Abstract
Posttranslational modification through palmitoylation regulates protein localization and function. In this study, we identify a role for the Drosophila melanogaster palmitoyl transferase Huntingtin-interacting protein 14 (HIP14) in neurotransmitter release. hip14 mutants show exocytic defects at low frequency stimulation and a nearly complete loss of synaptic transmission at higher temperature. Interestingly, two exocytic components known to be palmitoylated, cysteine string protein (CSP) and SNAP25, are severely mislocalized at hip14 mutant synapses. Complementary DNA rescue and localization experiments indicate that HIP14 is required solely in the nervous system and is essential for presynaptic function. Biochemical studies indicate that HIP14 palmitoylates CSP and that CSP is not palmitoylated in hip14 mutants. Furthermore, the hip14 exocytic defects can be suppressed by targeting CSP to synaptic vesicles using a chimeric protein approach. Our data indicate that HIP14 controls neurotransmitter release by regulating the trafficking of CSP to synapses.
Introduction
The precise localization of proteins to intracellular and plasma membrane domains is important for their proper function. Some intracellular proteins that play roles at the plasma membrane are modified by the addition of long-chain fatty acids that mediate protein targeting to the membrane (Dunphy and Linder, 1998). For example, the cotranslational attachment of myristic acid to an N-terminal glycine or the addition of fatty acids such as palmitate by thioester linkage to cysteine residues target proteins to specific subcellular compartments (Smotrys and Linder, 2004).
Palmitate is a 16-carbon saturated fatty acid that is attached to proteins posttranslationally. This modification increases the hydrophobicity of the protein, thereby facilitating interactions with lipid bilayers. In neurons, at least 32 proteins have been shown to be palmitoylated. These include channels, cell adhesion molecules, and signaling proteins as well as the presynaptic proteins cysteine string protein (CSP), synaptotagmin I (Syt I), SNAP25, neuronal synaptobrevin (n-Syb), and Huntingtin (Htt; el-Husseini Ael and Bredt, 2002; Smotrys and Linder, 2004). Unlike myristoylation and isoprenylation, which are permanent modifications, the palmitoylation of proteins is regulated via a cycle of enzymes that add and remove palmitate (Smotrys and Linder, 2004). The reversibility of palmitoylation of neuronal proteins is likely to be an important regulatory event, as patients with mutations in palmitoyl-protein thioesterase 1 (PPT1), which removes palmitate, exhibit a debilitating and early onset neuronal degeneration leading to childhood death (Vesa et al., 1995).
Recently, palmitoyl transferases (PATs) were discovered in Saccharomyces cerevisiae (Lobo et al., 2002; Roth et al., 2002). The defining feature of this family is the presence of a cysteine-rich domain (CRD) with an Asp-His-His-Cys (DHHC) motif, and it is required for PAT activity both in vitro and in vivo. Proteins with DHHC-CRD are conserved from yeast to mammals. Genetic and biochemical studies have identified substrates for several of the seven DHHC proteins in S. cerevisiae (Lobo et al., 2002; Roth et al., 2002), and proteomic analyses have expanded the repertoire of substrates for this family of enzymes significantly (Roth et al., 2006). Although the number of DHHC proteins in yeast is limited, flies contain 20 and mammals 23 DHHC proteins. Their tissue distribution and subcellular localization has been recently documented (Ohno et al., 2006). However, the in vivo role of these PATs in neurons has not been established.
In this study, we report a novel genetic screen that allowed us to identify mutations that affect neurotransmitter release. This screen led to the identification of Drosophila melanogaster htt-interacting protein 14 (hip14), a protein that has not been implicated previously in synaptic transmission but has been shown to palmitoylate Htt in vertebrates (Yanai et al., 2006). The synaptic defects associated with the loss of hip14 show striking similarities with the loss of csp (Zinsmaier et al., 1994). In addition, CSP as well as SNAP25 are mislocalized in hip14 mutants, and palmitoylation of CSP is critical for its synaptic localization. In contrast, Syt I and postsynaptic density 95 (PSD-95)/Discs large (DLG), which can be modified by HIP14 in vitro (Huang et al., 2004), are localized properly in hip14 mutants. Based on biochemical data and functional rescue assays, our data not only point to the importance of palmitoylation in synaptic function but also reveal CSP as a novel and one of the main targets for HIP14 in vivo.
Results
Isolation of mutations in complementation group 3L1
To identify novel proteins that affect synaptic transmission, we performed a forward genetic screen on chromosome arm 3L using the ey-FLP system (Stowers and Schwarz, 1999; Newsome et al., 2000). In previous 2L, 2R, and 3R chromosome arm screens, we selected flies in the F1 generation with aberrant phototactic properties, and, of the progeny, we isolated mutants with abnormal electroretinograms (ERGs), thereby measuring the ability of photoreceptors (PRs) to activate postsynaptic cells (Verstreken et al., 2003, 2005; Mehta et al., 2005). Although neurotransmission mutants isolated from prior ey-FLP screens initially displayed reduced phototaxis in the F1 generation, many behaved normally in subsequent generations despite retaining their ERG defects (Fabian-Fine et al., 2003; Verstreken et al., 2003, 2005; Koh et al., 2004). Therefore, we modified the screening strategy and omitted the phototaxis assay. Thus, F1 flies were directly subjected to ERG recordings. Of the 49,017 screened F1 flies, 1,077 were crossed and rescreened in the F2 generation. 77 were eventually retained as mutants with abnormal ERGs. Complementation tests with mutants previously shown to affect synaptic transmission identified two new alleles of csp (Zinsmaier et al., 1994) and one new allele of SNAP (Ordway et al., 1994), demonstrating the specificity of the strategy. Upon further complementation analysis, we identified eight novel complementation groups with two or more alleles. Here, we describe 3L1, a complementation group with three alleles: 3L11, 3L12, and 3L13. As shown in Fig. 1 A, all 3L1 mutants show lack of on-off transients but exhibit normal depolarization. By isolating mutants with these ERG defects, we and others have been able to identify genes that affect synaptic function or development (Zinsmaier et al., 1994; Stowers et al., 2002; Verstreken et al., 2003).
Figure 1.
Characterization of the ey-FLP screen phenotypes of 3L1 mutants. (A) ERGs of controls (y w ey-FLP GMR-lacZ; FRT80B/RpS174 P{w + } FRT80B) and 3L1 mutants (y w ey-FLP GMR-lacZ; 3L11, 3L12, or 3L13 FRT80B/RpS174 P{w + } FRT80B). The positions of on and off transients are indicated by gray arrowheads. (B and C) Confocal images showing control (y w ey-FLP GMR-lacZ; FRT80B/RpS174 P{w + } FRT80B) and 3L1 mutant (y w ey-FLP GMR-lacZ; 3L12 FRT80B/RpS174 P{w + } FRT80B) adult brains stained with mAb 24B10. (D and E) EM of control (D) and 3L12 mutant (E) PR terminals in the lamina (for genotypes, see A). Some of the capitate projections (cp) and mitochondria (m) are indicated. (F) Quantification of capitate projection distribution in controls and 3L11, 3L12, and 3L13 mutants. Error bars represent SEM. Bars: (B and C) 50 μm; (D and E) 1 μm.
The ERG phenotype suggests that the PRs fail to properly transmit signals to postsynaptic neurons. This may be caused by (1) functional impairment of the neurotransmission machinery or (2) aberrant synapse formation. To test whether developmental defects are present at the light microscopy level, we first stained adult brains with the PR-specific antibody against chaoptin, mAb 24B10 (Fujita et al., 1982). The Drosophila compound eye consists of ∼800 units, each comprised of eight PR cells that project into the lamina (R1–R6) or into two separate layers in the medulla (R7–R8; Fig. 1 B). In control flies, mAb 24B10 staining reveals a highly regular array of terminals in the medulla (Fig. 1 B). Similarly, 3L1 mutant PRs display a regular array of terminals in the medulla and no obvious defects in axonal targeting (Fig. 1 C). To determine whether R1–R6 PRs display defects in synapse formation, we also analyzed synaptic terminals in control and 3L1 mutant laminae by transmission EM (TEM). In controls, R1–R6 terminals are organized into units called cartridges, with each cartridge containing six PR terminals surrounding the postsynaptic lamina monopolar cell bodies (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200710061/DC1). Notably, the cartridge organization is preserved in the mutants (Fig. S1), indicating no obvious axonal sorting or targeting defects in the laminae. Thus, the ERG defects are likely caused by aberrant synaptic transmission rather than morphological disturbances.
TEM of PR synapses in the lamina also allows us to assess some ultrastructural features, including the number of mitochondria, number of active zones, and size, number, and shape of glial cell invaginations or capitate projections. The capitate projections are thought to be centers of endocytosis in PR terminals (Fabian-Fine et al., 2003). Interestingly, as shown in Fig. 1 (D–F; and Fig. S1), capitate projections are often shallow in mutants compared with controls, suggesting that mutations in 3L1 may affect synaptic function in the visual system.
3L1 encodes Drosophila hip14
To identify the 3L1 gene, we mapped the lesions in the mutants using P-element meiotic recombination (Zhai et al., 2003). Rough mapping placed 3L1 in the 72A–C cytological interval and showed that the mutations fail to complement Df(3L)brm11 (Fig. 2 A). Meiotic fine mapping mapped 3L1 between KG00222 and EY12630. Sequencing of CG6017 showed that 3L11 contains a Cys452Tyr mutation, 3L12 contains a 26-bp deletion leading to a premature stop codon at position 312, and 3L13 has a premature stop codon at position 432 (Fig. 2 B). CG6017 encodes a homologue of the human hip14 or DHHC17 and yeast Akr1p (ankyrin repeat-containing protein; Singaraja et al., 2002). Therefore, we renamed 3L11 as hip141, 3L12 as hip142, and 3L13 as hip143. Because Df(3L)brm11 is the smallest deletion that removes CG6017, we tested the three hip14 alleles in trans to the deficiency to determine the severity of these mutations. All trans-heterozygous combinations die as pharate adults. The notion that all alleles are similar in severity over a deficiency suggests that hip141, hip142, and hip143 are all either null or severe hypomorphic alleles (Fig. 2 C). However, hip142 and hip143 homozygotes die as third instar larvae, indicating that the chromosomes carrying these alleles may carry other lethal mutations.
Figure 2.
Identification of 3L1. (A) P-element fine mapping. Numbers separated by a backslash indicate recombinants out of total flies scored. Recombination distance in centiMorgans for the two closest P elements is indicated. A deficiency that complements (green) and one that fails to complement (red) are shown. The calculated mapping location based on P-element mapping of 3L1 is shown as a red star, and the coding regions of nearby genes (black boxes) are shown. The region used for rescue is indicated by the blue bar. (B) Intron-exon structure of CG6017 and protein structure of HIP14. The start codon is marked in green, and the stop codon in indicated in red. The position of the N- or C-termEGFP of the genomic rescue constructs are indicated by the green inserts. HIP14 contains five ankyrin repeats (green boxes), five TMDs (yellow boxes), and a DHHC-CRD domain (blue box). The molecular nature of the three hip14 alleles is indicated. (C) Lethal stage of hip14 mutant combinations. (D) Lethality rescue of hip14 mutant combinations by genomic and cDNA constructs expressed using elav-GAL4. Columns are divided such that the yellow portion indicates the result from the genomic construct, and white indicates the result from the cDNA. (C and D) L3, animals do not survive beyond the third instar larval stage; P, death during late pupal stage; ND, not tested; R, rescued lethality.
To determine whether the lethality and functional defects of the hip14 alleles can be rescued by CG6017, we generated a genomic rescue construct (Fig. 2 A). This construct rescues the lethality and ERG phenotypes associated with all trans-heterozygous mutants tested as well as hip141 homozygous animals (Fig. 2 D). Together, these results provide compelling evidence that lesions in hip14 are solely responsible for the defects observed in 3L1 alleles.
HIP14 is conserved from yeast to humans over the entire length of the protein, sharing 50.8% identity with its human homologue and 31.3% identity with the yeast protein (Singaraja et al., 2002). Drosophila hip14 encodes a 71-kD protein with five ankyrin repeats, a DHHC-CRD typically found in PATs, and five transmembrane domains (TMDs), suggesting it is an integral membrane protein (Singaraja et al., 2002; Huang et al., 2004). Recently, a study has shown that the DHHC family of proteins functions as PATs, including the yeast homologue of hip14, Akr1p (Roth et al., 2002). In Drosophila, at least 20 genes share a common 50-residue zinc finger–like sequence, which contains the DHHC-CRD. No analyses of mutants of any of these proteins in nematodes, flies, or mice have been reported.
HIP14 is localized pre- and postsynaptically
Both mouse and human HIP14 are strongly expressed in the brain and heart, and immuno-EM studies suggest that HIP14 localizes to the Golgi as well as to diverse vesicular structures present in the soma, axon, and dendrites of neurons (Singaraja et al., 2002; Huang et al., 2004). To determine the expression pattern and subcellular localization of HIP14 in Drosophila, we generated N- and C-terminal GFP-tagged genomic constructs (Fig. 2 B), as we failed to generate a specific antibody. Both GFP-HIP14 constructs rescue the lethality defects of hip14 mutants, suggesting that GFP-HIP14 is functional.
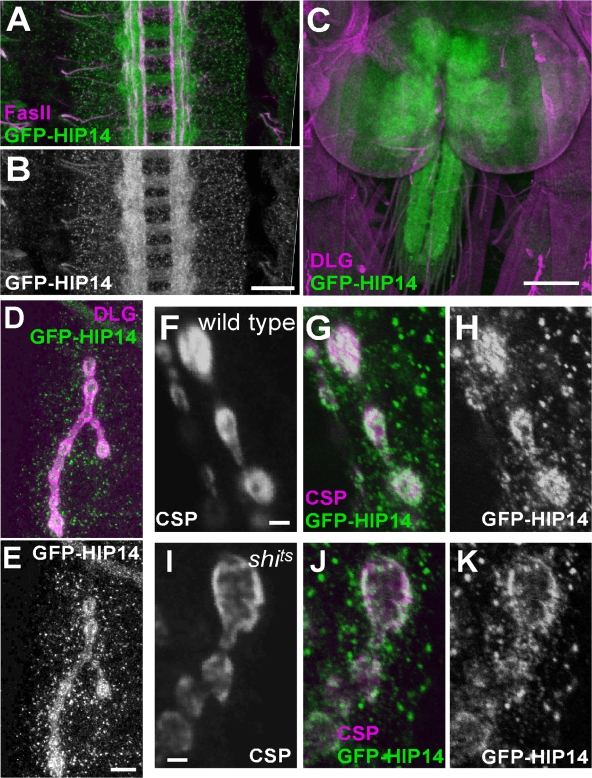
In stage 13–15 embryos, GFP-HIP14 is observed in the central nervous system (CNS) neuropil (Fig. 3, A and B). At the third instar larval stage, GFP-HIP14 is expressed in the ventral nerve cord (VNC) and is enriched in the neuropil (Fig. 3 C and Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200710061/DC1). At the neuromuscular junction (NMJ), GFP-HIP14 is localized to the pre- and postsynaptic regions (Figs. 3, D and E; and S2 B) and colocalizes extensively with presynaptic markers such as CSP (Fig. 3, F–H) and n-Syb (Fig. S2 C), suggesting that HIP14 is a presynaptic protein. Interestingly, similar to synaptic vesicle (SV)–associated proteins, HIP14 relocalizes to the presynaptic membrane upon SV depletion in stimulated shibirets1 mutants at the restrictive temperature (Fig. 3, I–K). These data indicate that HIP14 localization at the synapse is dynamic. Combined with the fact that HIP14 harbors five TMDs, the data suggest, but do not demonstrate, that HIP14 is a vesicle and presynaptic membrane–associated protein. Also, note that punctate GFP-HIP14 signals are also present in muscles (Fig. 3, D and E). However, GFP-HIP14 fails to colocalize with the Golgi marker Lava Lamp (Papoulas et al., 2005) in muscles, indicating that these punctae are not associated with Golgi structures. In summary, our analysis of the expression pattern of GFP-HIP14 indicates that HIP14 is enriched in the nervous system throughout development but is also present in other tissues.
Figure 3.
Localization of HIP14. Confocal images from genomic GFP-tagged hip14 transgenic animals. (A and B) Stage 14 embryo (y w; P{w + NtermGFP-hip14 + }) with GFP-HIP14 (green) and Fasciclin II (magenta). The GFP-HIP14 (green) channel is shown separately in B. (C) Third instar larval (y w; P{w + NtermGFP-hip14 + }) CNS with GFP-HIP14 (green) and DLG (magenta). (D and E) Third instar larvae NMJ boutons in genomic GFP-tagged hip14 transgenic animals (y w; P{w + NtermGFP-hip14 + }) with GFP-HIP14 (green) and DLG (magenta) to label the synaptic areas. The green channel is shown separately in E. (F–H) Third instar larval NMJ boutons in wild type (y w, shibirets/+; P{w + NtermGFP-hip14 + }/+) with GFP-HIP14 (green) and CSP (magenta) at ambient temperature. Both channels are separately shown in F and H. Images are single confocal sections. (I–K) Third instar larval NMJ boutons in shibirets mutant (y w, shibirets/Y; P{w + NtermGFP-hip14 + }/+) at restrictive temperature or 32°C with GFP-HIP14 (green) and CSP (magenta). Both channels are separately shown in I and K. Images are single confocal sections. Bars: (A and B) 20 μm; (C) 100 μm; (D and E) 10 μm; (F–K) 2 μm.
NMJs develop normally in hip14 mutants
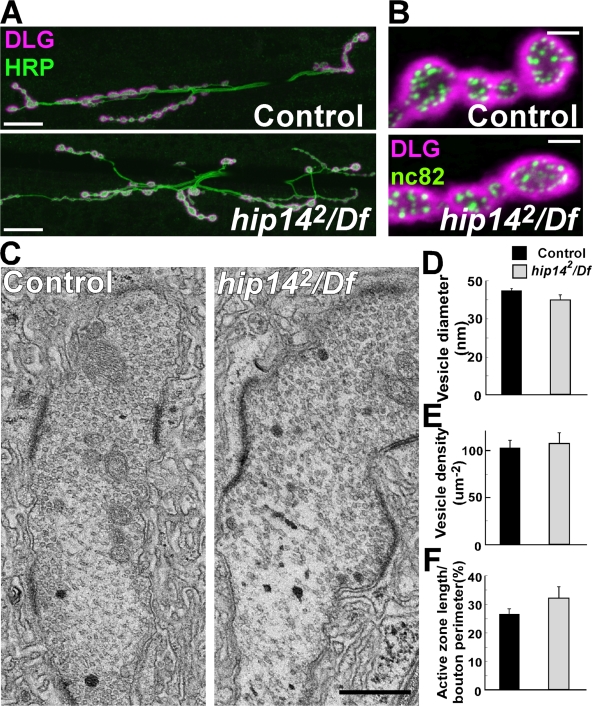
Enhancer screens for genes involved in axon guidance and/or synaptogenesis at the Drosophila NMJ identified CG6017/hip14 (Kraut et al., 2001), suggesting that hip14 may play a role in growth cone guidance and/or synaptogenesis. However, our analysis of the hip14 PR projections failed to uncover any obvious morphological defects (Figs. 1, B and C; and S1). To test whether this is also the case at NMJs, we examined their morphology with pre- and postsynaptic markers, including DLG/PSD-95 (Lahey et al., 1994), the presynaptic membrane marker HRP (Jan and Jan, 1982), and Bruchpilot, an active zone component (Kittel et al., 2006). However, similar to the PRs, we did not observe aberrant morphological features at the NMJs with any of these markers in hip14 mutants (Fig. 4, A and B).
Figure 4.
NMJ morphology is normal in hip14 mutants. (A) Confocal images of the third instar larval NMJ on muscles 6 and 7 labeled with anti-HRP (green) to mark all neuronal membranes and DLG (magenta) to label the pre- and postsynaptic regions in control (y w; FRT80B) and hip142/Df mutants (y w ey-FLP GMR-lacZ; hip142 FRT80B/Df(3L)brm11). (B) mAb nc82 (Bruchpilot) and DLG labeling of boutons of control (y w; FRT80B) and hip142/Df mutants (y w ey-FLP GMR-lacZ; hip142 FRT80B/Df(3L)brm11). (C) Ultrastructure of NMJ boutons in control (y w; FRT80B) and hip142/Df mutants (y w ey-FLP GMR-lacZ; hip142 FRT80B/Df(3L)brm11). (D–F) Quantification of synaptic features. Error bars represent SD. Bars: (A) 20 μm; (B) 2 μm; (C) 0.5 μm.
To determine whether there are ultrastructural defects, we performed TEM experiments of third instar NMJ boutons. We find that the mutants exhibit normal SV morphology and density, normal T-bar morphology, active zone length, and structure of the subsynaptic reticulum when compared with control animals (Fig. 4, C–F). These data indicate that similar to lamina synapses, NMJ synapses form properly in hip14 mutants and that hip14 does not contribute to axon guidance or synaptogenesis in Drosophila. The data also suggest that there is no defect in endocytosis, as many Drosophila endocytic mutants analyzed to date exhibit a reduced number of vesicles and/or aberrantly sized vesicles (Zhang et al., 1998; Guichet et al., 2002; Verstreken et al., 2003; Koh et al., 2004).
HIP14 is required for proper SV exocytosis
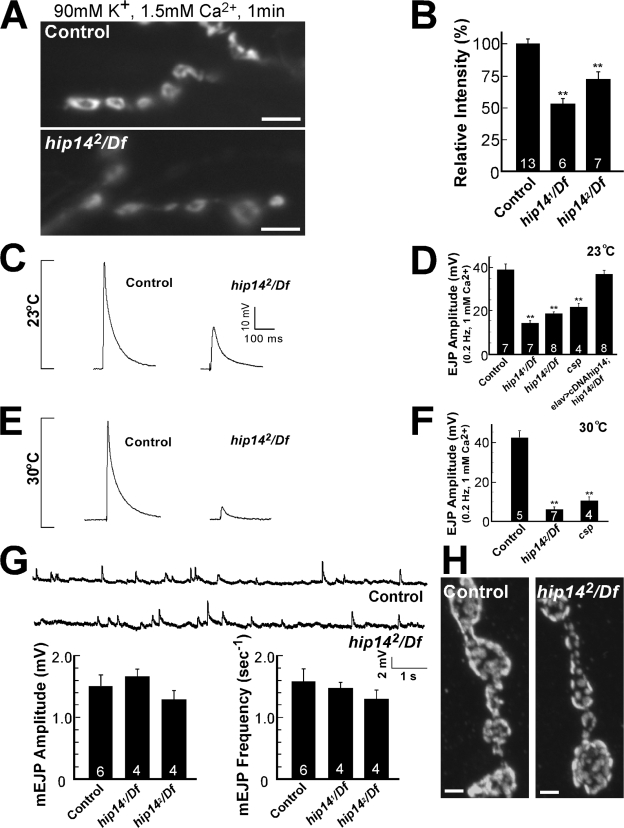
To assess the synaptic defects in hip14 mutants, we used the third instar NMJ. To determine whether HIP14 regulates SV cycling at the synapse, we performed live imaging of vesicle cycling with FM 1-43 (Ramaswami et al., 1994). FM 1-43 is nonfluorescent in aqueous environments, but, when bound to membranes, its fluorescence quantum yield increases. Thus, newly endocytosed vesicles in the presence of FM 1-43 are fluorescently labeled by the dye, providing a quantitative measure of vesicle cycling. As shown in Fig. 5 A, when controls are stimulated for 1 min with 90 mM K+ in the presence of FM 1-43, synapses are brightly labeled, indicating efficient vesicle retrieval from the membrane during stimulation. However, hip14 mutants take up less dye (∼50–70% of control; Fig. 5 B), suggesting a defect in vesicle cycling. Unlike many endocytic mutants analyzed in Drosophila (Verstreken et al., 2003; Koh et al., 2004), EM analysis of hip14 mutants did not show a reduction in SV density (Fig. 4 C), suggesting that the reduced FM 1-43 uptake is not likely to be caused by a defect in endocytosis.
Figure 5.
Evoked neurotransmitter release is impaired in hip14 mutants. (A and B) FM 1-43 dye uptake on controls (y w; FRT80B), hip141/Df mutants (y w ey-FLP GMR-lacZ; hip141 FRT80B/Df(3L)brm11), and hip142/Df mutants (y w ey-FLP GMR-lacZ; hip142 FRT80B/Df(3L)brm11). (A) Preparations were stimulated for 1 min in the presence of 4 μM of dye, 90 mM KCl, and 1.5 mM Ca2+ to label the exo-endo cycling pool. (B) Quantification of the labeling intensity of FM 1-43 shown in A. (C and E) Sample EJPs recorded in 1 mM of external Ca2+ at 0.2 Hz in controls and hip142/Df. Bath temperature was kept at 23 (C) or 30°C (E). (D and F) Quantification of EJP amplitudes recorded at 23°C are shown in D for all genotypes, including csp mutants (cspu1/cspx1) and cDNA rescue by presynaptic expression (elav-GAL4/+; UAS-cDNAhip14/+; hip142/Df(3L)brm11). Quantification for controls, hip142/Df, and csp mutants at 30°C is shown in F. Recordings were performed for 1 min, and 12 EJP amplitudes were averaged per recording. (G) Frequency and amplitude of miniature synaptic currents in hip14 mutants. Miniature EJPs were recorded in the presence of 5 μM tetrodotoxin in 0.5 mM Ca2+. (H) GluRIII immunostaining at NMJs of control and hip142/Df animals. (B, D, F, and G) The number of animals tested is indicated in the bars. Error bars represent SEM. **, P < 0.01 (t test). Bars: (A) 5 μm; (H) 2 μm.
To determine whether exocytosis is impaired at hip14 synapses, we stimulated mutant and control motor nerves at 0.2 Hz in 1 mM Ca2+ and recorded excitatory junctional potentials (EJPs) from the muscle. The EJP amplitudes in hip14 mutants during low frequency stimulation are ∼33–40% of controls (wild type, 39.4 ± 2.4 mV; hip141, 13.2 ± 1.4 mV; hip142, 18.9 ± 0.6 mV) at 23°C (Fig. 5, C and D), indicating that these mutants have a severe defect in evoked release. Furthermore, EJPs recorded from hip14 mutants at 30°C (0.2 Hz and 1 mM Ca2+) are severely reduced (wild type, 42.8 ± 3.7 mV; hip142, 5.9 ± 1.5 mV; Fig. 5, E and F), suggesting that the reduced EJP amplitudes in hip14 mutants are temperature sensitive, similar to what is observed in csp mutants (Fig. 5, D and F; Zinsmaier et al., 1994). These data demonstrate that HIP14 is required for proper exocytosis.
We also recorded miniature EJP amplitude and frequency. Both parameters are similar to controls (Fig. 5 G), suggesting normal transmitter loading and postsynaptic glutamate receptor clustering. To further assess postsynaptic integrity, we labeled control and mutant synapses for GluRIII, a subunit found in all glutamate receptor clusters (Petersen et al., 1997). GluRIII staining in hip14 mutants is not obviously different from controls (Fig. 5 H). These data suggest that HIP14 does not affect postsynaptic receptor clustering or function.
Because HIP14 is expressed both pre- and postsynaptically at the Drosophila NMJ (Fig. 3, D–H), we assessed whether the overexpression of HIP14 in neurons (elav-GAL4; UAS-cDNAhip14) was able to rescue the mutant phenotypes. When overexpressed in hip14 mutants, UAS-cDNAhip14 not only rescues the lethality but also restores the EJP amplitude in hip14 mutants to control levels (wild type, 39.4 ± 2.4 mV; elav-GAL4/+; UAS-cDNAhip14/+; hip142/Df, 37.1 ± 1.7 mV; Fig. 5 D). Collectively, these data indicate that the reduced EJP amplitudes observed in hip14 mutants stem from impaired presynaptic function and demonstrate an essential function for the gene in the nervous system.
CSP and SNAP25 are not properly localized at hip14 NMJs
Based on in vitro assays, mammalian HIP14 and the yeast homologue Akr1p are PATs (Babu et al., 2004; Huang et al., 2004) and are able to covalently attach palmitic acid to cysteine residues, thereby regulating the localization of various proteins. Although HIP14 is able to palmitoylate many neuronal proteins, including PSD-95, GAD43 (growth-associated protein 43), Syt I, SNAP25, and Htt in cell culture systems (Huang et al., 2004), the role and substrate specificity of HIP14 in neurons remain to be determined.
Therefore, we examined the levels and protein distribution of presynaptic components that are known to be palmitoylated, including PSD-95/DLG, Syt I, n-Syb, SNAP25, and CSP (el-Husseini Ael and Bredt, 2002) in control and hip14 mutant boutons. We stained NMJs with anti-DLG, which strongly labels the postsynaptic membrane as well as the most peripheral component of the presynaptic membrane (Lahey et al., 1994). We did not observe any differences between control and mutant NMJs (Fig. 4 A), indicating that DLG may not be a target for HIP14 in Drosophila. Therefore, we were able to use DLG to counterstain boutons that were labeled with Syt I, n-Syb, CSP, and SNAP25. As shown in Fig. 6 (A, B, and E), the levels and protein distributions of Syt I and n-Syb are similar in hip14 when compared with controls. However, the levels of CSP and SNAP25 are strongly reduced in the NMJ boutons of hip14 mutants (28 ± 5.2% of control for CSP and 29 ± 2.4% of control for SNAP25; Fig. 6, C–E). These data suggest that HIP14 is important for the proper synaptic localization of CSP and SNAP25 but not for Syt I and n-Syb.
Figure 6.
CSP and SNAP25 are mislocalized in hip14 mutants. (A–D) Confocal images showing labeling of control (y w; FRT80B; left) and hip142/Df mutant (y w ey-FLP GMR-lacZ; hip142 FRT80B/Df(3L)brm11; right) boutons on muscle 4 segment A4 for Syt I (A), n-Syb (B), CSP (C), and SNAP25 (D; green). (E) Quantification of labeling intensity for synaptic markers shown in A–D. **, P < 0.01 (t test). Error bars represent SEM. The number of animals tested is indicated in the bars. (F) Western blots of larval brain extracts of controls, hip141/Df, and hip142/Df using antibodies against CSP and SNAP25. Protein loading normalized using anti-actin labeling. (G and H) Confocal images showing labeling of control (G) and hip142/Df mutant (H) VNC with CSP (green) and DLG (magenta). The green channel is shown separately in the bottom panel. n, neuropil; c, cell body. (I) Hydroxylamine treatment of hip14 mutant brains. Larval brain extracts of controls (lanes 1 and 2) and hip142/Df (lanes 3 and 4) were treated with hydroxylamine (lanes 2 and 4) or Tris (lanes 1 and 3). After treatment, proteins were subjected to SDS-PAGE and immunoblotted with CSP antibodies. Bars: (A–D) 10 μm; (G and H) 20 μm.
To test whether the reduction in CSP and SNAP25 at synapses can be attributed to a decrease in the expression or mislocalization of these proteins, we performed a Western blot using third instar larval brain tissue. Compared with controls, the amount of SNAP25 in mutant brain is not reduced on the Western blot (Fig. 6 F), suggesting that SNAP25 is translated properly but is mislocalized in the absence of HIP14. On the other hand, the level of CSP in mutant brains is somewhat reduced (Fig. 6 F). Interestingly, the molecular mass of CSP in hip14 mutants is ∼6–7 kD less than in controls. This suggests that some posttranslational modification of CSP does not occur properly in hip14 mutants.
To determine whether the localization of CSP and SNAP25 is altered in hip14 mutants, we examined its expression in the larval VNC, where neuronal cell bodies reside. In hip14 mutants, CSP and SNAP25 are much more diffuse and more prominently localized in neuronal cell bodies when compared with controls, in which CSP and SNAP25 are more enriched in the neuropil (Fig. 6, G and H; and Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200710061/DC1). This indicates that CSP and SNAP25 are not properly targeted to synapses in hip14 mutants. Together, these data show that CSP and SNAP25 are dependent on HIP14 for their proper localization at synapses, a process that may depend on the HIP14 PAT activity.
Palmitoylation of CSP requires HIP14
The previous data are consistent with the hypothesis that HIP14 palmitoylates CSP. CSP carries 11–13 palmitoylation sites (van de Goor and Kelly, 1996), and these posttranslational modifications are necessary for trafficking the protein from the ER in cultured cells (Greaves and Chamberlain, 2006). To assess the palmitoylation state of CSP in hip14 mutants, we compared wild-type CSP with CSP that is stripped of its palmitate residues through chemical treatment with hydroxylamine, a compound that has been shown to efficiently cleave posttranslational fatty acyl thioester groups of proteins (van de Goor and Kelly, 1996). As shown in Fig. 6 I (lane 1), wild-type CSP isolated from control third instar larvae migrates as a doublet at 38 kD. The protein isolated from wild-type animals was then treated with hydroxylamine (Fig. 6 I, lane 2) and compared with CSP from hip14 mutants (Fig. 6 I, lane 3). The chemically depalmitoylated wild-type CSP (Fig. 6 I, lane 2) as well as treated and untreated CSP isolated from hip14 mutants (Fig. 6 I, lane 4) all migrate at exactly the same molecular mass, ∼6 kD smaller than the wild-type protein, which is similar to a mutant CSP (serine string protein [SSP]) in which the 11 palmitoylated cysteines were replaced with serines (Arnold et al., 2004). These data indicate that CSP is indeed palmitoylated (van de Goor and Kelly, 1996; Chamberlain and Burgoyne, 1998) and, more importantly, that all or most palmitate residues of CSP are lacking in hip14 mutants.
Overexpressed CSP in hip14 mutants does not localize to NMJs and fails to rescue exocytic defects
In addition to the severe reduction of CSP at hip14 synapses, our electrophysiological data show that hip14 mutants exhibit exocytic defects that are temperature sensitive (Fig. 5, C–F). These phenotypes are reminiscent of those associated with the complete loss of function of CSP, which exhibits about a 50% reduction in evoked response at 23°C and an almost complete loss of synaptic transmission at 30°C (Fig. 5, D and F; Zinsmaier et al., 1994). Furthermore, the csp- and hip14-associated phenotypes are also very different from the rather mild electrophysiological defects associated with the loss of SNAP25, probably because of redundancy with SNAP24 (Vilinsky et al., 2002). Thus, the protein localization data, the biochemical data, and the electrophysiological data are all consistent with the hypothesis that much of the phenotype in hip14 mutants is caused by a defect in CSP function, and the defect in evoked release observed in hip14 mutants may be predominantly caused by a failure of unmodified CSP to be localized to the synapse.
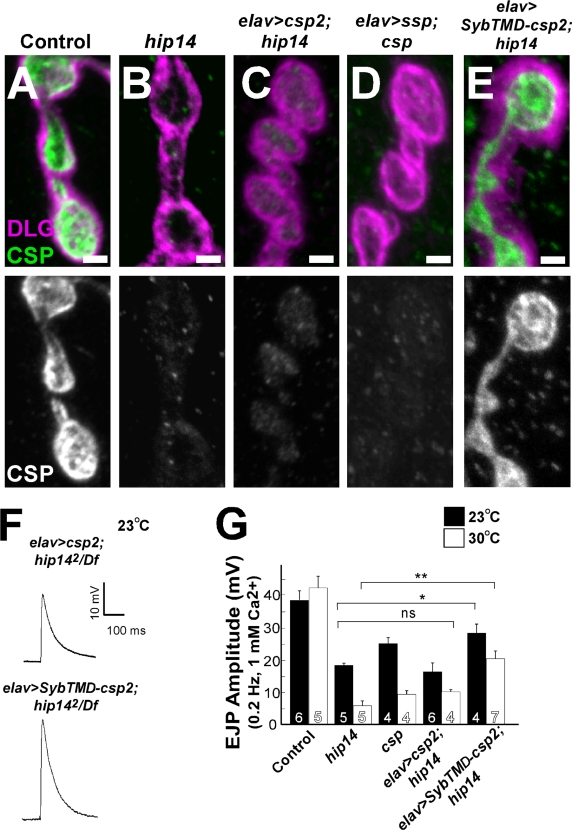
To explore whether trafficking of CSP to synapses is solely or predominantly dependent on palmitoylation by HIP14, we overexpressed CSP neuronally in hip14 mutants. We overexpressed wild-type UAS-CSP2, one of the three Drosophila isoforms expressed in neurons (Nie et al., 1999), which was previously shown to rescue csp-null mutants. In controls, CSP2 overexpression induced a fourfold up-regulation of CSP at the third instar larval NMJ relative to control animals (Fig. S4, A and B; available at http://www.jcb.org/cgi/content/full/jcb.200710061/DC1). However, in hip14 mutants, the levels of CSP at the boutons are not or are at most minimally up-regulated when compared with hip14 mutants that do not overexpress CSP (Fig. 7, A and C). These data again suggest that the decrease of CSP at NMJ boutons in hip14 mutants is not caused by translational repression and that any CSP targeting from the cell body to the synapses depends on HIP14.
Figure 7.
Chimeric but not wild-type CSP rescues the localization of CSP and the exocytic defects in hip14 mutants. (A–E) Confocal images showing labeling of larval filets with CSP (green) and DLG (magenta) to indicate the synaptic areas. Genotypes: control (y w; FRT80B; A), hip142/Df mutant (y w ey-FLP GMR-lacZ; hip142 FRT80B/Df(3L)brm11; B), elav>csp2; hip14 (elav-GAL4/+; P{w + UAS-csp2}/+; hip142 FRT80B/Df (3L)brm11; C), elav>ssp; csp (elav-GAL4/+;; P{w + UAS-csp-11c/s}, cspu1/cspx1; D), and elav>SybTMD-csp2; hip14 (elav-GAL4/+; P{w + UAS-SybTMD-csp}/+ hip142 FRT80B/Df(3L)brm11; E). CSP labeling is separately shown on the bottom panels. (F) Sample EJPs recorded in 1 mM Ca2+ at 0.2-Hz stimulation at 23°C when wild-type CSP2 or chimeric CSP is overexpressed in hip142/Df neurons. (G) Quantification of EJP amplitudes are recorded in 1 mM Ca2+ at 23°C (black bars) and at 30°C (white bars) in controls, hip14 mutant, csp mutants (w, cspu1/cspx1), CSP2 overexpressed neuronally in hip14 mutant background, and SybTMD-CSP2 overexpressed neuronally in hip14 mutant background. For recordings at 23°C (black bars), hip142/Df was used, whereas at 30°C (white bars), the hip142/hip141 allelic combination was used. Recordings were performed for 1 min at 0.2 Hz, and 12 EJP amplitudes were averaged per recording. *, P < 0.05; **, P < 0.01 (t test). Error bars represent SEM. The number of animals tested is indicated in the bars. Bars, 2 μm.
To further determine whether the palmitoylation of CSP is necessary for targeting of the protein to the synapse, we examined whether SSP (Arnold et al., 2004) localizes to NMJ boutons. Similar to wild-type CSP in hip14 mutants, SSP does not or very poorly localizes to NMJ boutons when expressed in csp-null mutants (Fig. 7 D). SSP is produced but localizes to the cell body of the neurons (Fig. S3 B), which is similar to wild-type CSP overexpressed in hip14 mutants. Collectively, these data further indicate that the palmitoylation of CSP is necessary for targeting the protein to the synapse.
Chimeric CSP localizes to synapses and significantly rescues hip14 mutants
Because CSP cannot be targeted to NMJ boutons in hip14 mutants, even when overexpressed, we generated a chimeric CSP in which full-length CSP2 is sandwiched between the N-terminal n-Syb sequence (111 bp) and the C-terminal n-Syb TMD (SybTMD; 234 bp) to forcibly target CSP to SVs at synapses. We expressed this chimeric CSP neuronally in csp-null mutants using elav-GAL4. Interestingly, the SybTMD-CSP2 protein is not only able to rescue the lethality of csp mutants but also the paralytic phenotype at 30°C even though it is expressed at lower levels than the wild-type CSP2 (Fig. S4, B–D). These data indicate that SybTMD-CSP2 is expressed and functional and further suggest that vesicle tethering of CSP is important for the function of CSP.
To test whether SybTMD-CSP2 localizes to NMJ synapses independently of HIP14, we determined its localization in hip14 mutants. As shown in Fig. 7 E and in contrast to wild-type CSP2 in hip14 mutants (Fig. 7 C), the chimeric CSP at third instar larvae boutons is easily detectable. This difference in immunostaining is not caused by elevated levels of expression of SybTMD-CSP2 when compared with wild-type CSP (Fig. S4, A–D).
Because SNAP25 may be a substrate for the CSP chaperone complex, the mislocalization of SNAP25 in hip14 mutants may be caused by the mislocalization of CSP and not necessarily because of defective palmitoylation. However, our data suggest that this is not the case, as the SNAP25 levels are still reduced in hip14 mutants that express the chimeric CSP that localizes to NMJs (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200710061/DC1). Thus, HIP14-mediated palmitoylation of SNAP25 seems critical for its synaptic localization.
If the exocytic defects in hip14 mutants largely stem from the mislocalization of CSP (Fig. 5, C–F), SybTMD-CSP2 should be able to at least partially rescue the exocytic defects in hip14 mutants. Indeed, when SybTMD-CSP2 is expressed neuronally in hip14 mutants, the EJPs are significantly rescued at 23°C (Fig. 7, F and G). Furthermore, at 30°C, EJPs in hip14 mutants recovered from 5 to 50% of control (Fig. 7 G). In contrast, wild-type CSP2 expression in hip14 neurons does not show significant rescue of the EJP phenotype (Fig. 7, F and G). These data indicate that a significant portion of the neurotransmission defects in hip14 mutants result from depletion of CSP at the synapse and further indicate the importance of palmitoylation and vesicle association for the normal function of CSP.
Discussion
Posttranslational modification by palmitate is critical for the proper localization and function of numerous proteins (Smotrys and Linder, 2004). The enzymes that mediate the addition of palmitate to proteins, PATs, were originally studied in S. cerevisiae (Roth et al., 2002) and are characterized by the presence of a CRD with an embedded DHHC motif that mediates the PAT activity of enzymes. Although DHHC-CRD proteins are conserved from yeast to mammals, no mutations in these proteins have been documented in any other model organism. Here, in an unbiased genetic screen to identify genes that affect synaptic transmission, we have identified mutants in hip14, a DHHC-CRD protein structurally conserved from yeast to man (Singaraja et al., 2002). Characterization of these mutants suggests that HIP14 plays an important role in synaptic function by mediating the palmitoylation and proper targeting of specific presynaptic proteins.
The importance of proper protein palmitoylation in the nervous system has been suggested by several findings. For instance, patients with lesions PPT1, a depalmitoylating enzyme, suffer from an early onset neurodegeneration that leads to childhood death (Vesa et al., 1995). Loss of PPT1 in Drosophila has also been associated with a synaptic dysfunction based on genetic interaction experiments (Buff et al., 2007). In addition, the presence of expanded repeats in pathogenic Htt protein has been shown to render it less amenable to palmitoylation, enhancing the formation of inclusion bodies (Yanai et al., 2006). However, the consequences on synaptic transmission of aberrant or absent palmitoylation in neurons have not been investigated in vivo.
Mammalian HIP14 is expressed ubiquitously but is most prominently present in the brain (Singaraja et al., 2002). Here, we find that GFP-HIP14 is expressed throughout the nervous system during development and in third instar larvae but is strongly enriched in the neuropil. Furthermore, the presynaptic expression of HIP14 rescues the lethality and phenotype of hip14 mutants, suggesting that it plays a critical role in presynaptic function. This is further substantiated by the observations that the miniature EJP amplitude and frequency are similar to wild type and that GluRIII localization is normal. All DHHC proteins, with the exception of yeast Ynl155W, are integral membrane proteins with four or more TMDs (Mitchell et al., 2006). The DHHC-CRD motif is typically located between two TMDs and is predicted to face the cytosol (Politis et al., 2005). Human HIP14 was shown to be localized at the Golgi and in cytoplasmic vesicles marked with SNAP25 in cultured neurons or transfected cells (Huang et al., 2004). We find that HIP14 mostly colocalizes with CSP and n-Syb at the synapse. Because HIP14 relocalizes to the presynaptic membrane upon SV depletion and harbors several TMDs, these data are consistent with HIP14 being an SV and presynaptic membrane-associated protein.
There are 20 DHHC proteins in Drosophila, raising issues about the protein specificity of each DHHC protein family member. Because there are numerous DHHC proteins, it is likely that each protein has one or a few specific target substrates. This may be unsurprising, as palmitoylation is a highly regulated event, and DHHC proteins are likely to be targeted to very specific cellular compartments, where they act locally on few targets. A previous mammalian study in culture systems shows that HIP14 is a PAT and suggests that candidate targets include PSD-95/DLG, Htt, Syt I, SNAP25, GAD43, and GAD65 (Huang et al., 2004). In addition, RNAi knockdown analyses in neurons have provided evidence that HIP14 modulates palmitoylation-dependent protein trafficking of PSD-95, GAD65, and Htt (Huang et al., 2004; Yanai et al., 2006). However, the role of HIP14 in neuronal activity and synaptic transmission had not been documented.
We find that HIP14 directly regulates the localization of two important presynaptic proteins: CSP and SNAP25. However, we did not observe any difference in the localization of DLG, n-Syb, and Syt I. We focused on CSP for several reasons. First, the loss of SNAP25 in Drosophila causes very mild exocytic defects. It has been proposed that this is the result of the redundant function of SNAP24 (Vilinsky et al., 2002). Second, the electrophysiological phenotype associated with the loss of HIP14 strongly resembles the unique temperature-sensitive phenotype associated with the loss of CSP (Zinsmaier et al., 1994), indicating that CSP is one of the main targets of HIP14. Third, it was previously shown that CSP is heavily palmitoylated and that this posttranslational modification is required in vivo (van de Goor and Kelly, 1996; Arnold et al., 2004). Collectively, these data suggest that much of the phenotype we observe in hip14 mutants is caused by the loss of CSP function. However, note that although the functional defects in hip14 strongly resemble those seen in csp mutants, the developmental defects, including the reduction in bouton number in csp mutants (Bronk et al., 2005), are not observed in hip14, indicating that palmitoylation- independent roles for CSP in neurons also exist.
The SV-associated protein CSP is critical for regulating neurotransmitter release and has been shown to prevent neuronal degeneration (Fernandez-Chacon et al., 2004; Zinsmaier et al., 1994). CSP binds the chaperone heat-shock cognate protein Hsc70 and regulates its ATP activity, and this cooperativity is required for regulated neurotransmitter release at synaptic terminals (Bronk et al., 2001). These data have led to the proposal that CSP is a chaperone that renatures nerve terminal proteins that misfold during the continuous operation of the SV cycle, a view which is supported by genetic studies in mice and Drosophila (Zinsmaier et al., 1994; Fernandez-Chacon et al., 2004). Binding experiments suggest that the protective effect may be mediated by CSP interacting with candidate substrates such as the SV protein n-Syb and the plasma membrane protein Syntaxin (Zinsmaier and Bronk, 2001).
CSP possesses a cysteine-rich region containing a high density of cysteine residues (14 in a span of 24 amino acids in mammals). Most of these cysteines are palmitoylated, a process proposed to be necessary for the secretion of CSP from the ER (Chamberlain and Burgoyne, 1998; Greaves and Chamberlain, 2006). Our data indicate that HIP14 is the PAT for CSP and that in the absence of HIP14, CSP is retained in the cell body. Furthermore, SSP is also retained in the cell body when overexpressed in neurons (Fig. S3 B). Thus, these data are consistent with a role for HIP14 in regulating the proper synaptic targeting of CSP. The observation that a chimeric CSP rescues much of the exocytic defects associated with the loss of hip14 mutants provides compelling evidence that a main function of HIP14 is the palmitoylation of CSP. This palmitoylation is most likely not only required in the cell body for proper targeting of CSP to the synaptic terminals; indeed, synaptic HIP14 may also play a role in a palmitoylation cycle occurring at synapses.
The loss of CSP and the expression of expanded Htt protein have both been shown to be associated with synaptic defects in exocytosis and neuronal degeneration (Zinsmaier et al., 1994; Fernandez-Chacon et al., 2004; Romero et al., 2007). Interestingly, our preliminary analyses suggest that wild-type human Htt and 128Q-expanded human Htt proteins are both mislocalized in flies lacking HIP14 (unpublished data). Thus, Htt also appears to be palmitoylated by HIP14 in Drosophila, which is similar to what has been observed in mice (Huang et al., 2004). Although we have not yet explored the effect of hip14 mutations on neuronal degeneration, it will be interesting to explore the relationship between HIP14, CSP, and Htt in the processes of synaptic transmission and neuronal degeneration in more detail.
Materials and methods
Genetics and molecular biology
Control animals are isogenized (y w ey-FLP GMR-lacZ; FRT80B) unless otherwise indicated. 3L11, 3L12, and 3L13 mutants (y w ey-FLP GMR-lacZ;3L1x FRT80B/TM6B, Tb) were isolated from an ey-FLP ethane methyl sulfonate screen as described previously (Verstreken et al., 2003) with modifications. cspx1 mutants and UAS-ssp (w;; P{w + UAS-csp-11c/s}, cspu1) flies were provided by K. Zinsmaier (University of Arizona, Tucson, AZ). P-element stocks and deficiencies were obtained from the Bloomington Drosophila Stock Center (Bellen et al., 2004; Parks et al., 2004), and 3L1 mapping was performed as described previously (Zhai et al., 2003).
We made a genomic rescue construct by PCR amplifying the 6.5-kb hip14 region from bacterial artificial chromosome clone AC093499. The fragment was cloned into the SalI restriction site of pP{CaSpeR-4} and sequenced. A cDNA construct was made by PCR amplifying hip14 from expressed sequence tag clone LD10758. The fragment was cloned into NotI and XbaI sites of pP{UAST} and sequenced.
To generate genomic GFP-tagged constructs, we first integrated an NheI site just before the ATG start codon (NtermGFP-HIP14) or after the hip14 stop codon (CtermGFP-HIP14) by site-directed mutagenesis (Stratagene). PCR-amplified EGFP sequence was cloned into the NheI site.
We generated chimeric n-SybTMD-CSP constructs by PCR amplifying 111-bp N-terminal and 234-bp C-terminal n-syb sequences from pP{UAST}-syb-GFP and the full-length CSP2 from pP{UAST}-csp2 (provided by K. Zinsmaier). In the next round of PCR, we fused them to generate N-terminal-Syb-csp2-C-terminal-Syb chimeric (SybTMD-csp2). After sequencing, SybTMD-csp2 was cloned into pP{UAST} at NotI and XbaI.
P{w + UAS-SybTMD-csp2} and P{w + UAS-csp2} were expressed using elav-GAL4. For analyses of CSP localization and physiology of third instar larvae, we generated elav-GAL4/+; P{w + UAS-SybTMD-csp2} hip142 FRT80B/Df(3L)brm11 and elav-GAL4/+; hip142 FRT80B P{w + UAS-csp2}/Df(3L)brm11.
Immunohistochemistry and Western blotting
For staining third instar larvae and adults, brains were dissected in modified HL3 solution (110 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 5 mM Hepes, 30 mM sucrose, 5 mM trehalose, and 10 mM MgCl2, pH 7.2) and fixed in 4% formaldehyde for 20 min. Tissue was washed with PBS and permeabilized with 0.4% Triton X-100. For shibirets experiments, third instar larvae were dissected in modified HL3 solution, incubated at 32°C for 5 min, and stimulated in prewarmed modified HL3 solution with 90 mM KCl and 1.5 mM Ca2+ for 5 min. After stimulation, the samples were fixed in 4% formaldehyde for 20 min. Labeling was performed according to standard protocols. Samples were mounted in Vectashield (Vector Laboratories) and were imaged using a confocal microscope (LSM 510; Carl Zeiss, Inc.) with the accompanying LSM5 software (Carl Zeiss, Inc.) at RT. Identical settings were used for control and experimental samples. In Fig. 1 (B and C), a 40× 1.3 NA lens was used. In Figs. 3 (A, B, and D–K), 4 (A and B), 5 H, 6 (A–D, G, and H), 7, S2 (B and C), S4 (A–C), and S5, a 63× 1.4 NA differential interference contrast plan-Neofluor oil-immersion lens (Carl Zeiss, Inc.) was used. For Figs. 3 C, S 2 A, and S3 A, a 10× 0.45 NA lens without oil was used. Captured images were processed using Amira 2.2 (Mercury Computer Systems) followed by Photoshop 7.0 (Adobe). Samples for Western blots were prepared by dissecting third instar larval brains and extracting proteins in radioimmunoprecipitation assay buffer with proteinase inhibitors on ice. Samples were then boiled in sample buffer for 5 min.
Antibodies that recognize the following proteins were used at the indicated dilutions for immunohistochemistry/Western blotting: Syt I at 1:5,000/1:10,000 (Littleton et al., 1993), n-Syb at 1:200/1:1,000, CSP at 1:100/1:1,000 (Zinsmaier et al., 1994), and SNAP25 at 1:200/1:1,000 (Vilinsky et al., 2002). The following antibodies were used only for immunohistochemistry: DLG (mouse mAb 4F3) at 1:50 (Parnas et al., 2001), DLG (rabbit polyclonal; provided by K. Choi, Baylor College of Medicine, Houston, TX) at 1:500, HRP (rabbit; Jackson ImmunoResearch Laboratories) at 1:200, mouse mAb 24B10 (Futsch) at 1:50, mouse mAb nc82 (Bruchpilot) at 1:100 (Kittel et al., 2006), Fasciclin II (mouse mAb 1D4) at 1:10 (Zito et al., 1997), GluRIII rabbit polyclonal (provided by A. DiAntonio, Washington University, St. Louis, MI) at 1:5,000 (Marrus et al., 2004), and GFP (rabbit; Invitrogen) at 1:500. AlexaFluor488- (Invitrogen) and Cy3 (Jackson ImmunoResearch Laboratories)-conjugated secondary antibodies were used at 1:250. HRP-conjugated antibodies (Jackson ImmunoResearch Laboratories) were used at 1:2,500 for Western blotting. Western blots were developed with ECL reagents (PerkinElmer).
Quantification
For synaptic protein quantification, anti-DLG (rabbit or mouse) was used to outline type I boutons (Lahey et al., 1994). Boutons were scanned with z steps of 0.5 μm. Using Amira software, the DLG-stained type I boutons in each confocal slice were highlighted, and the mean pixel intensity of all slices of the highlighted boutons of each NMJ were computed. Background fluorescence in muscle areas adjacent to the boutons was quantified similarly, and the background was subtracted from the bouton values to yield the mean intensity of labeling in the boutons. The mean value from at least three mutant NMJs was then expressed as a percentage of the corresponding control value.
ERG assay
ERGs were performed as described previously (Fabian-Fine et al., 2003; Verstreken et al., 2003); flies were immobilized with one eye and part of the thorax in a small drop of Elmer's school glue on a microscope slide. For the 3L screen, ∼50–100 male F1 flies were subjected to two ERG recordings. Flies with mutant ERGs were liberated and subsequently mated.
Electrophysiology and FM 1-43 dye uptake
For FM 1-43 dye uptake experiments and electrophysiological recordings, third instar larvae were dissected in modified HL3 without Ca2+, and motor neurons were cut. Larvae were stimulated in modified HL3 solution at ambient temperature with 4 μM FM 1-43, 90 mM KCl, and 1.5 mM Ca2+ for 1 min and were washed with modified HL3 solution. Images were captured using a microscope (Axioskop; Carl Zeiss, Inc.) with Axiovision 4.2 software (Carl Zeiss, Inc.), a 40× 0.75-W water immersion lens (Carl Zeiss, Inc.), and a camera (MRm; Carl Zeiss, Inc.) at RT. The intensity of FM 1-43 was analyzed and quantified as described previously (Verstreken et al., 2007).
EJPs and miniature EJPs recorded the membrane potential of muscle 6 using sharp 90–110 MΩ electrodes as described previously (Koh et al., 2004). The temperature of the preparations was controlled as described previously (Koh et al., 2004). The data were analyzed by Clampfit (MDS Analytical Technologies) for EJPs and by Mini Analysis Program 6.0.3 (Synaptosoft) for miniature EJPs.
TEM
TEM of PRs and NMJ boutons was performed as described previously (Verstreken et al., 2003). Images were captured using a transmission electron microscope (model 1010; JEOL) with a camera (US1000; Gatan) and digital micrograph. For the PRs and NMJ boutons, 3,000× and 8,000× magnifications were used, respectively. For statistical analyses, cross sections of 10 cartridges or boutons from three different animals were measured using ImageJ (National Institutes of Health).
Depalmitoylation assay
Third instar larvae from control and mutant animals were homogenized in lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 5 mM EDTA, pH 7.4, and 2% Triton X-100). After centrifugation at 16,000 g, the proteins in the soluble fraction were precipitated by the chloroform-methanol precipitation method. The pellet was resuspended in SDS lysis buffer (50 mM Tris-Cl, 5 mM EDTA, and 4% SDS) and treated with 150 mM NaCl and 2% Triton X-100 with or without 1 M hydroxylamine, pH 7.0, at ambient for 2 h. After hydroxylamine treatment, samples were boiled in SDS sample buffer and subjected to SDS-PAGE.
Online supplemental material
Fig. S1 shows that control and 3L1 mutant PR terminals in the lamina are correctly organized. Fig. S2 shows that CtermGFP-HIP14 is enriched in the CNS and synaptic terminal similar to NtermGFP-HIP14 and that GFP-HIP14 is colocalized with the SV marker n-Syb at NMJs. Fig. S3 shows the mislocalization of SNAP25 in hip14 mutants (A) and SSP in csp mutants (B) at the VNC. Fig. S4 shows the expression level of CSP2 and SybTMD-CSP2. Fig. S5 shows that SNAP25 levels are not restored in hip14 mutant larvae that overexpress SybTMD-CSP2, indicating that the mislocalization of SNAP25 in hip14 mutants is not caused by CSP reduction at the synaptic terminal. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200710061/DC1.
Supplemental Material
Acknowledgments
We are grateful to the Bloomington Stock Center, the Developmental Studies Hybridoma Bank, K. Zinsmaier, K. Choi, A. DiAntonio, and D.L. Deitcher for reagents. We thank N. Giagtzoglou, C-K. Yao, Y.Q. Lin, and other members of the Bellen laboratory for comments. We thank Yuchun He for injections to generate the transgenic lines. Confocal microscopy was supported by the Mental Retardation and Developmental Disabilities Research Center at Baylor College of Medicine.
P. Verstreken was supported by an R.L. Kirchstein National Research Service award, a Marie Curie Excellence Grant (MEXT-CT-2006-042267), the Research Fund Katholieke Universiteit Leuven, and Vlaams Instituut voor Biotechnologie. C.V. Ly was supported by a National Research Service Award, and H.J. Bellen is a Howard Hughes Medical Institute Investigator.
P. Verstreken's present address is Vlaams Instituut voor Biotechnologie, Department of Molecular and Developmental Genetics, Katholieke Universiteit Leuven, Center for Human Genetics, 3000 Leuven, Belgium.
Abbreviations used in this paper: CNS, central nervous system; CRD, cysteine-rich domain; CSP, cysteine string protein; DLG, Discs large; EJP, excitatory junctional potential; ERG, electroretinogram; HIP14, Htt-interacting protein 14; Htt, Huntingtin; NMJ, neuromuscular junction; n-Syb, neuronal synaptobrevin; PAT, palmitoyl transferase; PPT1, palmitoyl-protein thioesterase 1; PR, photoreceptor; PSD-95, postsynaptic density 95; SSP, serine string protein; SV, synaptic vesicle; Syt I, synaptotagmin I; TEM, transmission EM; TMD, transmembrane domain; VNC, ventral nerve cord.
References
- Arnold, C., N. Reisch, C. Leibold, S. Becker, K. Prufert, K. Sautter, D. Palm, S. Jatzke, S. Buchner, and E. Buchner. 2004. Structure-function analysis of the cysteine string protein in Drosophila: cysteine string, linker and C terminus. J. Exp. Biol. 207:1323–1334. [DOI] [PubMed] [Google Scholar]
- Babu, P., R.J. Deschenes, and L.C. Robinson. 2004. Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J. Biol. Chem. 279:27138–27147. [DOI] [PubMed] [Google Scholar]
- Bellen, H.J., R.W. Levis, G. Liao, Y. He, J.W. Carlson, G. Tsang, M. Evans-Holm, P.R. Hiesinger, K.L. Schulze, G.M. Rubin, et al. 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 167:761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronk, P., J.J. Wenniger, K. Dawson-Scully, X. Guo, S. Hong, H.L. Atwood, and K.E. Zinsmaier. 2001. Drosophila Hsc70-4 is critical for neurotransmitter exocytosis in vivo. Neuron. 30:475–488. [DOI] [PubMed] [Google Scholar]
- Bronk, P., Z. Nie, M.K. Klose, K. Dawson-Scully, J. Zhang, R.M. Robertson, H.L. Atwood, and K.E. Zinsmaier. 2005. The multiple functions of cysteine-string protein analyzed at Drosophila nerve terminals. J. Neurosci. 25:2204–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buff, H., A.C. Smith, and C.A. Korey. 2007. Genetic modifiers of Drosophila palmitoyl-protein thioesterase 1-induced degeneration. Genetics. 176:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, L.H., and R.D. Burgoyne. 1998. The cysteine-string domain of the secretory vesicle cysteine-string protein is required for membrane targeting. Biochem. J. 335:205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy, J.T., and M.E. Linder. 1998. Signalling functions of protein palmitoylation. Biochim. Biophys. Acta. 1436:245–261. [DOI] [PubMed] [Google Scholar]
- el-Husseini Ael, D., and D.S. Bredt. 2002. Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3:791–802. [DOI] [PubMed] [Google Scholar]
- Fabian-Fine, R., P. Verstreken, P.R. Hiesinger, J.A. Horne, R. Kostyleva, Y. Zhou, H.J. Bellen, and I.A. Meinertzhagen. 2003. Endophilin promotes a late step in endocytosis at glial invaginations in Drosophila photoreceptor terminals. J. Neurosci. 23:10732–10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Chacon, R., M. Wolfel, H. Nishimune, L. Tabares, F. Schmitz, M. Castellano-Munoz, C. Rosenmund, M.L. Montesinos, J.R. Sanes, R. Schneggenburger, and T.C. Sudhof. 2004. The synaptic vesicle protein CSP alpha prevents presynaptic degeneration. Neuron. 42:237–251. [DOI] [PubMed] [Google Scholar]
- Fujita, S.C., S.L. Zipursky, S. Benzer, A. Ferrus, and S.L. Shotwell. 1982. Monoclonal antibodies against the Drosophila nervous system. Proc. Natl. Acad. Sci. USA. 79:7929–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, J., and L.H. Chamberlain. 2006. Dual role of the cysteine-string domain in membrane binding and palmitoylation-dependent sorting of the molecular chaperone cysteine-string protein. Mol. Biol. Cell. 17:4748–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichet, A., T. Wucherpfennig, V. Dudu, S. Etter, M. Wilsch-Brauniger, A. Hellwig, M. Gonzalez-Gaitan, W.B. Huttner, and A.A. Schmidt. 2002. Essential role of endophilin A in synaptic vesicle budding at the Drosophila neuromuscular junction. EMBO J. 21:1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., A. Yanai, R. Kang, P. Arstikaitis, R.R. Singaraja, M. Metzler, A. Mullard, B. Haigh, C. Gauthier-Campbell, C.A. Gutekunst, et al. 2004. Huntingtin- interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 44:977–986. [DOI] [PubMed] [Google Scholar]
- Jan, L.Y., and Y.N. Jan. 1982. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc. Natl. Acad. Sci. USA. 79:2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel, R.J., C. Wichmann, T.M. Rasse, W. Fouquet, M. Schmidt, A. Schmid, D.A. Wagh, C. Pawlu, R.R. Kellner, K.I. Willig, et al. 2006. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 312:1051–1054. [DOI] [PubMed] [Google Scholar]
- Koh, T.W., P. Verstreken, and H.J. Bellen. 2004. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 43:193–205. [DOI] [PubMed] [Google Scholar]
- Kraut, R., K. Menon, and K. Zinn. 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11:417–430. [DOI] [PubMed] [Google Scholar]
- Lahey, T., M. Gorczyca, X.X. Jia, and V. Budnik. 1994. The Drosophila tumor suppressor gene dlg is required for normal synaptic bouton structure. Neuron. 13:823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton, J.T., M. Stern, K. Schulze, M. Perin, and H.J. Bellen. 1993. Mutational analysis of Drosophila synaptotagmin demonstrates its essential role in Ca(2+)-activated neurotransmitter release. Cell. 74:1125–1134. [DOI] [PubMed] [Google Scholar]
- Lobo, S., W.K. Greentree, M.E. Linder, and R.J. Deschenes. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268–41273. [DOI] [PubMed] [Google Scholar]
- Marrus, S.B., S.L. Portman, M.J. Allen, K.G. Moffat, and A. DiAntonio. 2004. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J. Neurosci. 24:1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, S.Q., P.R. Hiesinger, S. Beronja, R.G. Zhai, K.L. Schulze, P. Verstreken, Y. Cao, Y. Zhou, U. Tepass, M.C. Crair, and H.J. Bellen. 2005. Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron. 46:219–232. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.A., A. Vasudevan, M.E. Linder, and R.J. Deschenes. 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47:1118–1127. [DOI] [PubMed] [Google Scholar]
- Newsome, T.P., B. Asling, and B.J. Dickson. 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 127:851–860. [DOI] [PubMed] [Google Scholar]
- Nie, Z., R. Ranjan, J.J. Wenniger, S.N. Hong, P. Bronk, and K.E. Zinsmaier. 1999. Overexpression of cysteine-string proteins in Drosophila reveals interactions with syntaxin. J. Neurosci. 19:10270–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, Y., A. Kihara, T. Sano, and Y. Igarashi. 2006. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta. 1761:474–483. [DOI] [PubMed] [Google Scholar]
- Ordway, R.W., L. Pallanck, and B. Ganetzky. 1994. Neurally expressed Drosophila genes encoding homologs of the NSF and SNAP secretory proteins. Proc. Natl. Acad. Sci. USA. 91:5715–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas, O., T.S. Hays, and J.C. Sisson. 2005. The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat. Cell Biol. 7:612–618. [DOI] [PubMed] [Google Scholar]
- Parks, A.L., K.R. Cook, M. Belvin, N.A. Dompe, R. Fawcett, K. Huppert, L.R. Tan, C.G. Winter, K.P. Bogart, J.E. Deal, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. [DOI] [PubMed] [Google Scholar]
- Parnas, D., A.P. Haghighi, R.D. Fetter, S.W. Kim, and C.S. Goodman. 2001. Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron. 32:415–424. [DOI] [PubMed] [Google Scholar]
- Petersen, S.A., R.D. Fetter, J.N. Noordermeer, C.S. Goodman, and A. DiAntonio. 1997. Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 19:1237–1248. [DOI] [PubMed] [Google Scholar]
- Politis, E.G., A.F. Roth, and N.G. Davis. 2005. Transmembrane topology of the protein palmitoyl transferase Akr1. J. Biol. Chem. 280:10156–10163. [DOI] [PubMed] [Google Scholar]
- Ramaswami, M., K.S. Krishnan, and R.B. Kelly. 1994. Intermediates in synaptic vesicle recycling revealed by optical imaging of Drosophila neuromuscular junctions. Neuron. 13:363–375. [DOI] [PubMed] [Google Scholar]
- Romero, E., P. Verstreken, C.L. Li, G.-H. Cha, R. Hughes, H.J. Bellen, and J. Botas. 2007. Genetic suppression of neurodegeneration and neurotransmitter release abnormalities caused by expanded full-length huntingtin in the absence of nuclear import. Neuron. In press. [DOI] [PMC free article] [PubMed]
- Roth, A.F., Y. Feng, L. Chen, and N.G. Davis. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A.F., J. Wan, A.O. Bailey, B. Sun, J.A. Kuchar, W.N. Green, B.S. Phinney, J.R. Yates III, and N.G. Davis. 2006. Global analysis of protein palmitoylation in yeast. Cell. 125:1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaraja, R.R., S. Hadano, M. Metzler, S. Givan, C.L. Wellington, S. Warby, A. Yanai, C.A. Gutekunst, B.R. Leavitt, H. Yi, et al. 2002. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 11:2815–2828. [DOI] [PubMed] [Google Scholar]
- Smotrys, J.E., and M.E. Linder. 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73:559–587. [DOI] [PubMed] [Google Scholar]
- Stowers, R.S., and T.L. Schwarz. 1999. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 152:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers, R.S., L.J. Megeath, J. Gorska-Andrzejak, I.A. Meinertzhagen, and T.L. Schwarz. 2002. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 36:1063–1077. [DOI] [PubMed] [Google Scholar]
- van de Goor, J., and R.B. Kelly. 1996. Association of Drosophila cysteine string proteins with membranes. FEBS Lett. 380:251–256. [DOI] [PubMed] [Google Scholar]
- Verstreken, P., T.W. Koh, K.L. Schulze, R.G. Zhai, P.R. Hiesinger, Y. Zhou, S.Q. Mehta, Y. Cao, J. Roos, and H.J. Bellen. 2003. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 40:733–748. [DOI] [PubMed] [Google Scholar]
- Verstreken, P., C.V. Ly, K.J. Venken, T.W. Koh, Y. Zhou, and H.J. Bellen. 2005. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 47:365–378. [DOI] [PubMed] [Google Scholar]
- Verstreken, P., T. Ohyama, and H.J. Bellen. 2007. FM 1-43 labeling at the Drosophila neuromuscular junction. Methods Mol. Biol. In press. [DOI] [PMC free article] [PubMed]
- Vesa, J., E. Hellsten, L.A. Verkruyse, L.A. Camp, J. Rapola, P. Santavuori, S.L. Hofmann, and L. Peltonen. 1995. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 376:584–587. [DOI] [PubMed] [Google Scholar]
- Vilinsky, I., B.A. Stewart, J. Drummond, I. Robinson, and D.L. Deitcher. 2002. A Drosophila SNAP-25 null mutant reveals context-dependent redundancy with SNAP-24 in neurotransmission. Genetics. 162:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, A., K. Huang, R. Kang, R.R. Singaraja, P. Arstikaitis, L. Gan, P.C. Orban, A. Mullard, C.M. Cowan, L.A. Raymond, et al. 2006. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 9:824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, R.G., P.R. Hiesinger, T.W. Koh, P. Verstreken, K.L. Schulze, Y. Cao, H. Jafar-Nejad, K.K. Norga, H. Pan, V. Bayat, et al. 2003. Mapping Drosophila mutations with molecularly defined P element insertions. Proc. Natl. Acad. Sci. USA. 100:10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Y.H. Koh, R.B. Beckstead, V. Budnik, B. Ganetzky, and H.J. Bellen. 1998. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 21:1465–1475. [DOI] [PubMed] [Google Scholar]
- Zinsmaier, K.E., and P. Bronk. 2001. Molecular chaperones and the regulation of neurotransmitter exocytosis. Biochem. Pharmacol. 62:1–11. [DOI] [PubMed] [Google Scholar]
- Zinsmaier, K.E., K.K. Eberle, E. Buchner, N. Walter, and S. Benzer. 1994. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 263:977–980. [DOI] [PubMed] [Google Scholar]
- Zito, K., R.D. Fetter, C.S. Goodman, and E.Y. Isacoff. 1997. Synaptic clustering of Fascilin II and Shaker: essential targeting sequences and role of Dlg. Neuron. 19:1007–1016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.