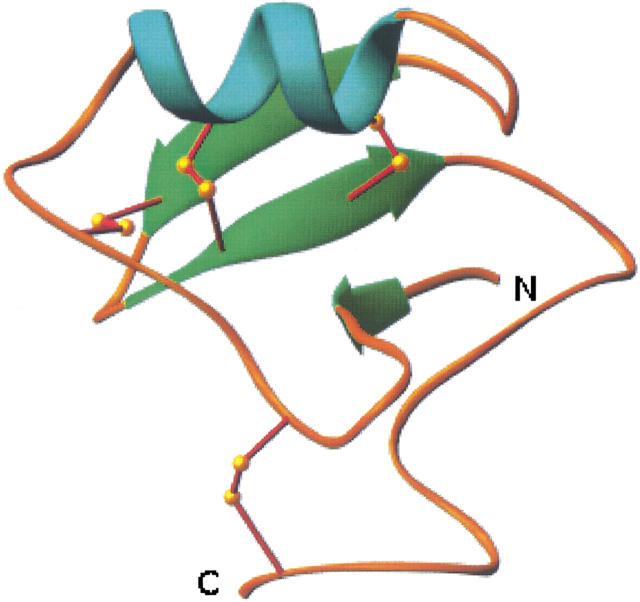
CsE-v2 is a compact globular protein with approximate dimensions of 25 × 32 × 36 Å (Fig. 2 ▶). CsE-v2 contains one α helix (residues 23–30) and a three-stranded antiparallel β sheet (residues 3–4, 38–41, and 46–49). The α helix is connected to the middle strand of the β sheet by a pair of disulfide bonds involving Cys 25-Cys 46 and Cys 29-Cys 48. The longer outer strand of the β sheet is linked to the long loop prior to the α helix by a disulfide bond between Cys 16 and Cys 41. The fourth disulfide bond between Cys 12 and Cys 65 limits the flexibility of the C terminus. There is one antiparallel β bulge that involves residues Ser 37, Tyr 38, and Glu 49, and there are 10 β turns. Pro 59 adopts a cis conformation, which has been reported in most β scorpion toxin structures.
Fig. 2.
Ribbon diagram of CsE-v2. The N and C termini are labeled. (Prepared with RIBBONS [Carson 1987].)
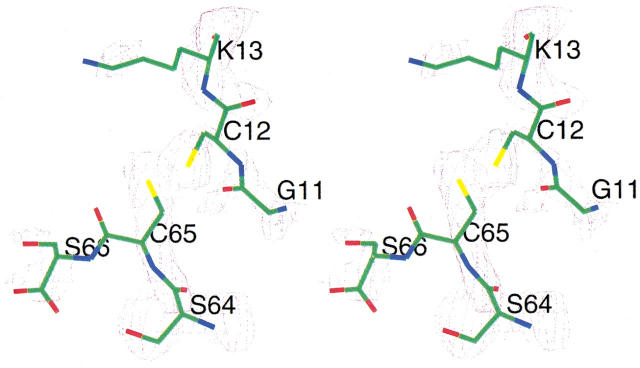
The high-resolution crystal structure of CsE-v3 revealed an error in the published C-terminal sequence; the correct sequence is Ser 64-Cys 65 rather than Cys 64-Ser 65 (Zhao et al. 1992). Similarly, the solution structure of the homologous toxin CsE-v1 (Lee et al. 1994) showed a C-terminal sequence of Ser 64-Cys 65. The published sequence of CsE-v2 lists the three C-terminal residues as Ser 64-Ser 65-Cys 66 (Babin et al. 1974). However, the high-resolution structure of CsE-v2 clearly shows that the correct C-terminal sequence is Ser 64-Cys 65-Ser 66 (Fig. 3 ▶), which is homologous to CsE-v1 and CsE-v3.
Fig. 3.
Portions of CsE-v2 with the superimposed difference electron density contoured at 1.0 σ. The electron density map was calculated with coefficients (Fo−Fc) and αcalc phases from the refined 2.2-Å resolution structure, but residues Cys12, Cys65, and Ser66 were not included in the calculation of phases. (Prepared with CHAIN [Sack 1988].)
Intermolecular hydrogen bonds and crystal packing in the two structures
The asymmetric unit of the large-cell crystal form contains two molecules of CsE-v2 related by a noncrystallographic translation approximately parallel to the C-axis. The distance between the centers of mass is 43.5 Å, and the closest contact along the C-axis between the two molecules is ∼8.4 Å. This approximate doubling of the C-axis converts the 32 screw axis into a 31 screw axis, which results in a change of space group from P3221 to P3121, even though the packing of molecules in the unit cell remains nearly the same. The rotation angle required to bring the two molecules into alignment is 3.5°. This rotation between the two molecules is sufficient to change the intermolecular hydrogen-bonding scheme relative to the molecule in the small cell. Although most of the differences are rather small, they are obviously sufficient to lead to an alternate crystal-packing scheme and consequently the doubled C-axis and different space group.
The CsE-v2 molecule in the small cell contains five intermolecular hydrogen bonds (Table 3). Molecules A and B in the large cell contain seven and five intermolecular hydrogen bonds, respectively. The hydrogen bond between symmetry-related hydroxyl oxygen atoms of Tyr 58 is the only bond that is present in all three molecules. Two other hydrogen bonds in the small-cell structure have counterparts in molecule A of the large-cell structure: Lys 13-Ser 66 and Tyr 42-Pro 61. In molecule B of the large-cell structure, the hydrogen bonds corresponding to Lys 13-Ser66 and Tyr 42-Pro 61 are not present. Also, the hydrogen bond between the carbonyl oxygen of Cys 65 and OG of Ser 66 is much shorter in molecule B compared to molecule A.
Table 3.
Differences in intermolecular contacts
| Distances (Å) | ||||||
| Residue | Atom | Residue | Atom | Small cell | Large cell mol. A | Large cell mol. B |
| Glu2 | OE2 | Lys27 | NZ | 3.30 | 2.99 | 2.99 |
| Asn7 | ND2 | Glu28 | OE1 | 2.61 | 3.23 | 3.61 |
| Ser9 | OG | Glu28 | OE2 | 3.35 | 2.93 | 2.73 |
| Lys13 | NZ | Ser66 | OT | 2.71 | 3.00 | 3.28 |
| Glu21 | OE2 | Lys32 | NZ | 3.08 | 4.36 | 3.95 |
| Tyr42 | OH | Pro61 | O | 2.96 | 2.98 | 3.94 |
| Tyr58 | OH | Tyr58 | OH | 2.26 | 2.55 | 2.51 |
| Tyr58 | OH | Ser66 | OG | 3.35 | 2.85 | 2.77 |
| Cys65 | O | Ser66 | OG | 3.28 | 3.12 | 2.47 |
Discussion
Table 1 lists the long-chain scorpion toxin structures with coordinates that have been deposited. Eight of these are X-ray structures, and the other six were determined by nuclear magnetic resonance (NMR). Figure 1 ▶ is a sequence alignment of CsE-v2 with these long-chain toxins. The toxins with the most sequence homology to CsE-v2 are CsE-v1 (86%) and CsE-v3 (95%). The largest structural difference among the three variant toxins occurs in the C-terminal loop that includes residues 59–65. In particular, the movement of Cys 65 (∼1.0 Å compared to CsE-v1 and CsE-v3) is required to accommodate the additional C-terminal residue Ser 66 in the crystal structure. It is interesting that the disulfide bond involving Cys 65 in CsE-v3 has two alternate conformations, but in the CsE-v2 structure there is no electron density to suggest an alternate conformation.
Comparison of the three variant toxins with the other three β toxins of known structure reveals a one-residue insertion at position nine of the variant sequences compared to CsE-I and a one-residue insertion at position 21 compared to Tityus serrulatus toxin (Ts1). The Centruroides noxius (Cn2) toxin has no deletions or insertions relative to the three variant toxins, and it shows more structural homology to CsE-v2 than CsE-I or Ts1.
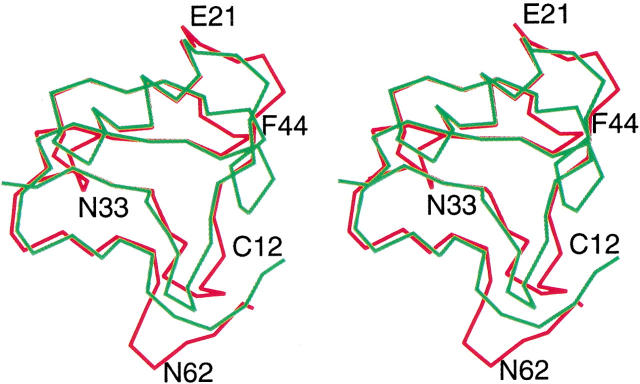
CsE-v2 has much less sequence homology with the α toxins, although CsE-V shows intermediate homology to CsE-v2 compared to other α and β toxins. Comparison of the variant toxins with α toxins reveals four insertions and one deletion, as well as an additional residue at the N terminus of the α toxins (Fig. 4 ▶). The first insertion occurs at position nine in the CsE-v2 sequence, where there is an extra serine residue in the three variant toxins. In the α toxins, residues 8–11 form a type I β turn, but in the variant structures, the extra residue transforms this loop into two overlapping β turns (residues 7–10 and 8–11). The second insertion involves residues 19–21 in CsE-v2, which are part of the loop just prior to the beginning of the α helix. In the three variant toxins, the first two residues in this insertion are Leu-Gly, and the third residue is either Lys or Glu. The third insertion occurs at residue 32–33 in CsE-v2 and is Lys-Asn in all three variant toxins. This sequence is part of two interlocking tight turns involving residues 31–34 and 32–35. These turns have no counterparts in the α toxins. The fourth insertion occurs at positions 63–64 and is Lys-Ser in all three variant toxins. This insertion causes the largest conformational change with respect to α toxins. Residues 60–63 form a tight turn, which again has no counterpart in the α toxins. This segment of the polypeptide chain is rotated ∼90° with respect to the same region in the α toxins. The extra N-terminal residue in the α toxins is usually Val, and it assumes a similar conformation in each protein. On the other hand, the conformations of the extra residue(s) at the C terminus (usually His) are quite different and do not suggest any type of common motif. The only deletion in the variant toxins relative to α toxins occurs between Ala 43 and Phe 44, which is in the loop between the second and third β strands. All of the α toxins except CsE-V have five additional residues at this location; CsE-V has two extra residues.
Fig. 4.
Comparison of α and β toxins. The Androctonus toxin (green) has been superimposed on CsE-v2 (red). Representative residues in CsE-v2 corresponding to insertions or deletions relative to α toxins are labeled. (Prepared with CHAIN [Sack 1988].)
There are four disulfide bonds in the scorpion toxins, and several of the structures show disorder of one or two of these bonds. CsE-v2 has two conformations for the Cys 29-Cys 48 bond, which was also observed in the CsE-v3 structure. The structure of CsE-v3 has an additional disordered disulfide bond between Cys 12 and Cys 65; the corresponding disulfide bonds in the toxins from Androctonus australis (AaHII) and Leiurus quinquestriatus hebraeus (LqqIII) are similarly disordered. The conformation of the disulfide bond with the higher occupancy in CsE-v3 is the same assumed by the corresponding cysteine residues in CsE-v2.
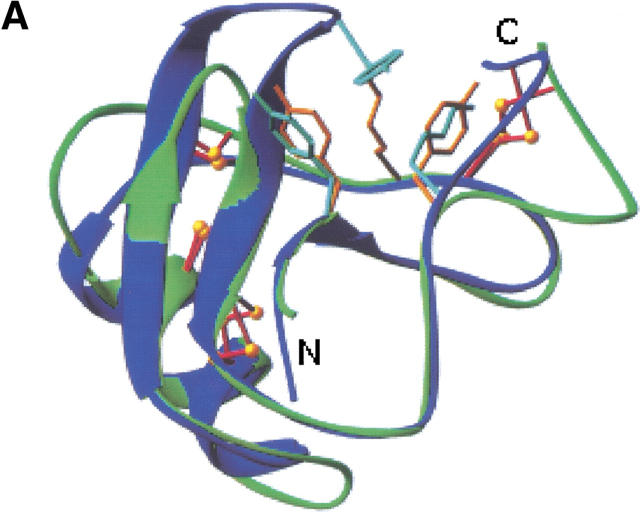
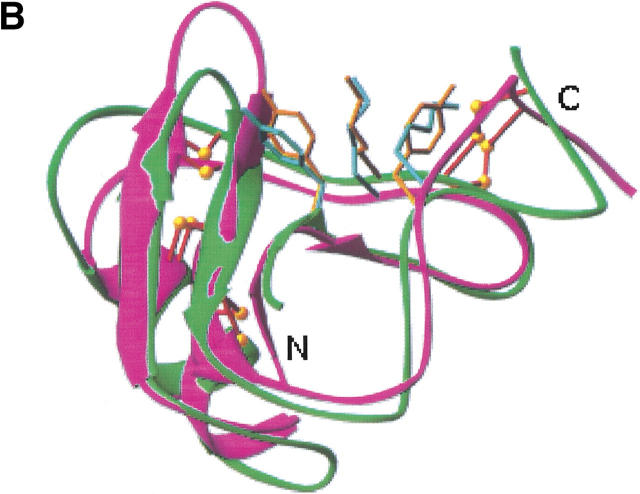
One common structural feature that has been emphasized in all the scorpion toxins is the presence of a three-amino-acid cluster of two aromatic residues and one positively charged residue (Fig. 5 ▶). This three-amino-acid cluster is found in all known toxin structures, except for CsE-V and B. judaicus (Bj-xtrIT), which is clearly in a different structural class of neurotoxins. However, different portions of the sequence in the α and β toxins contribute the residues that form this three-amino-acid cluster. It has been shown that modification of this basic residue (Lys 58 or Arg 58 in the α toxins; Lys 12 in CsE-I and Ts1 or Lys 13 in Cn2 and the variant toxins) causes a marked loss of toxicity. Bj-xtrIT has no basic residue in either of these positions.
Fig. 5.
Comparison of the aromatic/basic patch in α and β scorpion toxins. (A) CsE-v2, green; AaH2, blue. Orange residues (CsE-v2) are Tyr-4, Lys-13, and Tyr-58; cyan residues (AaH2) are Tyr-5, Tyr-42, and Lys-58. (B) CsE-v2, green; CsE-V, magenta. Cyan residues (CsE-V) are Tyr-5, Lys-13, and Lys-55. (Prepared with RIBBONS [Carson 1987].)
The mean temperature factors for the main- and side-chain atoms in Lys 13 are the lowest for any lysine residue in the CsE-v2. The side chain is anchored by two hydrogen bonds. Lys 13 NZ forms a hydrogen bond with one of the C-terminal carbonyl oxygen atoms (Ser 66 OT), and there is also a water molecule that is hydrogen-bonded to Lys 13 NZ and Asp 43 O. A similar hydrogen bond involving water and Asp 43 was identified in the CsE-v3 structure (Zhao et al. 1992).
In the β toxins the aromatic residues correspond to Tyr 4 and either Tyr 58 in the variant toxins or Trp in the corresponding position for Ts1, CsE-I, and Cn2. In all α toxins except CsE-V, the tyrosine residues in this cluster (Tyr 5 and Tyr 42) are invariant. However, in CsE-V the residue corresponding to the position of the second aromatic residue is Lys 55, which makes the importance of an aromatic residue in this position uncertain. Similarly, Tyr 5 is conserved in Bj-xtrIT, but there is no aromatic residue for the second position.
Although all of the structural studies have emphasized the importance of a basic residue with two aromatic residues, the position of the basic residue relative to the aromatic residues varies considerably (Fig. 4 ▶). The position of Tyr 4 is relatively invariant in all toxins. In the β toxins, this aromatic residue is the last in a series of aromatic residues that form a herringbone motif. In all of the α toxins (except CsE-V), there is an additional aromatic residue that extends the herringbone motif.
Structural alignment of the various α and β toxins suggests that a key distinguishing feature between the two classes is the length of the loop between the second and third β strands. The shorter loop in the β toxins exposes the critical Lys 13 side chain, whereas the longer loop in the α toxins buries the residue that corresponds to Lys 13 in the β toxins (Thr or Val, except for CsE-V, which is Lys). On the other hand, the conserved basic residue in α toxins (Arg 58 or Lys 58, except for CsE-V, which is Lys 55) remains exposed, even though access to it is much more limited.
Materials and methods
Crystallization and data collection
Full details of the preparation and crystallization of CsE-v2 have been described (Ealick et al. 1984). The space group of the small-cell crystals is P3221 with unit cell dimensions a = 48.8 Å and c = 43.7 Å, and there is one molecule in the asymmetric unit. The value of Vm (Matthews 1968) is 2.09, corresponding to a solvent volume fraction of 41%. Intensity data for the native crystals and a crystal soaked in a solution containing K2PtCl4 were collected at 23°C with a Picker FACS-1 diffractometer using an ω step-scan procedure and Ni-filtered CuKα radiation. One crystal was used for each data set. The reflections were divided into 2θ shells that each contained 100 reflections and were collected from high to low resolution. Complete sets of data, plus Friedel mates, were collected for the native protein and the Pt derivative. The Rsym for the native data using all Friedel mates was 0.05. To monitor and correct for decomposition effects, three standard reflections were measured periodically. During the collection of data, there was no significant decrease in the intensities of the standard reflections.
The space group of the large-cell crystals is P3121, and the unit cell dimensions are a = 48.8 Å and c = 87.2 Å. These crystals contain two toxin molecules per asymmetric unit. Data for the large-cell native and Pt-soaked crystals were collected at 22°C with a Nicolet X-100A area detector using CuKα radiation from a Rigaku RU-200 rotating anode generator operating at 40 kV and 100 mA. One crystal was used for the native data, and two crystals were used for the Pt data set. Oscillation frames covered 0.25° and were measured for 5 min. The detector was offset by 20°, and the crystal-to-detector distance was 12 cm. Although data extended slightly beyond 2.2 Å, it was truncated at 2.2 Å during merging and scaling. Indexing and integration of intensity data were carried out using the XENGEN processing programs (Howard et al. 1987). Table 2 gives statistics for the data processing. The overall Rsym is 0.074, and the Rsym is 0.095 for the highest resolution shell (2.20–2.34 Å). The data with l odd is systematically weaker than the l even data, especially at resolution <6 Å. Table 4 gives an analysis of Fobs versus resolution and l index for the large cell.
Table 4.
Analysis of Fobs versus resolution and l index for the large cell
| Value of l | Data range | Number of reflections | Average Fobs |
| Even | ∞6.0 Å | 180 | 28.53 |
| Odd | ∞6.0 Å | 179 | 6.24 |
| Even | 6.0–3.0 Å | 1146 | 24.66 |
| Odd | 6.0–3.0 Å | 1128 | 11.28 |
| Even | 3.0–2.2 Å | 1912 | 11.93 |
| Odd | 3.0–2.2 Å | 1915 | 7.97 |
Structure determination and refinement
The crystal structure of the small cell was solved first, using the molecular replacement routines in XPLOR (Brünger et al. 1987). For the cross-rotation search, the 1.2-Å crystal structure of CsE-v3 (Protein Data Bank [PDB] entry 2SN3; Zhao et al. 1992) was used as the search model. A cross-rotation function was calculated using a radius of integration of 18.5 Å and data between 12 Å and 4 Å resolution. The 198 peaks generated by the rotation search were subjected to Patterson correlation (PC) refinement (Brünger 1990), treating the entire CsE-v3 molecule as a rigid body. The model was rotated by the angles corresponding to the best rotation solution, and translation functions were calculated for each of the enantiomorphic space groups using data from 12 Å to 4 Å. The higher translation function T values and R-factors indicated that the correct space group was P3221.
The Pt atom positions in the small-cell Pt derivative were determined from a difference Patterson map and verified from difference Fourier maps using phases calculated with the model structure. Calculations were performed using the CCP4 package (Collaborative Computational Project, Number 4 1994). Single isomorphous replacement (SIR) phases were calculated with MLPHARE (Otwinoski 1991). The overall figure-of-merit was 0.40, using all data to 3.0-Å resolution (1311 reflections). To reduce bias in the subsequent refinement, electron density maps were calculated using SIR phases, as well as calculated phases from the molecular replacement solution combined with the SIR phases (Read 1986). Prior to calculation of electron density maps, the phases were improved by solvent-flattening techniques. Manual rebuilding was done by using the computer program CHAIN (Sack 1988).
Refinement was by simulated annealing using XPLOR (Brünger et al. 1987, 1990) with the stereochemical parameter files defined by Engh and Huber (1991). No σ cutoff was applied to the data. Five percent of the data was randomly selected and removed prior to refinement for analysis of the free R factor (Brünger 1992). The progress of the refinement was guided by the decrease in both the conventional and free R factors.
There are three differences in the published amino acid sequences for CsE-v2 and CsE-v3, plus one additional residue at the C terminus of CsE-v2. However, the high-resolution crystal structure of CsE-v3 revealed an error in the published C-terminal sequence of CsE-v3 (Babin et al. 1974); the correct sequence is Ser 64-Cys 65 rather than Cys 64-Ser 65. A similar correction was made in the CsE-v1 structures determined by NMR. Because the overall sequence of CsE-v2 is highly homologous to variants 1 and 3, it seemed likely that there was also an error in its published sequence. Therefore, all five of these residues in CsE-v2 (7, 10, 27, 64, and 65) were changed to alanine for the initial rigid body refinement. Using all data in the resolution range 8.0–2.8 Å, the R factor was 0.42. The side-chain density for these five residues was clearly seen in 2Fo-Fc maps using calculated phases alone or calculated phases combined with SIR phases. The disulfide bond between Cys 12 and Cys 65 was obvious, confirming that the published sequence by Babin et al. (1974) was incorrect. Refinement using simulated annealing reduced the R factor to 0.27, at which point maps showed density for Ser 66. Subsequent refinement included all 66 residues.
An overall B factor for the initial model was calculated and used for each atom in the initial stages of refinement. A flat bulk-solvent model was generated (Jiang and Brünger 1994), and the refinement using all data gave an R factor of 0.25 and free R factor of 0.30. Group B-factor refinement was then carried out with one B factor for each residue. Finally, based on an analysis of the free R factor, individual B factors were included in the final refinement. The final R factor is 0.229 (1528 reflections in the working set), and the free R factor is 0.281 (81 reflections in the test set).
The crystal structure of the large cell was determined using the 2.8-Å crystal structure of CsE-v2 in the small cell as the search model. The cross-rotation function showed only one large peak corresponding to no rotation; in other words, the orientation of the molecules in the large cell is essentially the same as in the small cell. The model was placed in the cell, and translation functions were calculated for each of the enantiomorphic space groups using data from 12 Å to 4 Å. The R factors and translation function T values for these solutions confirmed the space group P3121. The position of the second molecule in the asymmetric unit was derived simply by translating the first molecule parallel to the C-axis by half of the length of the axis. After rigid body refinement of this model, the rotation angle between the two molecules was 2.0°. However, simulated annealing refinement decreased the R factor very little, and the free R factor actually increased. Inspection of the model against the 2Fo-Fc and SIR maps showed several incorrect side chains, and seven residues in each molecule were in very poor density. Therefore, residues 1, 20–21, 53, 61–62, and 66 for each molecule were omitted, and rigid body refinement was repeated. After this refinement, the rotation angle between the two molecules had increased to 3.6°. The missing residues were then rebuilt, and noncrystallographic symmetry restraints were imposed throughout the remaining refinement. To reduce bias in the refinement, phases from the Pt derivative were used to calculate SIR maps, as well as combined with the calculated phases to produce electron density maps.
Refinement was by simulated annealing as described above. Weights for the noncrystallographic restraints were based on analysis of the free R factor over multiple refinement runs in which the noncrystallographic weight was varied. In the final refinement, weights of 50 and 25 kcal/mole/Å2 were used for main-chain atoms (CA, C, N, and O) and side-chain atoms, respectively.
Early in the course of the refinement, we noted that there was significant electron density in each molecule between Cys 29 and Cys 48, in addition to the originally modeled disulfide bond. In each case this density could be modeled by an alternate conformation of the disulfide bond between these two residues. None of the other disulfide bonds showed electron density suggestive of alternate conformations. Coordinates of the alternate sulfur positions and B factors for the sulfur atoms were included in the refinement, but the occupancies were fixed at 0.5. The same strategy for B-factor refinement was used as described above, except that the B factors between atoms related by noncrystallographic symmetry were also restrained. A flat-bulk solvent model was also generated (Jiang and Brünger 1994).
Water molecules were added by using a routine from XPLOR that searches for peaks in the 2Fo-Fc map and checks distance criteria for reasonable hydrogen-bond donors and acceptors. Of the 27 water molecules included in refinement, 22 are related by noncrystallographic symmetry. The other 5 water molecules are associated with molecule A. The final R factor is 0.255 (6098 reflections in the working set), and the free R factor is 0.287 (362 reflections in the test set). The relatively high values for R and free R are probably due to two factors: (1) the use of noncrystallographic symmetry and (2) the systematic weakness of l odd reflections, especially at resolution <3 Å (Table 4). The atomic coordinates and structure factors for both crystal forms of CsE-v2 have been deposited with the PDB (1JZA and 1JZB).
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.39202.
References
- Babin, D.R., Watt, D.D., Goos, S.M., and Mlejnek, R.V. 1974. Amino acid sequences of neurotoxic protein variants from the venom of Centruroides sculpturatus Ewing. Arch. Biochem. Biophys. 164: 694–706. [DOI] [PubMed] [Google Scholar]
- Barton, G. J. 1993. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6: 37–40. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1990. Extension of molecular replacement: A new search strategy based on Patterson correlation refinement. Acta Crystallogr. 46: 46–57. [Google Scholar]
- Brünger, A.T. 1992. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355: 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Kuriyan, J., and Karplus, M. 1987. Crystallographic R factor refinement by molecular dynamics. Science 235: 458–460. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Krukowski, J., and Erickson, J. 1990. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. 46: 585–593. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1987. Ribbon models of macromolecules. J. Mol. Graph. 5: 103–106. [Google Scholar]
- Catterall, W.A. 1979. Binding of scorpion toxin to receptor sites associated with sodium channels in frog muscle, correlation of voltage-dependent binding with activation. J. Gen. Physiol. 74: 357–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50: 760–763. [DOI] [PubMed] [Google Scholar]
- Couraud, F., Rochat, H., and Lissitzky, S. 1980. Binding of scorpion neurotoxins to chick embryonic heart cells in culture and relationship to calcium uptake and membrane potential. Biochemistry 19: 457–462. [DOI] [PubMed] [Google Scholar]
- Crest, M., Jacquet, G., Gola, M., Zerrouk, H., Benslimane, A., Rochat, H., Mansuelle, P., and Martin-Eauclaire, M.F. 1992. Kaliotoxin, a novel peptidyl inhibitor of neuronal BK-type Ca(2+)-activated K+ channels characterized from Androctonus mauretanicus mauretanicus venom. J. Biol. Chem. 267: 1640–1647. [PubMed] [Google Scholar]
- Ealick, S.E., Cook, W.J., Fontecilla-Camps, J.C., Suddath, F.L., Bugg, C.E., and Watt, D.D. 1984. Preliminary X-ray investigation of CsE-v2 scorpion toxin from Centruroides sculpturatus Ewing: Evidence of a reversible transition between crystal forms. J. Biol. Chem. 259: 12081–12083. [PubMed] [Google Scholar]
- Engh, R.A. and Huber, R. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. 47: 392–400. [Google Scholar]
- Galvez, A., Gimenez-Gallego, G., Reuben, J.P., Roy-Contancin, L., Feigenbaum, P., Kaczorowski, G.J., and Garcia, M.L. 1990. Purification and characterization of a unique, potent, peptidyl probe for the high conductance calcium-activated potassium channel from venom of the scorpion Buthus tamulus. J. Biol. Chem. 265: 11083–11090. [PubMed] [Google Scholar]
- Gimenez-Gallego, G., Navia, M.A., Reuben, J.P., Kaczorowski, G.J., and Garcia, M.L. 1988. Purification, sequence, and model structure of charybdotoxin, a potent selective inhibitor of calcium-activated potassium channels. Proc. Natl. Acad. Sci. 85: 3329–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.-L., Li, H.-M., Zeng, Z.-H., Liu, X.-Q., Wang, M., and Wang, D.-C. 1999. Crystal structures of two α-like scorpion toxins: Non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel. J. Mol. Biol. 292: 125–135. [DOI] [PubMed] [Google Scholar]
- He, X.-L., Deng, J.P., Wang, M., Zhang, Y., and Wang, D.-C. 2000. Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 Å. Acta Crystallogr. 56: 25–33. [DOI] [PubMed] [Google Scholar]
- Housset, D., Habersetzer-Rochat, C., Astier, J., and Fontecilla-Camps, J.C. 1994. Crystal structure of toxin II from the scorpion Androctonus australis Hector refined at 1.3 Å resolution. J. Mol. Biol. 238: 88–103. [DOI] [PubMed] [Google Scholar]
- Howard, A.J., Gilliland, G.L., Finzel, B.C., Poulos, T.L., Ohlendorf, D.H., and Salemme, F.R. 1987. The use of an imaging proportional counter in macromolecular crystallography. J. Appl. Crystallogr. 20: 383–387. [Google Scholar]
- Jablonsky, M.J., Watt, D.D., and Krishna, N.R. 1995. Solution structure of an old world-like neurotoxin from the venom of the new world scorpion Centruroides sculpturatus Ewing. J. Mol. Biol. 248: 449–458. [DOI] [PubMed] [Google Scholar]
- Jablonsky, M.J., Jackson, P.L., Trent, J.O., Watt, D.D., and Krishna, N.R. 1999. Solution structure of a beta-neurotoxin from the new world scorpion Centruroides sculpturatus Ewing. Biochem. Biophys. Res. Comm. 254: 406–412. [DOI] [PubMed] [Google Scholar]
- Jiang, J.-S. and Brünger, A.T. 1994. Protein hydration observed by X-ray diffraction; solvation properties of penicillopepsin and neuraminidase crystal structures. J. Mol. Biol. 243: 100–115. [DOI] [PubMed] [Google Scholar]
- Jover, E., Couraud, F., and Rochat, H. 1980. Two types of scorpion neurotoxins characterized by their binding to two separate receptor sites on rat brain synaptosomes. Biochem. Biophys. Res. Comm. 95: 1607–1614. [DOI] [PubMed] [Google Scholar]
- Landon, C., Sodano, P., Cornet, B., Bonmatin, J., Kopeyan, C., Rochat, H., Vovelle, F., and Ptak, M. 1997. Refined solution structure of the anti-mammal and anti-insect LqqIII scorpion toxin: Comparison with other scorpion toxins. Proteins 28: 360–374. [DOI] [PubMed] [Google Scholar]
- Lee, W., Jablonsky, M.J., Watt, D.D., and Krishna, N.R. 1994. Proton nuclear magnetic resonance and distance geometry/simulated annealing studies on the variant-1 neurotoxin from the new world scorpion Centruroides sculpturatus Ewing. Biochemistry 33: 2468–2475. [DOI] [PubMed] [Google Scholar]
- Li, H.M., Wang, D.-C., Zeng, Z.-H., Jin, L., and Hu, R.Q. 1996. Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 Å. J. Mol. Biol. 261: 415–431. [DOI] [PubMed] [Google Scholar]
- Luzzati, V. 1952. Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr. 5: 802–810. [Google Scholar]
- Matthews, B.W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33: 491–497. [DOI] [PubMed] [Google Scholar]
- Narahashi, T., Shapiro, B.I., Deguchi, T., Scuka, M., and Wang, E.M. 1972. Effects of scorpion venom on squid axon membranes. Amer. J. Physiol. 222: 850–857. [DOI] [PubMed] [Google Scholar]
- Oren, D.A., Froy, O., Amit, E., Kleinberger-Doron, N., Gurevitz, M., and Shaanan, B. 1998. An excitatory scorpion toxin with a distinctive feature: An additional alpha helix at the C terminus and its implications for interaction with insect sodium channels. Structure 6: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Otwinoski, Z. 1991. Maximum likelihood refinement of heavy atom parameters. In Isomorphous Replacement and Anomalous Scattering (eds. W. Wolf, P.R. Evans, and A.G.W. Leslie), pp. 80–86. SERC Daresbury Laboratory, Warrington, UK.
- Pintar, A., Possani, L.D., and Delepierre, M. 1999. Solution structure of toxin 2 from Centruroides noxius Hoffmann, a β-scorpion neurotoxin acting on sodium channels. J. Mol. Biol. 287: 359–367. [DOI] [PubMed] [Google Scholar]
- Polikarpov, I., Junior, M.S.M., Marangoni, S., Toyama, M.H., and Teplyakov, A. 1999. Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins. J. Mol. Biol. 290: 175–184. [DOI] [PubMed] [Google Scholar]
- Possani, L.D., Martin, B.M., and Sveden, I. 1982. The primary structure of noxiustoxin: A K+ channel blocker peptide purified from the venom of the scorpion Centruroides noxius Hoffmann. Carlsberg Res. Commun. 47: 285–289. [Google Scholar]
- Read, R.J. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A42: 140–149. [Google Scholar]
- Romey, G., Chicheportiche, R., Lazdunski, M., Rochat, H., Miranda, F., and Lissitzky, S. 1975. Scorpion neurotoxin: A presynaptic toxin which affects both Na+ and K+ channels in axons. Biochem. Biophys. Res. Commun. 64: 115–121. [DOI] [PubMed] [Google Scholar]
- Sack, J.S. 1988. CHAIN—A crystallographic modeling program. J. Mol. Graph. 6: 244–245. [DOI] [PubMed] [Google Scholar]
- Tugarinov, V., Kustanovich, I., Zilberberg, N., Gurevitz, M., and Anglister, J. 1997. Solution structures of a highly insecticidal recombinant scorpion alpha-toxin and a mutant with increased activity. Biochemistry 36: 2414–2424. [DOI] [PubMed] [Google Scholar]
- Zhao, B., Carson, M., Ealick, S.E., and Bugg, C.E. 1992. Structure of scorpion toxin Variant-3 at 1.2 Å resolution. J. Mol. Biol. 227: 239–252. [DOI] [PubMed] [Google Scholar]