Abstract
DNA and histone synthesis are coupled and ongoing replication is required to maintain histone gene expression. Here, we expose S phase–arrested cells to the kinase inhibitors caffeine and LY294002. This uncouples DNA replication from histone messenger RNA (mRNA) abundance, altering the efficiency of replication stress–induced histone mRNA down-regulation. Interference with caffeine-sensitive checkpoint kinases ataxia telangiectasia and Rad3 related (ATR)/ataxia telangiectasia mutated (ATM) does not affect histone mRNA down- regulation, which indicates that ATR/ATM alone cannot account for such coupling. LY294002 potentiates caffeine's ability to uncouple histone mRNA stabilization from replication only in cells containing functional DNA-activated protein kinase (DNA-PK), which indicates that DNA-PK is the target of LY294002. DNA-PK is activated during replication stress and DNA-PK signaling is enhanced when ATR/ATM signaling is abrogated. Histone mRNA decay does not require Chk1/Chk2. Replication stress induces phosphorylation of UPF1 but not hairpin-binding protein/stem-loop binding protein at S/TQ sites, which are preferred substrate recognition motifs of phosphatidylinositol 3-kinase–like kinases, which indicates that histone mRNA stability may be directly controlled by ATR/ATM- and DNA-PK–mediated phosphorylation of UPF1.
Introduction
Metazoan evolutionary emergence has depended not only on genetic complexity and developmentally regulated gene expression but also on the establishment of high-fidelity systems that coordinate accurate DNA replication with appropriate chromatin assembly. DNA replication and histone protein synthesis are essential, finely balanced S phase events. Disturbances result in misregulation of gene expression, cell cycle arrest, and chromosome instability, which result in developmental failure (Meeks-Wagner and Hartwell, 1986; Han et al., 1987; Wyrick et al., 1999).
Eukaryotic DNA synthesis is governed by active replication origin numbers together with an intrinsic catalytic rate operating at a replication fork; this, in turn, creates specific demands for timely delivery of additional components, such as histones, required for chromatin assembly. DNA and histone synthesis are coupled. DNA synthesis inhibition causes rapid histone mRNA destabilization that results in histone synthesis shutdown (DeLisle et al., 1983; Heintz et al., 1983), which raises the possibility that checkpoints regulate histone mRNA levels. Histone mRNA destabilization depends on a hairpin at the mRNA 3′ end (Graves et al., 1987) that binds hairpin-binding protein (HBP)/stem-loop binding protein (SLBP; Wang et al., 1996; Martin et al., 1997; Zhao et al., 2004). Efficient histone mRNA destabilization also requires the RNA helicase UPF1 (Kaygun and Marzluff, 2005).
Checkpoints control cell cycle timing after genomic insult (Zhou and Elledge, 2000). DNA replication failure blocks further initiation from later-firing replication origins (Painter and Young, 1980), stabilizes arrested replisome components arising from previously fired origins (Dimitrova and Gilbert, 2000; Feijoo et al., 2001) and also blocks entry into mitosis (Rao and Johnson, 1970). Checkpoints are also involved in histone and S phase coordination. Rad53 is required for degradation of excess histone protein not packaged into chromatin (Gunjan and Verreault, 2003) and plays a role in histone metabolism and chromatin assembly (Emili et al., 2001; Sharp et al., 2005). Ataxia telangiectasia mutated (ATM)/ataxia telangiectasia and Rad3 related (ATR), which are phosphatidylinositol 3-kinase–like kinase (PIKK) family checkpoint kinases, respond to DNA damage. DNA double-strand breaks activate ATM and ATR is activated by aberrant DNA structures induced by UV light or DNA synthesis inhibitors (Abraham, 2001). Another PIKK, DNA-activated protein kinase (DNA-PK), is required for DNA double-strand break repair by nonhomologous end joining (NHEJ) and telomere maintenance. It is also required for ionizing radiation–induced down-regulation of histone H2B gene transcription, which reflects a potential role in a DNA damage response (Schild-Poulter et al., 2003). Chk1 and 2 are kinases activated by ATR/ATM with partially overlapping functions. Known Chk1 functions include prevention of premature mitosis (Zachos et al., 2005), activation of homologous recombination repair (Sorensen et al., 2005), and, in metazoans, activation of the origin firing and replisome integrity checkpoint (Feijoo et al., 2001; Zachos et al., 2005) operating downstream of ATR/ATM (Dimitrova and Gilbert, 2000).
Here, we investigated whether checkpoint components monitoring DNA replication, which effect the origin firing checkpoint, also control histone mRNA stability. DNA replication and histone mRNA stability are linked by two parallel and interdependent pathways, one sensitive to caffeine and the other to LY294002. It is likely that ATR is the mediator of the caffeine-sensitive pathway and our results argue strongly that DNA-PK is the LY294002-sensitive mediator. We show that DNA-PK is activated during replication stress and that it, together with a caffeine-sensitive protein, plays a critical role in the regulation of histone mRNA stability. Checkpoint regulation of histone mRNA decay occurring via either ATR or DNA-PK does not require either Chk1 or 2 activity. Our data are consistent with the notion that the cellular machinery controlling histone mRNA stability is a direct target of ATR and DNA-PK.
Results
Caffeine abrogates the replication origin firing checkpoint and inhibits histone mRNA decay induced by replication stress in HeLa cells
Small molecule inhibitors of the ATR–Chk1 pathway, such as caffeine, abrogate an S phase checkpoint controlling replication origin firing in CHO cells (Dimitrova and Gilbert, 2000; Feijoo et al., 2001), which are particularly amenable to protocols involving multiple cell cycle arrests. To investigate a potential linkage between known checkpoint pathways with a role in origin firing and the regulation of histone mRNA decay in response to replication stress, we investigated whether caffeine could abolish this checkpoint response in human cells amenable to analysis of histone mRNA decay.
Eukaryotic DNA replication takes place at discrete sites that may be visualized by pulse labeling cells with halogenated derivatives of deoxyuridine (dU; O'Keefe et al., 1992) and stained with labeled antibodies specific to each dU derivative (Dimitrova and Gilbert, 2000). The spatial pattern of replication sites reveals their temporal position within S phase, which allows the dynamics of groups of coordinately replicated chromosomal domains to be investigated.
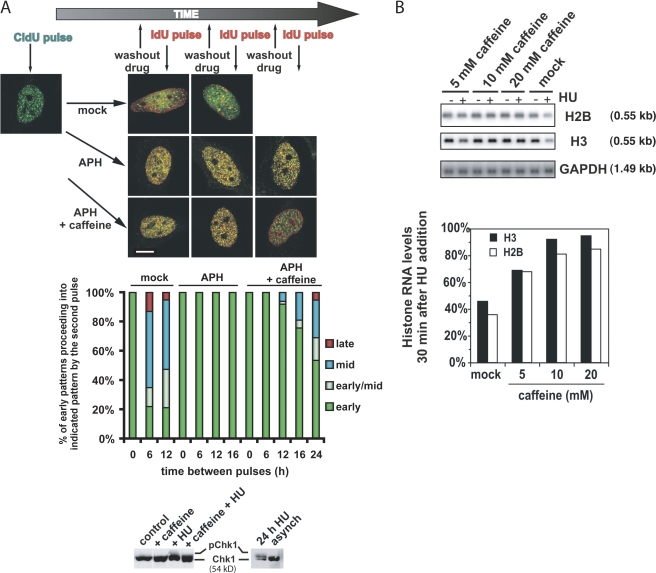
To determine if abrogation of ATR function in HeLa cells arrested in S phase would allow the initiation of replication at later replicating sites, asynchronously growing cells were briefly pulse labeled with chlorodeoxyuridine (CldU) and either left untreated (mock treated) or treated with the DNA polymerase inhibitor aphidicolin (APH) for 6–24 h in the absence or presence of 5 mM caffeine. Afterward, cells were washed free of inhibitors, briefly pulse labeled with iododeoxyuridine (IdU), and then fixed and stained with anti-CldU (Fig. 1 A, green) or anti-IdU (red) antibodies.
Figure 1.
Caffeine abrogates the replication checkpoint and inhibits histone mRNA decay induced by replication stress in HeLa cells. (A) Caffeine abrogates the replication checkpoint. (top) Asynchronous HeLa cells were pulsed with CldU for 20 min and incubated without drugs (mock), in the presence of APH alone, or in the presence of both APH and caffeine. At different times (typically 6–24 h) after the CldU pulse, cells were washed free of drugs and pulsed with IdU for 20 min. CldU or IdU incorporation was visualized by immunofluorescence confocal microscopy. Data are single optical sections showing the initial CldU pulse (green) incorporated into an early replication pattern and, at times thereafter, IdU pulses (red) either colocalized (yellow) with the CldU pulse in the presence of APH or being incorporated into progressively later replication patterns in mock-treated cells and in cells where the replication checkpoint has been abrogated by caffeine addition. Bar, 10 μm. (middle) Graph summarizes typical data and shows the fraction of those cells with an early replication pattern at the first pulse proceeding into the indicated pattern (early, early/mid, mid, or late) visualized by the second pulse after each treatment. (bottom) Nocodazole-arrested HeLa cells were released into drug-free medium for 14 h. The cells were either mock treated (control) or treated with 5 mM caffeine alone (+caffeine) for 1 h before being treated for a further 2 h either with (+HU and +caffeine +HU) or without (control and +caffeine) HU to induce replication stress. Cell lysates were then immunoblotted for Chk1. (B) Caffeine inhibits histone mRNA decay. Asynchronous HeLa cells were treated with the indicated caffeine concentration or mock treated. After 1 h, 2 mM HU or water was added and incubation was continued for 30 min followed by isolation of RNA. RNA was analyzed by Northern blotting using probes detecting histone H2B mRNA, H3 mRNA, or GAPDH mRNA. Histone mRNA levels were standardized with respect to GAPDH mRNA levels and the graph shows histone mRNA levels in HU-treated cells expressed as a percentage of the level in untreated cells.
Cells in early S phase at the time of the CldU pulse (with many sites of CldU incorporation distributed throughout euchromatin; Fig. 1 A, top left) initiated replication at mid–S phase replicating sites (identified by peripheral and perinucleolar staining in the IdU pulse) at 6 h and finally at late-replicating heterochromatin within 12 h (Fig. 1 A, mock). In APH-treated cells without added caffeine, IdU labeling largely colocalized with CldU at all times investigated, indicating that, as expected, there was no progression through S phase or initiation within later replicating domains under these conditions (Fig. 1 A, APH). In contrast, in cells treated with caffeine during exposure to APH, IdU was incorporated into progressively later-replicating domains and was accompanied by a lack of DNA synthesis from previously initiated sites (Fig. 1 A, APH + caffeine). Quantitation of data in Fig. 1 A indicates that caffeine abrogates the origin firing replication checkpoint in HeLa cells, albeit with slightly reduced kinetics of progression through S phase in comparison to untreated cells or compared with the same experiment in CHO cells (Dimitrova and Gilbert, 2000). Although the reason for this is unknown, it may be related to cell type differences in rates of recovery from replication arrest (Jackson, 1995). Immunoblotting of the ATR substrate Chk1 from cells treated with the replication inhibitor hydroxyurea (HU) was used to confirm ATR inhibition by 5 mM caffeine (Fig.1 A, bottom).
Treatment with either HU or APH also results in histone mRNA decay (Baumbach et al., 1987; Levine et al., 1987). To test if checkpoint components also regulate the stability of histone mRNA, we investigated caffeine's effect on replication stress–induced histone mRNA decay. Asynchronous HeLa cells were pretreated with a range of caffeine concentrations before HU addition. Subsequently, total RNA was isolated and analyzed by Northern blotting using probes that detect either histone H2B or H3 mRNA. As expected, HU addition alone initiated efficient destabilization of histone H2B and H3 mRNA, which, after 30 min, were reduced to ∼40% of control levels (Fig. 1 B, mock). Caffeine affected histone mRNA decay induced by replication stress. At a concentration where it inhibits ATR (Hall-Jackson et al., 1999) and the related PIKK ATM (Blasina et al., 1999) and abrogates the origin firing replication checkpoint (Fig. 1 A; Dimitrova and Gilbert, 2000), caffeine partially inhibited histone mRNA decay induced by replication stress with ∼70% of histone mRNA remaining after 30-min exposure to HU. Inhibition of replication stress–induced histone mRNA decay by caffeine was dose dependent and, in the presence of 20 mM caffeine, 85–95% of histone mRNA remained after 30 min of HU treatment. These results indicate that destabilization of histone mRNA induced by replication stress is sensitive to the PIKK inhibitor caffeine and suggest that PIKKs may be involved in the control of histone mRNA stability.
Inhibition of ATR signaling does not interfere with replication stress–induced histone mRNA decay
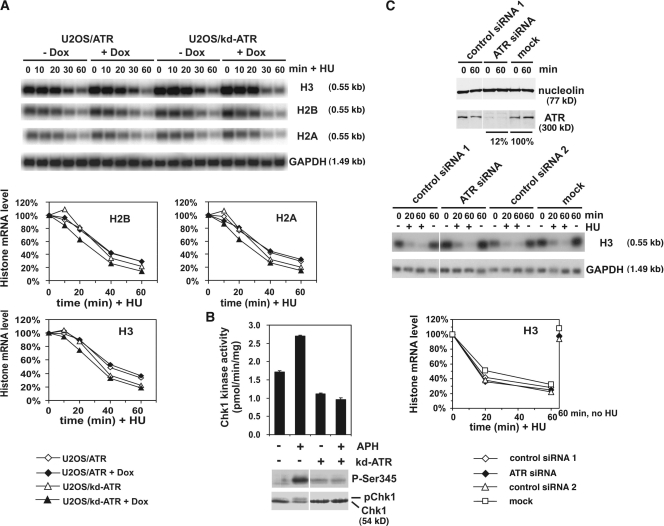
To investigate a specific role for ATR in regulating histone mRNA decay, we tested cells (U2OS/kinase dead [kd] ATR) that conditionally overexpress a kd form of ATR (D2475A) that acts in a dominant-negative manner to block normal ATR function (Nghiem et al., 2001). Control and doxycycline-treated cells overexpressing either kd-ATR or wild-type (wt) ATR were treated with HU for various times, total RNA was isolated, and histone H2A, H2B, and H3 mRNA levels were determined by Northern blotting (Fig. 2 A).
Figure 2.
Inhibition of ATR signaling does not interfere with replication stress–induced histone mRNA decay. (A) Time course of HU-induced decay of histone mRNA in cells expressing kd-ATR. Expression of wt-ATR or kd-ATR was either induced (+Dox) or not (−Dox) by treatment of U2OS/ATR and U2OS/kd-ATR cells with doxycycline. Cells were treated with 2 mM HU and RNA was isolated at indicated times after HU addition. Histone RNA was analyzed as in Fig. 1. Changes in histone H2A, H2B, and H3 mRNA levels were compared with the level at 0 min. (B) Inactivation of ATR signaling using cells expressing kd-ATR suppresses phosphorylation and activation of Chk1 in response to replication stress. Expression of kd-ATR was either induced (+kd-ATR) or not (−kd-ATR) by treatment of U2OS/kd-ATR cells with doxycycline or buffer. Cells were then treated with 50 μg/ml APH for 16 h to induce replication stress. Subsequently, cell lysates were subjected to a Chk1 immunoprecipitation kinase assay (top) or immunoblotted using Chk1 antibodies (bottom, anti–phospho-Ser345 and total Chk1 antibody). (C) Ablation of ATR by RNAi does not interfere with HU-induced histone mRNA decay. Asynchronous HeLa cells were transfected with siRNA targeting ATR, control siRNA 1 (targeting luciferase), control siRNA 2 (targeting a nonrelevant gene), or mock transfected. Cell lysates were immunoblotted for ATR and nucleolin (top). 32 h after transfection, cells were treated with or without 2 mM HU for 0, 20, or 60 min or left untreated for 60 min, and then lysed for RNA analysis (middle). Histone H3 mRNA levels were analyzed as in Fig. 1. Graph (bottom) shows changes in H3 mRNA levels compared with the level at 0 min. In parallel, cell lysates were immunoblotted for ATR (to confirm knockdown) and nucleolin (loading; left). Data are expressed as mean ± range for duplicate experiments.
In the absence of overexpressed protein, the pattern of histone mRNA decay was the same for all histone variants tested and, as expected, HU addition resulted in rapid (<60 min) destabilization of histone mRNA (Fig. 2 A). However, overexpression of kd-ATR (or wt-ATR) did not significantly affect either the rate or extent of decay of any of the histone mRNAs. Overexpression of kd-ATR in this cell line has been shown previously to interfere with ATR function. To confirm that kd-ATR overexpression did indeed interfere with ATR signaling, we determined the activity and phosphorylation state of Chk1 in response to replication stress. Chk1 phosphorylation and activation was completely suppressed in the presence of overexpressed kd-ATR but not wt-ATR (Fig. 2 B).
In an alternative approach, we used siRNA to deplete ATR in HeLa cells that had reduced ATR levels to ∼12% of control levels (Fig. 2 C, top). The rate and extent of histone H3 mRNA decay in ATR-depleted cells exposed to replication stress was almost identical to that in control or mock-treated cells (Fig. 2 C, middle and bottom). Collectively, the results in Fig. 2 indicate that ATR is not functionally limiting for the rate or extent of histone mRNA decay induced by replication stress.
Replication stress–induced histone mRNA decay occurs efficiently in cells lacking ATM
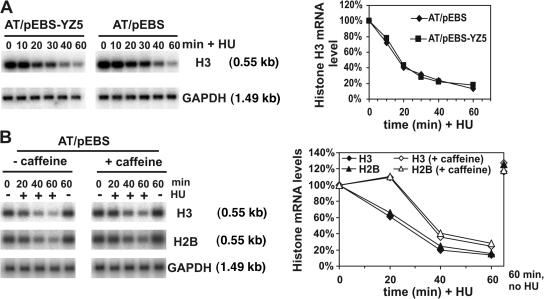
ATM is also a caffeine-sensitive component of a DNA damage checkpoint (Blasina et al., 1999). To establish if ATM was necessary for replication stress–induced histone mRNA decay, we compared histone mRNA decay induced by replication stress in ATM null cells (AT221JE-T) transfected either with an empty vector or a vector encoding wt-ATM (Fig. 3 A). Histone H3 mRNA decay in these cells occurred with similar kinetics to decay in HeLa and U2OS cells. Reintroduction of ATM had no effect on either the rate or extent of replication stress–induced histone H3 mRNA decay, which indicates that ATM, like ATR, is not functionally limiting for the rate or extent of histone mRNA decay induced by replication stress.
Figure 3.
Replication stress induces efficient, caffeine-sensitive histone mRNA decay in cells lacking ATM. (A) ATM is not required for histone mRNA decay induced by replication stress. AT22IJE-T/pEBS (AT/pEBS) cells lacking ATM and AT22IJE-T/pEBS-YZ5 (AT/pEBS-YZ5) cells complemented with ATM were treated with 2 mM HU for the times indicated, and RNA was isolated and analyzed by Northern blotting as in Fig. 1. Histone H3 mRNA levels were normalized to GAPDH mRNA levels and compared with the level at 0 min. (B) Caffeine inhibits histone mRNA decay in AT cells. AT/pEBS cells were treated with caffeine or mock treated. After 1 h, 2 mM HU or water was added and cells were incubated for the indicated times before RNA isolation and analysis. Graph shows changes in histone H2B and H3 mRNA levels compared with the level at 0 min.
The kinetics of mRNA decay induced by replication stress in ATM null cells is altered by caffeine but not ATR depletion
To test if ATM and ATR are functionally redundant, we first depleted ATR in ATM null cells using siRNA as in Fig. 2. Western blotting analysis confirmed that the level of ATR protein was reduced at least fivefold using this approach (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200708106/DC1). Depleting ATR to this extent results in a significant loss of replication checkpoint function (Casper et al., 2002; unpublished data). The rate and extent of replication stress–induced histone mRNA decay in these ATR-depleted ATM null cells was indistinguishable from that in control siRNA-treated cells, indicating that the absence of any effect of ATR ablation by siRNA on histone mRNA levels (Fig. 2 C) was not caused by redundancy between ATR and ATM and suggesting that if ATR plays an essential role in regulating histone mRNA abundance, these much-reduced levels of ATR are sufficient for this function in an ATM null background.
In an alternative approach to the question of ATR/ATM redundancy, we investigated the effects of adding caffeine to ATM null cells. ATM null cells were pretreated for 1 h with 5 mM caffeine before the induction of replication stress with HU. Interestingly, histone mRNA decay was inhibited by caffeine for up to 20 min after HU addition. Subsequently, decay resumed at a rate approximating that observed in mock-treated cells exposed to HU (Fig. 3 B). These data indicate that ATM null cells retain a caffeine-sensitive component that, when inhibited, results in reduced efficiency of histone mRNA destabilization during replication stress. Collectively with the data in Fig. S1, this suggests formally that either ATR may not be the only other caffeine-sensitive component or that the differences inherent in alternative approaches to inactivate ATR (instantaneous inhibition by caffeine compared with siRNA-induced ATR knockdown over 12–48 h) ultimately result in different kinetics of replication stress–induced histone mRNA destabilization.
The general PIKK family inhibitor wortmannin affects the efficiency of replication stress–induced histone mRNA decay
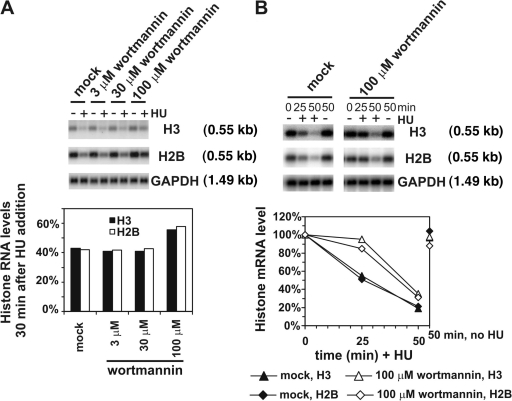
Our data using caffeine, a reversible inhibitor of a relevant protein kinase family, albeit at high concentration, implicated PIKKs in regulating the efficiency of histone mRNA decay after replication stress. To obtain additional evidence for such a role and identify at a molecular level the relevant target proteins, we investigated the effects of wortmannin and determined its effect on histone H2B and H3 mRNA decay. Wortmannin brings about the inhibition of PIKK family members by covalent inactivation. Although pretreatment with 3 or 30 μM wortmannin had little or no effect, 100 μM wortmannin significantly inhibited histone mRNA decay occurring during the first 30 min after HU addition (Fig. 4 A). To determine the kinetics of replication stress–induced histone mRNA decay, with or without wortmannin, histone mRNA H3, and H2B mRNA levels were measured 25 and 50 min after HU addition (Fig. 4 B). Normal histone mRNA decay was observed in mock-treated cells. Interestingly, histone mRNA decay was inhibited by 100 μM wortmannin for up to 25 min after HU addition. Subsequently, decay resumed at a rate approximating that observed in mock-treated cells exposed to HU. These data suggest that PIKK family members do play a role in controlling histone mRNA decay and that they modulate the kinetics of this process by delaying the onset of replication stress–induced histone mRNA decay.
Figure 4.
Wortmannin induces a delay in replication stress–induced histone mRNA decay. (A) Concentration dependence of delay in histone mRNA decay induced by wortmannin. Asynchronous HeLa cells were treated with the indicated concentration of wortmannin or mock treated with DMSO. After 1 h, cells were treated with or without 2 mM HU and RNA was isolated after a further 30 min for analysis by Northern blotting. Histogram shows histone mRNA levels in HU-treated cells compared with levels in corresponding non–HU-treated cells. (B) Kinetic analysis of delay on histone mRNA decay induced by wortmannin. Asynchronous HeLa cells were treated with 100 μM wortmannin or mock treated with DMSO. After 1 h, cells were treated with or without 2 mM HU. RNA was isolated at the indicated times after HU addition and isolated from untreated cells after 50 min. Graph shows the changes in histone mRNA levels compared with the level at 0 min.
DNA-PK is a component of the checkpoint mechanism sensing replication stress and controlling histone mRNA stability
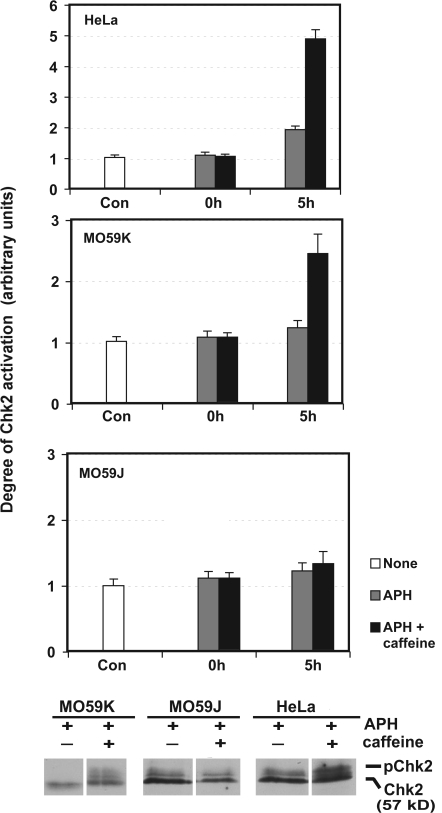
Exposing cells to replication stress results in activation of Chk1 and 2 (Feijoo et al., 2001). Activation of Chk1 is believed to be dependent on its phosphorylation by ATR (Fig. 2 B; Abraham, 2001), whereas the activation of Chk2 occurs independently of both ATR (not depicted) and ATM (Feijoo et al., 2001). To investigate more fully the effects of PIKK family inhibitors, we tested the effects of caffeine (Fig. 5 A) on replication stress–induced activation of Chk2 in HeLa cells. Interestingly, although caffeine abolishes replication stress–induced activation of Chk1 (not depicted; Feijoo et al., 2001), the presence of caffeine in cells experiencing replication stress resulted in a progressive increase in Chk2 phosphorylation and activation (Fig. 5) that was independent of ATM (not depicted). To investigate whether another PIKK could be responsible for the activation of Chk2 in response to replication stress, we examined the Chk2 response to caffeine in the DNA-PK–deficient cell line M059J alongside its DNA-PK wt counterpart M059K under conditions of replication stress. Exposure of M059K cells to replication stress resulted in the previously observed activation of Chk2 in response to caffeine (Fig. 5). In contrast, in DNA-PK–deficient cells (M059J), Chk2 was not activated under identical conditions. Together, these data indicate that Chk2 activation in response to replication stress is dependent on DNA-PK and suggest that activation of this pathway may be a consequence of a failure of ATR/ATM signaling.
Figure 5.
Inhibition of ATR/ATM signaling by caffeine results in a DNA-PK–mediated activation of Chk2. HeLa, MO59J (DNA-PK−), or MO59K (DNA-PK+) cells were either untreated (Con) or synchronized in metaphase by treatment with nocodazole for 14 h. Mitotic cells were removed and plated in fresh medium containing APH for 24 h (0 h). Caffeine or buffer was added and cells were incubated for an additional 5 h. Cell lysates were subjected to Chk2 immunoprecipitation kinase assay (top) or immunoblotted with Chk2 antibody (bottom). Chk2 band shifting (pChk2) is indicative of phosphorylation (Feijoo et al., 2001). Data are expressed as mean ± range for duplicate determinations.
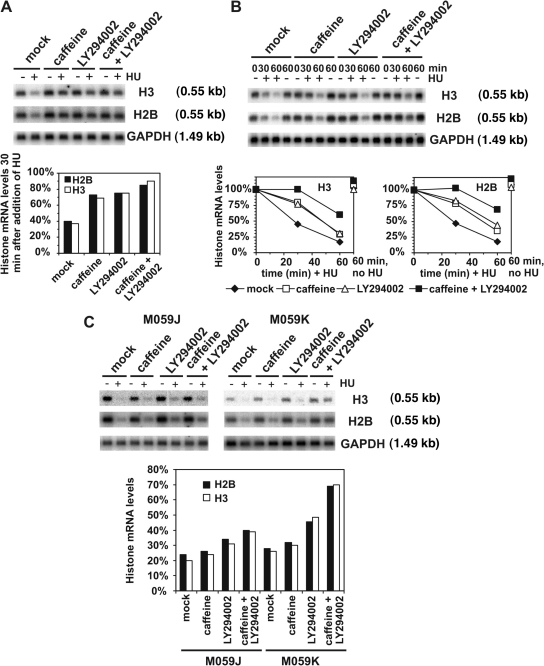
Concentrations of caffeine that inhibit ATR and ATM signaling in vivo are ineffective against DNA-PK (Sarkaria et al., 1999), although LY294002 has been reported to be a selective inhibitor of DNA-PK (Stiff et al., 2004). To investigate a role for DNA-PK in the regulation of replication stress–induced histone mRNA decay, we determined the effect of LY294002 on this process. Treatment of cells with 200 μM LY294002, a concentration that blocks DNA-PK function but does not affect ATR or ATM function (Stiff et al., 2004), inhibited histone H2B and H3 mRNA decay during the first 30 min after HU addition. As before, the addition of 5 mM caffeine also partially restored histone mRNA stability. Interestingly, the effects of caffeine and LY294002 combined were additive (Fig. 6 A). Cells exposed to 5 mM caffeine and 200 μM LY294002 together retained close to maximum levels of histone mRNA after a 30-min treatment with HU (Fig. 6 A). Kinetic analysis showed that, as for wortmannin, either caffeine or LY294002 alone significantly delayed the onset of histone H2B and H3 mRNA decay induced by replication stress (Fig. 6 B). Treatment with caffeine and LY294002 together completely abolished the rapid initiation of histone mRNA decay normally observed on imposition of replication stress. Decay resumed after ∼30 min in the presence of caffeine alone, LY294002 alone, or caffeine and LY294002 together (Fig. 6 B). However, even after extended periods of replication stress, the effects of caffeine and LY294002 combined remained additive, with mRNA levels remaining at ∼70% of control values after 1 h, which indicates that these compounds are affecting at least two distinct pathways regulating histone mRNA decay. Given the target specificity of caffeine and LY294002 on PIKK family members, these data strongly implicate DNA-PK in the control of replication stress–induced histone mRNA decay (Fig. 6 B).
Figure 6.
DNA-PK is a regulator of histone mRNA stability. (A) Asynchronous HeLa cells were treated with 5 mM caffeine alone, 200 μM LY294002 alone or 5 mM caffeine and 200 μM LY294002 together or mock treated with DMSO. After 1 h, cells were treated with or without 2 mM HU and RNA was isolated after an additional 30 min for analysis as in Fig. 1. The graph shows histone mRNA levels in HU-treated cells, with 100% defined as histone mRNA levels in non–HU-treated cells. (B) Asynchronous HeLa cells were treated with 5 mM caffeine alone, 200 μM LY294002 alone, or 5 mM caffeine and 200 μM LY294002 together or mock treated with DMSO. After 1 h, cells were treated with or without 2 mM HU, RNA was isolated at the indicated times, and all samples were analyzed by Northern blotting as in Fig. 1. Graphs show changes in histone mRNA levels compared with the level at 0 min. (C) M059J (DNA-PK−) and M059K (DNA-PK+) cells were treated with 5 mM caffeine alone, 200 μM LY294002 alone, or 5 mM caffeine and 200 μM LY294002 together or mock treated with DMSO. After 1 h, cells were treated with or without 2 mM HU and RNA was isolated after a further 1-h incubation and analyzed as in Fig. 1. The graph shows histone mRNA levels in HU-treated cells compared with histone mRNA levels in non–HU-treated cells.
To establish if histone mRNA decay in response to replication stress is indeed regulated by a DNA-PK–dependent pathway, we tested whether the additive effect of caffeine and LY294002 on inhibiting replication stress–induced histone mRNA decay was dependent on the presence of DNA-PK. We reasoned that if the effect of LY294002 is to potentiate the ability of caffeine to both delay the onset and inhibit the progressive destabilization of histone mRNA via the inhibition of DNA-PK, it follows that in cells lacking a functional DNA-PK signaling pathway, LY294002 would fail to inhibit histone mRNA decay in caffeine-treated cells exposed to replication stress. Therefore, M059J and M059K cells were pretreated with 5 mM caffeine alone, 200 μM LY294002 alone, or 5 mM caffeine and 200 μM LY294002 together or mock treated. All cells were treated with or without 2 mM HU and incubated for 1 h (to maximize the relative contribution of DNA-PK compared with a caffeine-sensitive checkpoint component), and RNA was isolated and analyzed by Northern blotting (Fig. 6 C). As before, HU induced significant mRNA decay, with 20% of histone H2B or H3 mRNA remaining after 1 h of HU treatment. At this time, the effect of caffeine on the extent of mRNA decay was minimal, as would be expected from the data in Fig. 6 B. Consistent with a role for DNA-PK in this pathway, LY294002 significantly increased the levels of histone mRNA remaining in M059K cells compared with M059J cells after 1 h of exposure to HU. Importantly, we found that in M059K but not M059J cells, LY294002 dramatically potentiated the effect of caffeine in inhibiting histone mRNA decay in response to replication stress. Combined treatment of M059K cells with LY294002 and caffeine severely reduced the extent of mRNA decay, with ∼70% of mRNA remaining after 1 h of exposure to HU. In comparison, M059J cells showed significantly less sensitivity to combined LY294002 and caffeine treatment (Fig. 6 C). Collectively, the data in Fig. 6 indicate that DNA-PK plays an important role in regulating histone mRNA stability.
Camptothecin induces single strand nicks that are converted into double-strand breaks during replication. DNA-PK has been implicated in the cellular response to camptothecin- induced lesions (Shao et al., 1999). We therefore investigated whether replication stress induced by camptothecin affected histone mRNA levels and the relative contributions (if any) of caffeine- and LY294002-sensitive pathways to the destabilization of histone message. We found that treatment of cells with 10 μM camptothecin induced a rapid disappearance of the histone message with a time course similar to that observed with HU. LY294002 alone had a significant negative effect on the efficiency of histone mRNA decay. Caffeine had a substantial effect in stabilizing the bulk of histone mRNA over the course of the experiment. Importantly, however, the combination of both caffeine and LY294002 was required to obtain maximal stabilization of histone mRNA (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200708106/DC1). These data indicate that in the case of either HU- or camptothecin-induced replication stress, at least two PIKK pathways are involved in histone mRNA decay.
Our data support roles in the regulation of histone mRNA stability for both a caffeine-sensitive pathway, presumably mediated by ATR (Fig. 1; Kaygun and Marzluff, 2005), and an LY294002-dependent pathway involving DNA-PK (Fig. 6). To gain further insight into the potential mechanisms by which histone mRNA abundance might be regulated by replication stress–induced signaling, we investigated the role of known downstream components of PIKK signaling pathways. Chk1 is a well-characterized mediator of ATR signaling during replication stress and we show here that Chk2 is a target for DNA-PK– dependent signaling (Fig. 5). Interfering with downstream effectors of ATR/ATM signaling abrogates the replication checkpoint controlling origin firing and replisome stability in both CHO (Feijoo et al., 2001) and HeLa cells (unpublished data). We therefore wished to address whether abrogation of Chk1 and/or Chk2 function interferes with histone mRNA decay during replication stress.
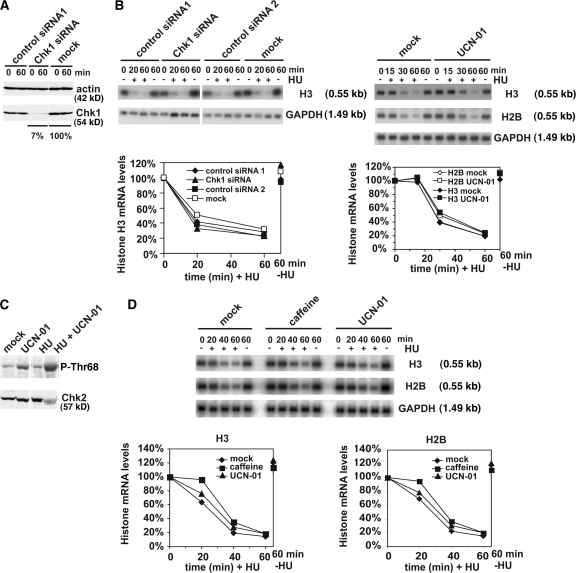
Depletion of Chk1 to very low levels using siRNA (Fig. 7 A; Rodriguez and Meuth, 2006) had no effect on replication stress–induced histone mRNA decay (Fig. 7 B, left). In an alternative approach, we used the Chk1 inhibitor UCN-01, which interferes with the replication checkpoint (Feijoo et al., 2001; unpublished data). Again, kinetics of replication stress–induced histone mRNA decay were largely unaffected by Chk1 inhibition (Fig. 7 B, right). These data show that Chk1 is not necessary for histone mRNA decay but do not exclude the possibility that it is involved in mediating a component of the caffeine-sensitive element of histone mRNA stability control. As shown here (Figs. 2, 5, and 6), it is conceivable that when this pathway is compromised, up-regulation of a DNA-PK–dependent pathway acts to control histone mRNA decay, potentially, though not necessarily, through Chk2. Indeed, Chk1 inhibition with UCN-01, like caffeine (Fig. 5), results in increased phosphorylation of Chk2 in response to replication stress (Fig. 7 C). To address this issue directly, we therefore used DLD-1 cells, which lack functional Chk2. Histone mRNA decay in response to replication stress in this cell line was very similar to that observed in all other lines tested, which indicates that Chk2 is not required for this process. Histone mRNA decay was, however, inhibited by 5 mM caffeine for ∼20 min after HU addition before a resumption at a rate approximating that observed in mock-treated cells exposed to HU (Fig. 7 D), which indicates that the ability of caffeine to interfere with the efficiency of histone mRNA decay was conserved. Histone mRNA decay was largely unaffected by the presence of UCN-01 in these cells, indicating that neither Chk1 nor Chk2 are functionally limiting for replication stress–induced histone mRNA decay. In an alternative approach, we also used the small molecule inhibitor debromohymenialdisine, which has been shown previously to inhibit both Chk1 and 2 in vivo (Curman et al., 2001). The imultaneous inhibition of both kinases did not affect the kinetics of HU-induced histone mRNA (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200708106/DC1), confirming that neither Chk1 nor Chk2 activity is functionally limiting for this process.
Figure 7.
Inhibition of Chk1 function results in increased phosphorylation of Chk2. Chk1 and 2 are not functionally limiting for the control of replication stress–induced histone mRNA decay. (A) Asynchronous HeLa cells were transfected with siRNA targeting Chk1 or control siRNA 1 (targeting luciferase) or mock transfected. Cell lysates were immunoblotted for Chk1 to confirm knockdown and actin (loading control). (B, left) Asynchronous HeLa cells were transfected with siRNA targeting Chk1, control siRNA 1 (targeting luciferase) or siRNA 2 (targeting a nonrelevant gene) or mock transfected. After 32 h, cells were treated with or without 2 mM HU for 0, 20, and 60 min or left untreated for 60 min and then lysed for RNA analysis. Histone H3 mRNA levels were analyzed as in Fig. 1. The graph shows changes in H3 mRNA levels compared with the level at 0 min. (B, right) Asynchronous HeLa cells were incubated with or without 300 nM of Chk1-selective inhibitor UCN-01. 1 h later, cells were incubated with or without 2 mM HU for the indicated times and lysed for RNA analysis as before. The graph shows changes in histone mRNA levels compared with the level at 0 min. (C) HeLa cells were synchronized in metaphase by treatment with nocodazole for 14 h and then released. After 14 h, cells were treated with or without 300 nM UCN-01 for 1 h, then 2 mM HU was added for a further 2 h. Cell lysates were immunoblotted with a phosphospecific Chk2 antibody (top) and a total anti-Chk2 antibody (bottom). (D) Asynchronous DLD-1 cells were incubated with or without 300 nM of Chk1-selective inhibitor UCN-01 or with or without caffeine. 1 h later, cells were incubated with or without 2 mM HU for the indicated times and lysed for RNA analysis as before. The graphs show changes in histone mRNA levels compared with the level at 0 min.
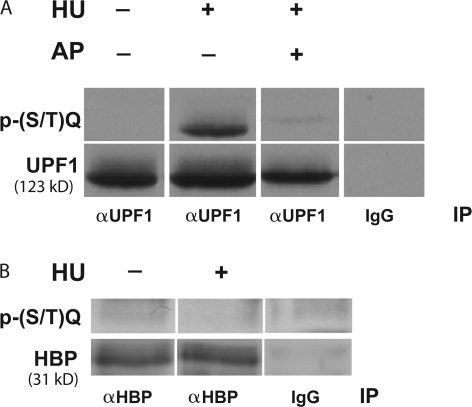
To address the possibility that components required for the efficient stabilization of histone mRNA are direct targets of PIKKs involved in sensing replication-induced stress, we took advantage of the availability of phosphospecific antibodies that recognize the substrate specificity motif (SQ and TQ) shared by PIKKs (O'Neill et al., 2000). HBP/SLBP is necessary for histone mRNA expression, is a rate-limiting component for histone pre-mRNA processing in cycling cells, and is known to be regulated by phosphorylation (Zheng et al., 2003; Zhao et al., 2004). UPF1, an RNA helicase involved in degradation of nonsense mRNAs and in mRNA stability control (Kim et al., 2005), has recently been implicated in the regulation of histone mRNA decay (Kaygun and Marzluff, 2005). UPF1 and HBP/SLBP both contain multiple S/TQ sites, which are substrate recognition motifs common to both ATR/ATM and DNA-PK (Hall-Jackson et al., 1999). Immunoblotting of UPF1 immunoprecipitates from HU- and mock-treated cell lysates indicated that phosphorylation of UPF1 at S/T-Q sites increased significantly in response to replication stress (Fig. 8 A). In contrast, no change in phospho-S/TQ immunoreactivity was observed either in whole cell lysates (not depicted) or HBP/SLBP immuno-precipitates from mock and HU-treated cells (Fig. 8 B).
Figure 8.
Replication stress results in increased phosphorylation of UPF1 but not HBP/SLBP on PIKK substrate motifs. Asynchronous HeLa cells were incubated with or without 2 mM HU for 16 h. (A) Cell lysates were subjected to immunoprecipitation using goat anti-UPF1 antibodies or IgG control, and immunoprecipitates were treated with AP or buffer before SDS-PAGE and immunoblotting with anti–phospho-S/TQ (top) and anti-UPF1 antibodies (bottom). (B) Cell lysates were subjected to immunoprecipitation using anti-HBP antisera and immunoprecipitates were immunoblotted with anti–phospho-S/TQ (top) and anti-HBP antiserum (bottom).
Discussion
A key mechanism in regulating the delivery of histone protein to newly synthesized DNA occurs via control of histone mRNA transcription and degradation (Schumperli, 1986; Marzluff and Duronio, 2002). In this paper, we provide evidence that at least two distinct PIKK signaling pathways participate in a checkpoint coupling DNA replication with histone mRNA abundance. Using caffeine and LY294002, selective inhibitors of distinct PIKKs, together with siRNA-mediated knockdown and cell lines lacking relevant gene products, we demonstrate a novel role for DNA-PK in the efficient destabilization of histone mRNA during replication stress. In addition to replicational slowing and fork arrest induced by HU, we find that another genotoxic stress, camptothecin, which induces lesions converted into double-strand breaks during replication, also causes efficient histone mRNA destabilization that could be completely blocked by inhibition using caffeine and LY294002 and, by implication, both ATR/ATM and DNA-PK signaling. These data indicate that both pathways contribute to the efficient regulation of histone abundance. However, it is possible that, depending on the genotoxic agent used, ATR/ATM and DNA-PK may not always be fully redundant with each other for histone mRNA down-regulation.
In contrast to the G1 checkpoint that blocks histone mRNA expression (Schild-Poulter et al., 2003), checkpoint components downstream of PIKKs are not required for regulating histone mRNA abundance during replication stress. We show that UPF1 but not HBP/SLBP is a direct target of replication stress– induced PIKK signaling, which suggests a potential mechanism by which coupling of DNA replication with histone mRNA abundance may operate.
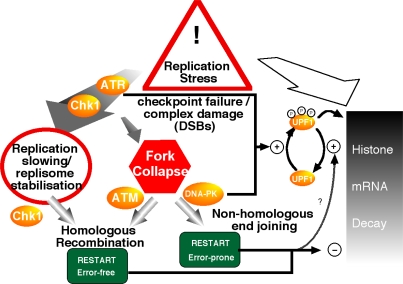
Exposure of cells to replication stress induces histone mRNA decay via a currently poorly understood pathway. The occurrence of replication stress arising from the presence of DNA damage results in a variety of checkpoint responses, which include suppression of new DNA synthesis via inhibition of replication origin firing, stabilization of slowed or arrested replication forks, and activation of DNA repair pathways to enable subsequent replication restart at stalled replication forks (Abraham, 2001; Lundin et al., 2002). Homologous recombination repair and NHEJ pathways both operate on lesions associated with arrested or collapsed replication forks to effect replication restart (Fig. 9; Arnaudeau et al., 2001; Lundin et al., 2002). In eukaryotes, homologous recombination repair requires the activation of ATR/ATM and the downstream effector Chk1 (Sorensen et al., 2005), whereas NHEJ requires the activity of the related PIKK DNA-PK. NHEJ is required for replication restart at forks where a DNA double-strand break has been generated but appears to be inessential for repair associated with slowed or arrested replication forks (Lundin et al., 2002). Such observations suggest that the extent of NHEJ in vivo may be linked to the efficiency of ATR signaling. Limitations of ATR signaling will result in higher levels of fork collapse (Dimitrova and Gilbert, 2000), resulting in activation of DNA-PK and consequent NHEJ.
Figure 9.
Model for the coordination of DNA replication and histone production in mammalian cells. Exposure of cells to replication stress induces histone mRNA decay via a currently poorly understood pathway (top right arrow). Replication stress results in stabilization of slowed or stalled replication forks via a pathway involving ATR and Chk1 (top left arrow). Replication restart from stalled replication forks occurs predominantly via a pathway involving homologous recombination. At some frequency, replication forks either encounter complex DNA damage or fail to be stabilized by the ATR–Chk1 pathway, which results in replication fork collapse, generating DNA double-strand breaks. These may be repaired either via ATM-dependent homologous recombination–induced replication restart or via NHEJ mediated by DNA-PK. Failure of ATR signaling (such as occurs in the presence of caffeine) will result in higher levels of fork collapse, generating an increased level of substrate for DNA-PK–mediated NHEJ. Coordinated regulation of histone mRNA decay by both ATR/ATM and DNA-PK during replication stress ensures that, irrespective of the extent to which each pathway operates in any given circumstance, the supply of histones will remain closely coupled to the demand required for the assembly of newly synthesized chromosomes. The RNA helicase UPF1 involved in histone mRNA decay (Kaygun and Marzluff, 2005) may act as an effector of ATR/DNA-PK signaling and induce histone mRNA decay. Upon relief from replication stress, histone mRNA stability is restored to normal levels presumably by a mechanism linked to the restart of replication forks and involving changes in UPF1 phosphorylation.
Our data support a model in which coordinate regulation of replication-dependent histone mRNA levels with DNA replication requires DNA-PK in addition to a caffeine-sensitive pathway (presumably mediated by ATR or ATM; Fig. 9). In vivo, the relative contribution from each signaling arm would be based on the nature of the DNA lesion generating replication stress and thus the relevant replication restart pathway. In these experiments, we have used caffeine to investigate and identify novel components of this checkpoint response by deliberately interfering with the efficiency of the ATR pathway and thus revealing the contribution of DNA-PK to the regulation of histone message abundance.
Caffeine, at concentrations believed to selectively and rapidly inhibit ATR/ATM, which blocked Chk1 phosphorylation and abrogated the origin firing checkpoint (Fig. 1 A), did partially affect histone mRNA decay in response to replication stress, supporting a role for ATR/ATM in this process. Maximal inhibition, however, required significantly higher caffeine concentrations well in excess of those required to interfere with predicted ATR/ATM-mediated checkpoint responses (Hall-Jackson et al., 1999; Sarkaria et al., 1999). This observation is consistent with the fact that all PIKK family members share a related kinase domain and those tested exhibit some sensitivity to inhibition by this xanthine alkaloid (Sarkaria et al., 1999). Importantly, low doses of caffeine or UCN-01, which affect replisome stability via inhibition of ATR–Chk1 signaling resulting in replication fork collapse (Dimitrova and Gilbert, 2000; Feijoo et al., 2001), gave rise to elevated Chk2 phosphorylation that was dependent on the presence of active DNA-PK. Such data support the hypothesis that failure of, or interference with, ATR/ATM-dependent pathways results in elevated signaling through DNA-PK.
Consistent with this model, overexpression of kd-ATR or significant ablation of ATR function by siRNA either in wt or ATM null cells, which are experimental approaches in which cycling cells are exposed for prolonged periods to suboptimal levels of a functional checkpoint protein, had little or no effect on the efficiency of histone mRNA decay (Fig. 2). This was presumably because in such circumstances the elevated levels of resultant fork collapse result in increased NHEJ activity and thus activation of a DNA-PK–mediated mechanism.
Expression of a dominant-negative form of ATR at levels sufficient to efficiently block the phosphorylation and activation of a known substrate (Chk1) had no effect on replication stress–induced mRNA decay. Although this might be expected, it does contrast with results obtained with these cells elsewhere (Kaygun and Marzluff, 2005) in which overexpression of a kd form of ATR did affect the efficiency though not necessarily the overall extent of histone mRNA decay. Although reasons for this discrepancy are unknown, one possibility concerns differences in the extent of kd-ATR overexpression. Zhou and Elledge (2000) have suggested, given the relatedness between PIKKs, that overexpression of kd alleles of specific PIKKs might result in interference with other PIKK-dependent pathways. Here, overexpression of kd-ATR was optimized to block phosphorylation of Chk1. It is conceivable that a different overexpression protocol (Kaygun and Marzluff, 2005) may result in interference not only with ATR-mediated events but functions associated with other PIKKs also. Importantly, the data presented here do not exclude a role for ATR signaling in the regulation of histone mRNA decay but indicate that, in the cell types analyzed here, ATR is not functionally limiting for this process.
These data together with other published observations (Kaygun and Marzluff, 2005) strongly implicate PIKKs in an S phase checkpoint controlling the efficiency of histone mRNA decay in response to replication stress. Screening experiments using a broad range of small molecule inhibitors indicate that complete uncoupling of histone mRNA decay from replication stress may be achieved, and preliminary data indicate that active compounds do not directly target known checkpoint components (unpublished data). As suggested previously (Brown and Baltimore, 2003), cells lacking known checkpoint signaling function nonetheless may develop residual checkpoint responses for critical cellular pathways that may well involve further key signaling molecules in addition to ATR and DNA-PK. The linkage between replication and histone decay may be such a critical pathway.
UPF1 is predominantly associated with polyribosomes when hyperphosphorylated by hSMG1 (Yamashita et al., 2001) and histone mRNA decay is believed to be a cytoplasmic event, which suggests that checkpoint activation of histone mRNA decay might involve relaying a checkpoint signal emanating from arrested replication forks into the cytoplasm. Unlike their upstream activators (DNA-PK, ATR, and ATM), whose activation is dependent on their physical association with the originating DNA lesion, Chk1 and 2 are believed to relay checkpoint signals to cellular targets removed from the site of damage. However, in contrast to multiple other checkpoint signaling pathways, blocking Chk1 and 2 signaling had no effect on histone mRNA decay during replication stress.
Hyperphosphorylated UPF1 has, however, been found associated with chromatin, and this association increases in response to DNA damage (Azzalin and Lingner, 2006). Additionally, cells overexpressing HBP/SLBP show elevated levels of associated UPF after replication stress (Kaygun and Marzluff, 2005). Conceivably, direct phosphorylation of these components by PIKK family members such as ATR and DNA-PK may play a role in coordinating DNA replication and histone mRNA levels. Consistent with this notion, S/TQ phosphorylation increased in UPF1 though not HBP/SLBP during replication stress. These data together with previously published results (Kaygun and Marzluff, 2005) suggest a model in which elevated phosphorylation of UPF1 mediated by replication stress–induced PIKKs results in increased affinity of HBP/SLBP for UPF1 (either directly or indirectly), which in turn promotes the efficient destabilization of histone mRNA. Whatever the molecular details underpinning the coordination of replication with histone mRNA levels, it is likely that, in addition to UPF1 and PIKKs, other components are required to transduce signals emanating from DNA lesions in the nucleus to decay machinery residing in the cytoplasm. Future work will focus on establishing how this may be achieved.
Materials and methods
Cell lines and culture conditions
HeLa (Feijoo et al., 2001), ataxia telangiectasia (AT) fibroblasts (Ziv et al., 1997), DLD-1 (Bhattacharyya et al., 1995), and U2OS cells (Nghiem et al., 2001) were grown as described previously. Induction of ATR expression in U2OS cells was done by incubation with 1.5 μg/ml doxycycline for 3 d. DNA-PK–deficient (M059J) and –proficient cells (M059K) were grown in DME or F-12/DME medium supplemented with 10% (vol/vol) FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Unless stated otherwise, experiments were performed with asynchronous cells. Synchronized cells were arrested in metaphase by adding 40 ng/ml nocodazole (EMD) for 12–14 h. Mitotic cells were harvested by shake-off, collected by centrifugation, washed three times with PBS, and released into fresh medium.
Drug treatment
Caffeine (Sigma-Aldrich) was used, unless stated otherwise, at a final concentration of 5 mM; HU (Sigma-Aldrich) was used at 2 mM; APH (Sigma-Aldrich) at 50 μg/ml; and UCN-01 (National Cancer Institute) at 300 nM. Wortmannin and LY294002 (EMD) were used at the indicated concentrations. HU and APH were used in different experiments to induce replication stress. No differences were observed in the timing or magnitude of checkpoint responses in response to either agent.
RNA analysis
Total RNA was prepared using Trizol (Invitrogen) and analyzed by Northern blotting as described previously (Zhao et al., 2004). Human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA for probing Northern blots spanned nucleotides 2,041–3,239 of the open reading and was PCR amplified from genomic DNA. RNAs were visualized by autoradiography or with a phosphorimager (FLA3000; Fujifilm). The Aida 2.0 software 2D quantification tool (Raytest GmbH) was used for quantitation.
Cell lysates, immunoprecipitation, blotting, and kinase assays
Cell lysates for Chk1 and 2 activity, immunoprecipitation, phosphatase treatment, SDS-PAGE, and blotting were performed as described previously (Feijoo et al., 2001). Chk1 and 2 antibodies were generated as described previously (Feijoo et al., 2001) and used at 1 μg/ml. Rabbit polyclonal anti–phospho-Chk1 (Ser345), anti–phospho-Chk2 (Thr68), anti–phospho- S/TQ (Cell Signaling Technology), and rabbit anti-ATR (EMD) were used at 1:2,000. Monoclonal anti-actin (Sigma-Aldrich) and anti-nucleolin (Santa Cruz Biotechnology, Inc.) were used at 1:1,000. HRP-conjugated anti–rabbit (Sigma-Aldrich) and anti–mouse antibodies (Sigma-Aldrich) were used at 1:5,000. Rabbit anti-HBP (Zhao et al., 2004), rabbit anti-UPF1 (provided by N. Gehring [University of Heidelberg, Heidelberg, Germany] and M. Hentze [European Molecular Biology Laboratory, Heidelberg, Germany]), and goat anti-UPF1 (Bethyl Laboratories, Inc.) were used as described previously (Feijoo et al., 2001). AP was used at 10 U/μl for 1 h.
RNAi
RNAi was performed using ATR siRNA (target sequence AACCTCCGTGATGTTGCTTGA), Chk1 siRNA (target sequence AAGAAGCAGTCGCAGTGAAGA), control siRNA 1 targeting luciferase, and control siRNA 2 targeting an unrelated gene TAPP1 (target sequence GGTCAAGCCAGGGAACTTC) from Thermo Fisher Scientific. Cells were transfected with siRNA using oligofectamine (Invitrogen) according to the manufacturer's instructions. 48 h after transfection (unless otherwise stated), cells were treated with HU for the indicated times and RNA or protein samples were prepared.
Assay to monitor S phase progression
Asynchronous HeLa cells were pulsed with 30 μM CldU for 20 min, washed with PBS, and subsequently incubated for increasing lengths of time (typically 6, 12, or 16 h) either in the absence of drugs (mock treatment), in the presence of 50 μg/ml APH alone, or in the presence of both APH and 5 mM caffeine. Cells were then washed free of drugs with PBS and pulsed with 30 μM IdU for 20 min. Differential staining of DNA sites substituted with halogenated derivatives of dU was performed essentially as described previously (Feijoo et al., 2001) and >200 cells were scored for each data point. Images were acquired on a laser scanning confocal microscope (TCS SP1; Leica) using a Plan Apochromat 100× NA 1.4 oil-immersion objective (HCX). FITC and Texas red fluorescence was generated using the 488-nm line of the Ar laser and the 568-nm line of the Kr laser, respectively. Images were acquired using confocal software 2.61 (Leica) and assembled using Photoshop 7.0.1 (Adobe).
Online supplemental material
Fig. S1 shows that histone mRNA decay occurs normally in AT cells treated with ATR siRNA. Fig. S2 shows the effects of caffeine and LY294002 on camptothecin-induced histone mRNA decay. Fig. S3 shows that inhibition of Chk1 and 2 kinases using the broad-spectrum checkpoint kinase inhibitor debromohymenialdisine does not affect histone mRNA decay. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708106/DC1.
Supplemental Material
Acknowledgments
We are grateful to N. Gehring and M. Hentze for providing us with anti-UPF1 antibodies and to D. Sutton for excellent technical support.
Confocal microscopy was undertaken in the Sheffield Light Microscopy Facility funded by the Wellcome Trust (grant GR077544AIA). This paper was supported by funding from Cancer Research UK (grant C10338/A3855 to C. Smythe), Association for International Cancer Research (grant 99-2241 to C. Smythe), the Wellcome Trust (grant 076220 to B. Müller), and Tenovus Scotland (grant G03/9 to B. Müller).
Abbreviations used in this paper: APH, aphidicolin; AT, ataxia telangiectasia; ATM, AT mutated; ATR, AT and Rad3 related; CldU, chlorodeoxyuridine; DNA-PK, DNA-activated protein kinase; dU, deoxyuridine; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HBP, hairpin-binding protein; HU, hydroxyurea; IdU, iododeoxyuridine; kd, kinase dead; NHEJ, nonhomologous end joining; PIKK, phosphatidylinositol 3-kinase–like kinase; SLBP, stem-loop binding protein; wt, wild type.
References
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177–2196. [DOI] [PubMed] [Google Scholar]
- Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235–1245. [DOI] [PubMed] [Google Scholar]
- Azzalin, C.M., and J. Lingner. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16:433–439. [DOI] [PubMed] [Google Scholar]
- Baumbach, L.L., G.S. Stein, and J.L. Stein. 1987. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 26:6178–6187. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, N.P., A. Ganesh, G. Phear, B. Richards, A. Skandalis, and M. Meuth. 1995. Molecular analysis of mutations in mutator colorectal carcinoma cell lines. Hum. Mol. Genet. 4:2057–2064. [DOI] [PubMed] [Google Scholar]
- Blasina, A., B.D. Price, G.A. Turenne, and C.H. McGowan. 1999. Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 9:1135–1138. [DOI] [PubMed] [Google Scholar]
- Brown, E.J., and D. Baltimore. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper, A.M., P. Nghiem, M.F. Arlt, and T.W. Glover. 2002. ATR regulates fragile site stability. Cell. 111:779–789. [DOI] [PubMed] [Google Scholar]
- Curman, D., B. Cinel, D.E. Williams, N. Rundle, W.D. Block, A.A. Goodarzi, J.R. Hutchins, P.R. Clarke, B.B. Zhou, S.P. Lees-Miller, et al. 2001. Inhibition of the G2 DNA damage checkpoint and of protein kinases Chk1 and Chk2 by the marine sponge alkaloid debromohymenialdisine. J. Biol. Chem. 276:17914–17919. [DOI] [PubMed] [Google Scholar]
- DeLisle, A.J., R.A. Graves, W.F. Marzluff, and L.F. Johnson. 1983. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol. Cell. Biol. 3:1920–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, D.S., and D.M. Gilbert. 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili, A., D.M. Schieltz, J.R. Yates III, and L.H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 7:13–20. [DOI] [PubMed] [Google Scholar]
- Feijoo, C., C. Hall-Jackson, R. Wu, D. Jenkins, J. Leitch, D.M. Gilbert, and C. Smythe. 2001. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra–S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, R.A., N.B. Pandey, N. Chodchoy, and W.F. Marzluff. 1987. Translation is required for regulation of histone mRNA degradation. Cell. 48:615–626. [DOI] [PubMed] [Google Scholar]
- Gunjan, A., and A. Verreault. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 115:537–549. [DOI] [PubMed] [Google Scholar]
- Hall-Jackson, C.A., D.A. Cross, N. Morrice, and C. Smythe. 1999. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 18:6707–6713. [DOI] [PubMed] [Google Scholar]
- Han, M., M. Chang, U.J. Kim, and M. Grunstein. 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 48:589–597. [DOI] [PubMed] [Google Scholar]
- Heintz, N., H.L. Sive, and R.G. Roeder. 1983. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 3:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.A. 1995. S-phase progression in synchronized human cells. Exp. Cell Res. 220:62–70. [DOI] [PubMed] [Google Scholar]
- Kaygun, H., and W.F. Marzluff. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12:794–800. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K., L. Furic, L. Desgroseillers, and L.E. Maquat. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 120:195–208. [DOI] [PubMed] [Google Scholar]
- Levine, B.J., N. Chodchoy, W.F. Marzluff, and A.I. Skoultchi. 1987. Coupling of replication type histone mRNA levels to DNA synthesis requires stem-loop sequences at the 3′ end of the mRNA. Proc. Natl. Acad. Sci. USA. 84:6189–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin, C., K. Erixon, C. Arnaudeau, N. Schultz, D. Jenssen, M. Meuth, and T. Helleday. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 22:5869–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F., A. Schaller, S. Eglite, D. Schumperli, and B. Müller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W.F., and R.J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14:692–699. [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner, D., and L.H. Hartwell. 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 44:43–52. [DOI] [PubMed] [Google Scholar]
- Nghiem, P., P.K. Park, Y. Kim, C. Vaziri, and S.L. Schreiber. 2001. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. USA. 98:9092–9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, R.T., S.C. Henderson, and D.L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 116:1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, T., A.J. Dwyer, Y. Ziv, D.W. Chan, S.P. Lees-Miller, R.H. Abraham, J.H. Lai, D. Hill, Y. Shiloh, L.C. Cantley, and G.A. Rathbun. 2000. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J. Biol. Chem. 275:22719–22727. [DOI] [PubMed] [Google Scholar]
- Painter, R.B., and B.R. Young. 1980. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc. Natl. Acad. Sci. USA. 77:7315–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, P.N., and R.T. Johnson. 1970. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 225:159–164. [DOI] [PubMed] [Google Scholar]
- Rodriguez, R., and M. Meuth. 2006. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol. Biol. Cell. 17:402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria, J.N., E.C. Busby, R.S. Tibbetts, P. Roos, Y. Taya, L.M. Karnitz, and R.T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375–4382. [PubMed] [Google Scholar]
- Schild-Poulter, C., A. Shih, N.C. Yarymowich, and R.J.G. Hache. 2003. Down-regulation of histone H2B by DNA-dependent protein kinase in response to DNA damage through modulation of octamer transcription factor 1. Cancer Res. 63:7197–7205. [PubMed] [Google Scholar]
- Schumperli, D. 1986. Cell-cycle regulation of histone gene expression. Cell. 45:471–472. [DOI] [PubMed] [Google Scholar]
- Shao, R.G., C.X. Cao, H. Zhang, K.W. Kohn, M.S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J.A., G. Rizki, and P.D. Kaufman. 2005. Regulation of histone deposition proteins Asf1/Hir1 by multiple DNA damage checkpoint kinases in Saccharomyces cerevisiae. Genetics. 171:885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, C.S., L.T. Hansen, J. Dziegielewski, R.G. Syljuasen, C. Lundin, J. Bartek, and T. Helleday. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7:195–201. [DOI] [PubMed] [Google Scholar]
- Stiff, T., M. O'Driscoll, N. Rief, K. Iwabuchi, M. Lobrich, and P.A. Jeggo. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- Wang, Z.F., M.L. Whitfield, T.C. Ingledue III, Z. Dominski, and W.F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028–3040. [DOI] [PubMed] [Google Scholar]
- Wyrick, J.J., F.C. Holstege, E.G. Jennings, H.C. Causton, D. Shore, M. Grunstein, E.S. Lander, and R.A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 402:418–421. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., T. Ohnishi, I. Kashima, Y. Taya, and S. Ohno. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15:2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos, G., M.D. Rainey, and D.A. Gillespie. 2005. Chk1-dependent s-m checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol. Cell. Biol. 25:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., S. McKillop-Smith, and B. Müller. 2004. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 117:6043–6051. [DOI] [PubMed] [Google Scholar]
- Zheng, L., Z. Dominski, X.C. Yang, P. Elms, C.S. Raska, C.H. Borchers, and W.F. Marzluff. 2003. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 23:1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.B., and S.J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature. 408:433–439. [DOI] [PubMed] [Google Scholar]
- Ziv, Y., A. Bar-Shira, I. Pecker, P. Russell, T.J. Jorgensen, I. Tsarfati, and Y. Shiloh. 1997. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 15:159–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.