Abstract
The integrin family of heterodimeric transmembrane receptors mediates cell–matrix adhesion. Integrins often localize in highly organized structures, such as focal adhesions in tissue culture and myotendinous junctions in muscles. Our RNA interference screen for genes that prevent integrin-dependent cell spreading identifies Z band alternatively spliced PDZ-motif protein (zasp), encoding the only known Drosophila melanogaster Alp/Enigma PDZ-LIM domain protein. Zasp localizes to integrin adhesion sites and its depletion disrupts integrin adhesion sites. In tissues, Zasp colocalizes with βPS integrin in myotendinous junctions and with α-actinin in muscle Z lines. Zasp also physically interacts with α-actinin. Fly larvae lacking Zasp do not form Z lines and fail to recruit α-actinin to the Z line. At the myotendinous junction, muscles detach in zasp mutants with the onset of contractility. Finally, Zasp interacts genetically with integrins, showing that it regulates integrin function. Our observations point to an important function for Zasp in the assembly of integrin adhesion sites both in cell culture and in tissues.
Introduction
Integrin-mediated adhesion between the ECM and the cytoskeleton is crucial for tissue interactions during development. Integrins are heterodimeric single-pass transmembrane receptors consisting of α and β subunits found in all animals from sponges to humans (Hughes, 2001). The globular extracellular domains of both subunits contribute to binding of ECM ligands. The short cytoplasmic carboxyl-terminal domains of integrins lack intrinsic catalytic activity. They organize the actin cytoskeleton through adaptor proteins and signal by associating with protein kinases and GTPases (Giancotti and Tarone, 2003). Disruption of the ECM, integrins, or their cytoskeletal adaptors affects integrin-mediated adhesion. Loss of integrin function leads to cell-spreading defects, muscle detachment, and, in the human disease epidermolysis bullosa, the separation between epidermis and dermis (Bökel and Brown, 2002; Devenport et al., 2007).
Integrins typically localize in highly organized structures at sites of transmembrane linkage. The best characterized of these linkages is the focal adhesion found on mammalian fibroblasts in tissue culture (Burridge et al., 1988). In tissues, small adhesion sites mature during development into stable hemiadherens junctions that connect epithelia to the basement membrane and into myotendinous junctions that connect the tips of striated muscles to the ECM. In striated muscles, actin filaments are anchored to myotendinous junctions and to Z lines, which border the smallest functional unit of muscles, the sarcomere (Clark et al., 2002). Z lines are laterally connected to the ECM by costameres (Garamvölgyi, 1965; Pardo et al., 1983; Ervasti, 2003). Connecting Z lines to other Z lines and to the surrounding connective tissue ensures synchronous, uniform muscle contraction. The Z line–costamere complex is morphologically similar to myotendinous junctions and contains many of the same proteins, among them integrins, which make the connection of the Z line to the ECM at the costamere (Pardo et al., 1983; Volk et al., 1990; Reedy and Beall, 1993; Ervasti, 2003). In mice, Drosophila melanogaster, and Caenorhabditis elegans, integrins are required for sarcomere assembly and Z line formation (Volk et al., 1990; Bloor and Brown, 1998; Schwander et al., 2003; Lecroisey et al., 2007).
Tissue culture studies have revealed a large number of proteins implicated in intracellular signaling and adaptor functions at focal adhesions (Zaidel-Bar et al., 2007). How integrin adhesion sites form in vivo, however, is complex and the set of molecules required is not well defined. One class of proteins often found at focal adhesions and at related structures, such as myotendinous junctions, is the LIM domain family (Kadrmas and Beckerle, 2004). Most proteins containing LIM domains, notably the paxillin and zyxin families, have been implicated in cell adhesion and integrin signaling (Kadrmas and Beckerle, 2004). In contrast, the Alp/Enigma family, which is a group of proteins defined by an amino-terminal PDZ domain and one or three carboxyl-terminal LIM domains, maintains actin anchorage at the Z line of muscle cells together with α-actinin (Clark et al., 2002; te Velthuis et al., 2007). α-Actinin is a major component of Z lines. It cross-links antiparallel actin filaments from opposite sarcomeres. Flies lacking α-actinin die as first-instar larvae because of defects in Z line maintenance, yet they initially form normal striated muscles (Fyrberg et al., 1998; Dubreuil and Wang, 2000). Alp/Enigma family proteins cooperate with α-actinin in Z line maintenance. Alp and Cypher/Z band alternatively spliced PDZ-motif protein (ZASP), the best characterized members of the family, colocalize with α-actinin at Z lines and their PDZ domain directly interacts with the carboxyl terminus of α-actinin (Xia et al., 1997; Faulkner et al., 1999; Pomies et al., 1999; Zhou et al., 1999; Klaavuniemi et al., 2004). Mutations in Alp and Cypher/ZASP demonstrate their function in Z line maintenance. Mice that lack Alp or Cypher function develop fragmented Z lines and cardiomyopathy or congenital myopathy, respectively (Pashmforoush et al., 2001; Zhou et al., 2001). Likewise, mutations in ZASP, the human Cypher orthologue, result in dilated cardiomyopathy (Vatta et al., 2003; Arimura et al., 2004).
In this study, we analyze Zasp, the only member of the Alp/Enigma family in D. melanogaster, and identify novel roles for Alp/Enigma family proteins, both in tissue culture cells and flies. We show that Zasp is required for the formation of three different integrin adhesion sites: integrin adhesion sites in tissue culture and Z lines and functional myotendinous junctions in muscles.
Results
S2R+ cells are a model system to study integrin-mediated adhesion
To identify novel genes involved in integrin-mediated cell adhesion and spreading, we investigated two D. melanogaster cell lines, S2 and S2R+ cells, for their ability to uncover such genes using RNAi. Both cell lines are believed to be derived from embryonic hemocytes but exhibit differences in their ability to spread on substrates (Schneider, 1972; Yanagawa et al., 1998). In routine culture conditions, S2 cells are small and spherical, which is typical of unspread cells. However, they can spread when plated on the lectin concanavalin A. Concanavalin A–induced spreading is controlled by remodeling of the actin cytoskeleton upon binding of lectins to the polysaccharide side chains of plasma membrane proteins and lipids (Rogers et al., 2003). In contrast, S2R+ cells are large, flat, and strongly adherent even in the absence of concanavalin A or any other externally supplied ECM substrate. Incubation of S2R+ cells with mys (encoding βPS integrin) double-stranded RNA (dsRNA) disrupts cell spreading and causes rounding up, indicating that this ability to spread is integrin dependent (Kiger et al., 2003). We therefore sought to test whether S2 and S2R+ cells can be used to differentiate lectin-mediated cell spreading from integrin-mediated cell spreading.
As both cell lines express βPS integrin, we first assessed its subcellular localization in spreading S2R+ and S2 cells. In S2R+ cells stained with anti–βPS integrin antibody, we observed integrin staining typical of integrin adhesion sites, with bright foci along the cell edge and streaks in areas of potentially increased local forces (Fig. 1 A). In contrast, S2 cells spread on concanavalin A do not exhibit these integrin adhesion sites (Fig. 1 B). Instead of distinct foci and streaks, βPS integrin is exclusively localized intracellularly, most likely because S2 cells do not express the αPS1 and αPS2 integrin subunits (Gotwals et al., 1994), and without a heterodimerization partner, βPS integrin does not translocate to the plasma membrane. To further support the notion that we observed functional adhesion sites in S2R+ cells, we stained them with antibodies against other markers commonly found in focal adhesions, such as talin, vinculin, paxillin, and phosphotyrosine. In each case, we observed similar clusters at the cell edge in S2R+ cells but not in S2 cells (Fig. 1, C–F; and not depicted). We then determined whether talin, vinculin, and integrin localize to the same clusters. We observed colocalization of vinculin and αPS2 integrin as well as that of talin and βPS integrin (Fig. 1, G–L). These observations point to a major role for integrins in S2R+ cell spreading and indicate that S2 cells spread on concanavalin A by an integrin-independent mechanism.
Figure 1.
D. melanogaster S2R+ cells exhibit integrin adhesion sites. Integrins cluster in adhesion sites in S2R+ cells. (A–F) Anti–βPS integrin antibody staining of S2R+ (A) and S2 (B) cells spread on concanavalin A. βPS integrin localizes to bright foci and streaks at the cell edge of S2R+ cells. We observed similar clustering with anti–C. elegans vinculin antibody staining (C) and anti-talin antibody staining (E) in S2R+ but not in S2 cells (D and F). (G–I) Colocalization of anti–C. elegans vinculin (G) and anti–αPS2 integrin (H) antibody staining at the cell edge of S2R+ cells. Merge is shown in I. (J–L) Colocalization of anti-talin (J) and anti–βPS integrin (K) antibody staining. Merge is shown in L. Indicated areas are shown enlarged on the right. Bar, 15 μm.
We next tested if this difference in spreading can be exploited to screen for novel regulators of integrin-mediated cell spreading. We therefore compared cell spreading of S2 cells on concanavalin A and S2R+ cells in the absence of Abi, a known regulator of cytoskeletal remodeling that acts through SCAR, and rhea (encoding talin), the major linker of integrins to the actin cytoskeleton (Brown et al., 2002; Kunda et al., 2003; Rogers et al., 2003; Ginsberg et al., 2005). Although RNAi with Abi causes spreading defects, which are characterized by a star-shaped morphology, in both S2 and S2R+ cells, RNAi with rhea results in cell spreading defects exclusively in S2R+ cells (Fig. 2, A–F). The exclusive spreading defects of S2R+ cells in the absence of talin indicate that we can screen for novel genes involved in integrin regulation by comparing S2R+ and S2 cell spreading and that S2R+ cells are a suitable model system to study integrin-mediated adhesion.
Figure 2.
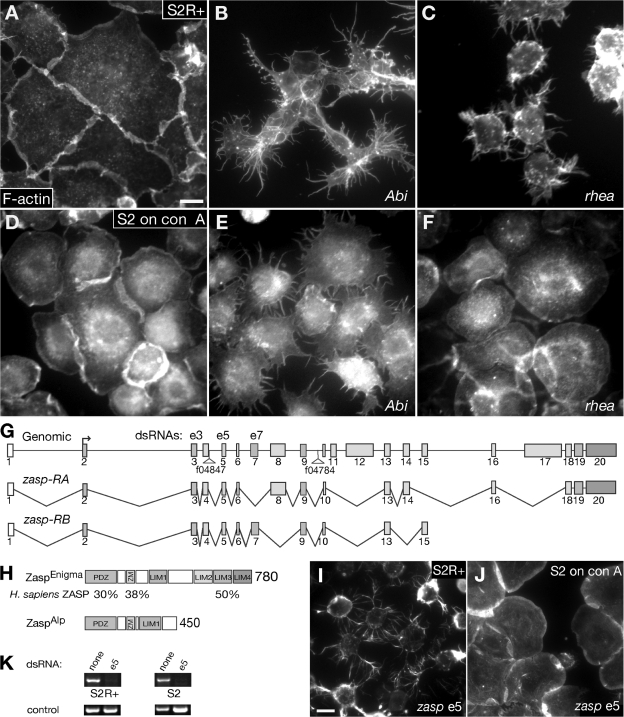
Zasp is required for integrin-dependent spreading of S2R+ cells. (A–F) S2R+ cells spreading without addition of external ligand (A–C) and S2 cells spreading on concanavalin A (D–F). A and D, no RNAi treatment; B and E, treatment with Abi RNAi; C and F, treatment with rhea (talin) RNAi. Cells were stained with Alexa 594–phalloidin for filamentous actin. Abi-depleted cells show star-shaped phenotypes in both cell lines (B and E), whereas talin depletion results in rounding up only in S2R+ cells (C). (G) Schematic presentation of the zasp gene. Translated exons are shown in gray and untranslated exons in white. piggyBac insertions used to generate the zaspΔ deletion and dsRNAs used to target the zasp gene are indicated. Only the two major splice variants are shown. (H) The D. melanogaster zasp gene encodes two major proteins. ZaspEnigma is the Enigma-like protein and ZaspAlp is the Alp-like protein. Numbers represent the amino acid length of each protein. Below three conserved domains, we show the percent identity between Zasp and its human orthologue. (I and J) zasp exon 5 RNAi targeting zasp-RA and zasp-RB. Cells are stained with Alexa 594–phalloidin to visualize the actin cytoskeleton. (I) S2R+ cells round up and exhibit many filopodia-like processes. (J) S2 cells spread on concanavalin A show no phenotype. (K) RT-PCR analysis of zasp dsRNA-treated cells compared with untreated ones. zasp mRNA (153 bp exon 5 amplicon) is depleted in both S2R+ and S2 cells. Control PCR was done with primers against an untargeted gene. Bars, 15 μm.
We then conducted a pilot screen for novel genes required for cell spreading with 72 candidate genes selected from a genome-wide screen for cell shape changes in S2R+ cells (unpublished data). We selected candidate genes with phenotypes potentially related to cell spreading defects such as a round, star-shaped, or rough-edged cell shape. Parallel RNAi treatment of S2 and S2R+ cells uncovered 12 genes that show an exclusive phenotype in S2R+ cells, suggesting they are specifically involved in integrin-mediated processes and not in general cytoskeletal remodeling or cell viability (Tables S1 and S2 and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200707045/DC1). As expected, this group contains the genes encoding βPS integrin and αPS2 integrin and genes with a well-characterized role in regulating integrin adhesion like talin and Rap1 GTPase. Among the novel genes we identified, we chose to focus on CG30084.
The Alp/Enigma family has a single member in D. melanogaster
CG30084 represents the single member of the D. melanogaster Alp/Enigma family of PDZ-LIM domain proteins. We named CG30084 zasp because the major predicted splice variant encodes a protein highly similar to human ZASP (Fig. 2 H; Faulkner et al., 1999). In C. elegans, the Alp and Enigma subfamilies are encoded as splice variants of a single gene (McKeown et al., 2006). To test if zasp encodes both Alp and Enigma splice variants, we sequenced 21 ESTs (Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200707045/DC1). Together with data from the Berkeley D. melanogaster Genome Project, we predict that zasp contains 20 exons and is transcribed into two major transcripts (Fig. 2, G and H). The majority of ESTs (20/29) encode Enigma-like proteins (zasp-RA). We also found three ESTs corresponding to the Alp subfamily (zasp-RB, available from GenBank/EMBL/DDBJ under accession no. EF221635). LIM1 is truncated in the Enigma-like protein ZaspEnigma, most likely resulting in only three functional LIM domains as in vertebrate Enigma proteins. Our sequence data confirm the predictions of a recent bioinformatics analysis (te Velthuis et al., 2007). LIM1 is most closely related to the LIM domain of the Alp subfamily, whereas LIM2–4 are most closely related to the LIM domains of the Enigma subfamily (te Velthuis et al., 2007).
Loss of Zasp disrupts integrin adhesion sites in S2R+ cells
Treating S2R+ cells with zasp dsRNA targeting exon 5, which depletes ZaspAlp and ZaspEnigma, results in severe spreading defects often associated with the formation of filopodia-like processes (Fig. 2 I). S2R+ cells round up similar to mys- or rhea-depleted cells. In contrast, we observed no phenotype in S2 cells (Fig. 2 J). The absence of zasp mRNA after dsRNA treatment in both cell types was verified by RT-PCR (Fig. 2 K). We observed an identical phenotype with dsRNA targeting exon 3, confirming the specificity of the spreading defect (unpublished data).
We next determined if integrin adhesion sites are affected in Zasp-depleted S2R+ cells. zasp exon 3 and exon 5 dsRNA-treated S2R+ cells show no integrin adhesion sites (unpublished data). Treating S2R+ cells with zasp exon 7 dsRNA, which only depletes ZaspAlp, strongly impairs integrin adhesion sites. In severe cases, integrin adhesion sites are either completely absent or hardly visible (Fig. 3 B). Loss of integrin adhesion sites is also observed in Zasp-depleted cells that are partially spread, indicating that this defect is not secondary to changes in cell shape (Fig. 3 C).
Figure 3.
Zasp localizes to integrin adhesion sites and Zasp depletion disrupts integrin adhesion sites. (A–C) Anti–βPS integrin antibody (green) and Alexa 594–phalloidin (red) costaining of wild-type S2R+ cells or cells treated with zasp exony 7 dsRNA (B and C). Typically, integrin adhesion sites are very small or absent (B). In milder cases, integrin adhesion sites are reduced in number and the cell retracts its edge between two integrin adhesion sites (C). (D–I) Anti–βPS integrin antibody (green) and anti-Zasp antibody (red) coimmunostaining of S2R+ (D–F) and S2 (G–I) cells spread on concanavalin A. Zasp colocalizes with βPS integrin in foci and streaks in S2R+ cells (D–F). Bar, 15 μm.
Finally, as loss of Zasp disrupts integrin adhesion sites, we wanted to know if Zasp colocalizes with βPS integrin in integrin adhesion sites. For this purpose, we raised an antibody against ZaspEnigma, which recognizes both ZaspAlp and ZaspEnigma. Like βPS integrin, Zasp localizes to the same bright foci and streaks at the cell edge (Fig. 3, D–F). Furthermore, the absence of any localized Zasp staining in S2 cells spread on concanavalin A (Fig. 3, G–I) argues that functional integrin heterodimers are required to localize Zasp in S2R+ cells. The distinct spreading defects in the absence of Zasp and its localization in integrin adhesion sites indicate that Zasp is a novel regulator of integrin-mediated cell spreading.
Zasp mRNA and protein expression is similar to that of βPS integrin
In mammals, most Alp/Enigma family members function in Z line maintenance (Xia et al., 1997; Faulkner et al., 1999; Pomies et al., 1999; Zhou et al., 1999, 2001; Pashmforoush et al., 2001). To learn more about Zasp function and its potential involvement in the formation of integrin adhesion sites, we investigated its expression profile during D. melanogaster embryogenesis. mRNA and protein expression largely overlap (Fig. 4). Preblastoderm-stage embryos show weak staining, indicating a maternal contribution (Fig. 4, A and B). Zygotically, Zasp is expressed in areas where βPS integrin is known to function (Hutson et al., 2003; Devenport and Brown, 2004; Narasimha and Brown, 2004), e.g., the leading edge during dorsal closure and the midgut during midgut fusion (Fig. 4, C–H). At late stages, Zasp expression is particularly strong in mesodermal tissues such as visceral, pharyngeal, and somatic muscles (Fig. 4, I and J).
Figure 4.
Zasp protein and mRNA expression patterns overlap during embryogenesis. RNA in situ hybridization with zasp full-length antisense mRNA (left) and anti-Zasp (right) antibody staining. (A) A low level of zasp mRNA, likely the maternal contribution, is visible in preblastoderm-stage embryos. (B) Zasp protein is also detected in a preblastoderm-stage embryo. (C and D) Zygotic mRNA and protein expression is first detected in the proctodeum (pr) and the midgut primordium (pm). (E and F) In stage-11 embryos, zasp mRNA and Zasp protein expression is predominant in the leading edge (le) of epidermal cells adjacent to the amnioserosa (as). (G) Dorsal view of a stage-12 embryo reveals mRNA localization in the midgut (mg) and in the leading edge. (H) Zasp protein is expressed in several rows of germ band cells next to the leading edge at stage 14. (I) Strong mRNA expression is visible in the midgut and pharyngeal muscles (phm) in a dorsal view of a stage-17 embryo. (J) Zasp protein expression is additionally visible in somatic muscles (sm) and visceral mesoderm (vm). Bars, 50 μm.
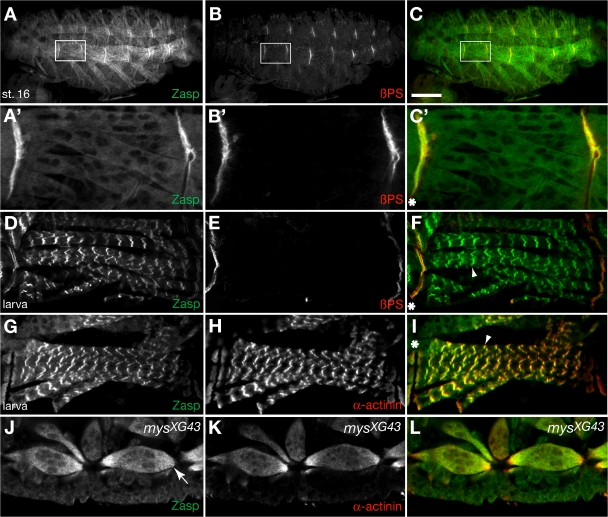
D. melanogaster αPS2 and βPS integrin subunits are enriched at myotendinous junctions where they function in the adhesion of muscles to the tendon matrix (Leptin et al., 1989; Brabant and Brower, 1993; Brown, 1994). Strikingly, Zasp is also enriched at myotendinous junctions (Fig. 5 A). Coimmunostaining with anti-Zasp and anti–βPS integrin antibodies reveals that Zasp tightly colocalizes with βPS integrin at myotendinous junctions in the embryo (Fig. 5, A′–C′). In larvae, Zasp still localizes to myotendinous junctions but facing the cytoplasmic side of βPS integrin (Fig. 5, compare D and E). Finally, the diffuse muscle staining in embryos is refined into the specific localization of Zasp into a repetitive line pattern (Fig. 5 D). To see if these lines correspond to Z lines, we analyzed Zasp distribution in relation to α-actinin, a well-known marker of Z lines (Saide et al., 1989). Zasp and α-actinin tightly colocalize at myotendinous junctions from stage 16 onward and later in Z lines, confirming that Zasp localizes to Z lines (Fig. 5, G–I; and not depicted). We also analyzed expression and subcellular localization of Zasp with an endogenous homozygously viable GFP-Zasp fusion (G00189; Morin et al., 2001). We verified the fusion by RT-PCR and dsRNA injection into embryos. Live imaging of GFP-Zasp shows the same localization as antibody staining, confirming the specificity of our antibody (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200707045/DC1).
Figure 5.
Zasp colocalizes with integrins at myotendinous junctions during embryonic development and with α-actinin at muscle Z lines. (A–C) Anti-Zasp antibody staining (A), anti–βPS integrin antibody staining (B), and merge of a stage-16 embryo (C). Zasp and βPS integrin colocalize at myotendinous junctions. Indicated areas are shown enlarged in A′–C′. (D–F) Anti-Zasp antibody staining (D), anti–βPS integrin antibody staining (E), and merge (F) of a first-instar larva. Note the slightly wider gap of Zasp staining compared with that of βPS integrin staining at the myotendinous junction. (G–I) Anti-Zasp antibody staining (G), anti–α-actinin antibody staining (H), and merge (I) of a first-instar larva. Note the tight colocalization of Zasp and α-actinin at myotendinous junctions and Z lines. (J–L) Anti-Zasp antibody staining (J), anti–α-actinin antibody staining (K), and merge (L) of a zygotic mysXG43 mutant embryo. Zasp and α-actinin no longer localize at the termini of detached muscles (J, arrow). Asterisks indicate myotendinous junctions. Arrowheads indicate Z lines. Bar, 50 μm.
To determine if integrins recruit Zasp to myotendinous junctions, we examined Zasp distribution in zygotic mysXG43 mutant embryos lacking βPS integrin. Zasp and α-actinin no longer localized to the tips of detached muscles, or did so only in a weak gradient (Fig. 5, J–L). In a mysXG43 maternal and zygotic mutant, Zasp is completely unlocalized (unpublished data), indicating that Zasp and α-actinin are recruited to myotendinous junctions by integrins. The expression data suggest that Zasp functions together with integrins and α-actinin in several morphogenetic processes.
zaspΔ mutants die as first-instar larvae
To investigate if Zasp indeed functions in the formation of integrin adhesion sites, we made a zasp mutant by recombining two flippase recognition target–bearing piggyBac elements inserted in introns four (f04847) and nine (f04784) of zasp (Fig. 2 G). We recovered three identical lethal lines deleting exons 5–9 and resulting in a frameshift. The deletion lines, whose identity was verified by PCR and Southern blotting, remove all major splice variants (Figs. 2 G and 6 A, Table S3, and not depicted). They have no zasp mRNA expression as shown by RT-PCR and RNA in situ hybridization with a full-length probe (Fig. 6, B–D). In addition, Zasp protein is absent in stage-17 zygotic mutants (Fig. 6, E and F). Finally, zasp mutants that are transheterozygous over a large deficiency show the same phenotypes as zasp homozygotes even though we observed a higher embryonic lethality (not depicted and Fig. 6 G). The combined data show that we created a zasp hypomorph, which we refer to as zaspΔ.
Figure 6.
zasp mutant embryos die as first-instar larvae. (A) Putative deletion lines were screened for the presence of residual piggyBac elements by means of PCR, using transposon-specific primers (RB(WH+) reverse and forward) in combination with genome-specific primers (exon 3 forward and 9 reverse). Amplification of both PCR products indicates the presence of both residual piggyBac elements and therefore a recombination and deletion event. Genomic DNA extracted from control flies (C, pBac{WH}f04847) is only amplified with the e3forward/RBreverse primers. 1- and 2-kb size markers are indicated. (B) Absence of zasp mRNA in two deletion lines was verified by RT-PCR using primers CACCATGGCCCAACCACAGCTGCTG and GCGCGCGTGATTCTTGCAG. Amplification of a 2.1-kb band (asterisk) is detected only in wild-type embryos (wt). (C and D) RNA in situ hybridization with a full-length antisense probe demonstrates absence of zasp mRNA in zaspΔ mutant embryos (D). (E and F) Anti-Zasp antibody staining reveals no obvious Zasp protein in zaspΔ mutant embryos (F). (G) Stage of death of zaspΔ mutants. (H) Developmental stage of zaspΔ mutant larvae was determined by the number of teeth on the mouth hooks, which increase with instar. Mouth hooks of zaspΔ9 mutant larvae (iv) look like those of first-instar wild-type larvae (i). Bar, 50 μm.
The majority of zaspΔ mutants die as first-instar larvae and 12% die as embryos. Most zaspΔ mutant larvae die within the first 24 h, coinciding with the onset of muscle contractility. Even though some larvae can live longer, they do not progress beyond the first-instar stage as shown by their small size and mouth hook morphology (Fig. 6 H).
Zasp is required for sarcomere assembly and recruits α-actinin to the Z line
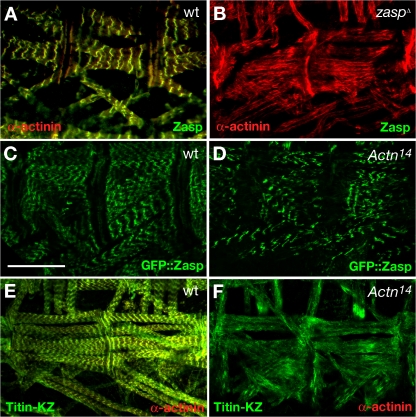
As muscle contractions were slower in zaspΔ mutant larvae than in wild-type larvae, we investigated sarcomere assembly in late–stage 17/first-instar larvae by comparing the actin organization in muscles of wild-type and zaspΔ mutants. zaspΔ mutants have lost the typical striated muscle pattern, indicating a Z line defect (Fig. 7, A and B). In mammalians, the main interaction partner of Zasp at the Z line is α-actinin. We therefore wanted to know if D. melanogaster Zasp interacts biochemically with α-actinin. Immunoprecipitation of wild-type extracts with anti-Zasp antibody pulls down α-actinin (Fig. 7 C). In contrast, immunoprecipitation of extracts from zaspΔ larvae or with preimmune serum does not pull down α-actinin, demonstrating the specificity of this interaction. Intriguingly, well-characterized α-actinin–null mutants, such as Actn14, still develop a striated muscle pattern and Z lines (Fyrberg et al., 1990, 1998; Dubreuil and Wang, 2000), indicating that Zasp plays a more important role in Z line assembly than α-actinin. α-Actinin has no maternal contribution (Perrimon et al., 1985); therefore, Actn-null mutant larvae should correspond to a complete loss of function. To better characterize these differences, we investigated sarcomere and Z line ultrastructure of wild-type, zaspΔ, and Actn14 mutant larvae by transmission electron microscopy (Fig. 7, D–G). In freshly hatched wild-type larvae, Z lines can be readily observed and are spaced at regular intervals (Fig. 7 D). In 1-d-old α-actinin mutant larvae, we still observed Z lines spaced at regular, but wider intervals than in the wild type, possibly because of detachment of actin fibers from the Z line (Fig. 7 E). These phenotypes correspond to previous observations (Fyrberg et al., 1998). In contrast, in freshly hatched zaspΔ mutant larvae, Z lines are either completely absent with only occasional accumulations of electron-dense material (Fig. 7 F) or Z line remnants are severely disorganized and irregularly spaced (Fig. 7 G). In addition, filaments are disorganized and no longer arranged in parallel arrays (Fig. 7, F and G).
Figure 7.
zaspΔ mutants develop no striated muscles and Z lines. (A) Late–stage 17 wild-type embryo stained with phalloidin to visualize actin fibers. The striated muscle pattern, which indicates sarcomere differentiation, is clearly visible (arrowhead). A sarcomere extends from the center of one block of actin staining to the next. (B) Late–stage 17 zaspΔ mutant embryo stained with phalloidin. No striation is evident. (C) Zasp physically interacts with α-actinin in larvae. Immunoprecipitation (IP) was conducted with preimmune serum or anti-Zasp antibody using wild-type and zaspΔ9 mutant larvae. Detection was performed by Western blotting (WB) with anti– α-actinin antibody. Only immunoprecipitation with anti-Zasp antibody with wild-type larvae coprecipitates α-actinin. Molecular mass is indicated in kilodaltons. (D–G) Ultrastructural analysis of first-instar larval sarcomeres and Z lines by electron microscopy. (D) Newly hatched wild type. (E) 1-d-old first-instar Actn14 mutant. (F and G) Newly hatched zaspΔ mutant. Black arrowheads indicate Z lines, Z line remnants, or electron-dense material. Bars: (A and B) 50 μm; (D–G) 1 μm.
To determine the mechanism of Zasp function at Z lines, we stained with anti–α-actinin antibody. zaspΔ mutant larvae lost α-actinin localization specifically at muscle Z lines, showing that Zasp is required to localize α-actinin to Z lines (Fig. 8, A and B). If Zasp is upstream of α-actinin, Zasp should still localize to Z lines in α-actinin mutants. In Actn14 larvae, which is a null allele (Fyrberg et al., 1990; Dubreuil and Wang, 2000), GFP-Zasp indeed still localizes to Z lines in a manner very similar to its wild-type distribution (Fig. 8, C and D). We finally tested the distribution of titin, which binds to α-actinin and has been proposed to form a ternary complex together with Zasp (Sorimachi et al., 1997; Young et al., 1998; Au et al., 2004). Titin no longer localizes to Z lines in Actn mutants (Fig. 8, E and F). All these phenotypes are completely penetrant and demonstrate that Zasp is required for Z line assembly.
Figure 8.
zaspΔ mutants do not recruit α-actinin to Z lines. (A) Wild-type larva stained with anti-Zasp antibody (green) and anti–α-actinin antibody (red). (B) In zaspΔ mutant larvae, α-actinin fails to localize to Z lines and instead appears to localize along the length of actin filaments. (C) Localization of endogenous GFP-Zasp (G00189) in fixed wild-type first-instar larva. (D) GFP-Zasp still localizes to Z lines in an Actn14-null mutant larva. (E and F) Anti–titin-KZ/anti–α-actinin coimmunostaining of wild-type (E) and Actn14 (F) mutant first-instar larva. Titin does not localize to Z lines in the Actn mutant. Bar, 50 μm.
Zasp mediates muscle attachment together with integrins
We next analyzed zaspΔ mutants for muscle detachment. We observed muscle-attachment defects from stage-16 embryos onward, as seen by detachment of muscle fibers from myotendinous junctions (Fig. 9, A and B). We also saw muscle patterning defects with muscles missing in some segments (unpublished data). Late–stage 17 embryos occasionally exhibit strong muscle detachment and rounding up of muscles (Fig. 9 C). This complete muscle detachment is progressive, as we observed a penetrance of 12% (n = 190) round muscles when we fixed embryos at stage 17, but almost complete penetrance when we fixed nonhatched embryos 24 h later (98%; n = 112). The phenotype is also progressive in larvae, where we observed 17% round muscle phenotypes (n = 174) when larvae were fixed at the end of first instar. This phenotype is similar to that of embryos lacking zygotic βPS integrin (mysXG43; Fig. 9 D), although it occurs later. βPS integrin still localizes to myotendinous junctions in zaspΔ mutants even when muscles round up, showing that Zasp is not required for integrin localization (Fig. 9 C′).
Figure 9.
zaspΔ mutants have muscle-attachment defects. Stage-17 embryos are stained with an antibody against muscle myosin heavy chain (MHC; green) to visualize somatic muscles and an antibody against βPS integrin (red) to visualize myotendinous junctions. (A) Wild-type embryo. (B) zaspΔ mutant embryo with mild muscle detachment. (C) zaspΔ mutant embryo with severe muscle detachment. (D) mysXG43 mutant embryo shown for comparison. Indicated areas are shown enlarged in A′–D′. Arrows indicate detached muscles. Asterisks indicate myotendinous junctions. Bar, 50 μm.
If Zasp is a crucial component of integrin adhesion sites, zaspΔ should also genetically interact with integrins. The αPS2 integrin subunit is only expressed in mesodermal cells, but not in epidermal cells of the myotendinous junction (Bogaert et al., 1987). To test for a genetic interaction, we used a hypomorphic mutation in αPS2 integrin (ifSEF) that shows weak muscle-attachment defects only at the last stage of embryogenesis. In ifSEF mutants, βPS integrin localization to myotendinous junctions is lost at stage 17, corresponding to the onset of the phenotype (Bloor and Brown, 1998; Devenport et al., 2007; Fig. 10, A and B). However, stage-16 ifSEF embryos show normal βPS integrin localization and no muscle phenotype (Fig. 10, C and D). Intriguingly, ifSEF mutants lacking one copy of zaspΔ display a much more severe muscle-detachment phenotype and, additionally, the phenotype appears at an earlier stage when βPS integrin is still localized to myotendinous junctions (Fig. 10, E, F, and I). This genetic interaction is specific to zaspΔ because we observed no enhancement with either of the two piggyBac elements used to generate the zaspΔ mutation (Fig. 10, G–I; and not depicted). The strong genetic interaction we observed between Zasp and αPS2 integrin shows that they act together in maintaining muscle attachment.
Figure 10.
Zasp genetically interacts with integrins. To determine the stage at which muscle attachment fails, embryos were labeled with antibodies against muscle myosin heavy chain (MHC; green) and βPS integrin (red). Merge is shown in the left panels. The right panels show βPS integrin. (A and B) Stage-17 ifSEF embryo showing mild muscle detachment. (C and D) Stage-16 ifSEF embryo showing wild-type muscle organization. (E and F) Stage-16 ifSEF; zaspΔ9/+ embryo showing severe muscle detachment. (G and H) Stage-17 ifSEF; f04784/+ embryos showing mild muscle detachment identical to ifSEF embryos. (I) Percentage of embryos showing muscle detachment. ND, not determined. Arrows indicate detached muscles. Asterisks indicate myotendinous junctions. Bar, 50 μm.
Discussion
We identified a novel regulator of cell–matrix adhesion, Zasp, in an RNAi screen for integrin-dependent cell spreading. We propose that Zasp mediates two related functions, one upstream of α-actinin organizing the Z line and the other downstream of integrins regulating assembly of functional adhesion sites.
In our screen, we compare cell spreading of S2 and S2R+ cells. S2R+ cells are S2 cells that have acquired novel traits over time, such as the ability to spread without externally added ECM ligands (Yanagawa et al., 1998). For our purposes, the only relevant difference between these two cell lines is the absence of αPS2 integrin from S2 cells. S2 cells transfected with αPS2 integrin spread and grow like S2R+ cells and, conversely, upon RNAi-mediated integrin depletion, S2R+ cells can be grown for weeks and look like S2 cells (Gotwals et al., 1994; unpublished data). S2R+ cells most likely secrete their own ECM ligand, which is similar to what has been reported for human fibroblasts (Grinnell and Feld, 1979). A good candidate may be Tenascin-m, which is an αPS2βPS ligand (Graner et al., 1998) and which causes rounding up of S2R+ cells when depleted by RNAi (Kiger et al., 2003). We observed putative integrin adhesion sites in S2R+ cells that look like integrin adhesion sites and are composed of focal adhesion proteins like talin and vinculin. We were unable to observe actin fibers attached to these adhesion sites, probably because actin bundling in S2R+ cells is not sufficient to allow visualization by fluorescence microscopy. The colocalization of integrin with vinculin and talin, the disruption of these sites in mutants affecting cell spreading, and the absence of adhesion sites in S2 cells spread on concanavalin A strongly argue that we observed functional integrin adhesion sites.
Our pilot RNAi screen uncovered 12 genes that show exclusive phenotypes in S2R+ cells upon depletion. Five of these genes are known to function in cell–matrix adhesion either directly or by regulating mesodermal gene transcription or RNA processing, which validates our approach (Brown, 1994; Artero et al., 1998; Brown et al., 2002; Huelsmann et al., 2006). Several classical focal adhesion proteins, such as vinculin or FAK, were not included in the pilot screen because they did not show a phenotype in the genome-wide screen. This is not too surprising given that some of them also do not have identifiable phenotypes in vivo (Alatortsev et al., 1997; Grabbe et al., 2004). Still, we have probably missed several genes because of the high variability of cell shapes in S2R+ cells, which is also evident in the high number of candidates in which we could not reproduce the originally observed cell shape change.
Integrin adhesion sites in cell culture are considered to be precursors of adhesion complexes found in tissues such as myotendinous junctions. In D. melanogaster S2R+ cells, Zasp localizes to integrin adhesion sites. The functional importance of this localization is shown by the loss of integrin adhesion sites upon Zasp depletion and the concomitant failure of cell spreading. Collectively, these results suggest that Zasp functions in organizing or maintaining integrin adhesion sites to allow cell spreading. Our fly data are in agreement with this conclusion, as we observed colocalization of Zasp and βPS integrin at myotendinous junctions and muscle detachment in zaspΔ mutant embryos, again demonstrating that Zasp plays a crucial role in integrin-mediated adhesion. The muscle-detachment phenotype is weaker than the tissue culture phenotype. This may be because of the Z line defect, which precludes normal muscle contractility. Alternatively, the weaker detachment phenotype may be because of a maternal contribution of Zasp rescuing earlier muscle-attachment defects, or because the myotendinous junction contains more components and is therefore less easily disrupted. We also observed defects in other tissues that require integrin function, such as wing blisters in zaspΔ clones and a genetic interaction of Zasp and integrin during wing formation, confirming the general role of Zasp in integrin adhesion (unpublished data). Vertebrate and C. elegans Alp/Enigma proteins have no reported function at integrin adhesion sites, but they are known to localize to focal adhesions and myotendinous junctions (Pomies et al., 1999; Pashmforoush et al., 2001; Henderson et al., 2003; McKeown et al., 2006). In two other studies, the authors observed no colocalization with vinculin at focal adhesions; however, the studies were not conducted in muscle cells but rather in CHO cells, endothelial cells, and blood platelets (Bauer et al., 2000; Klaavuniemi et al., 2004). Zasp is the only member of the Alp/Enigma family of PDZ-LIM domain proteins in D. melanogaster, whereas there are seven members of that family in vertebrates (Kadrmas and Beckerle, 2004). The lack of data for a function of vertebrate PDZ-LIM family members in integrin adhesion is therefore most likely owing to genetic redundancy.
Several lines of evidence argue that Zasp acts as a cytoskeletal adaptor downstream of integrins. First, Zasp is recruited to integrin adhesion sites in S2R+ cells and is recruited by βPS integrin to myotendinous junctions in the embryo. Second, even in zaspΔ embryos with strong muscle detachment, βPS integrin still localizes to myotendinous junctions or muscle tips. Third, during larval stages we observed Zasp on the cytoplasmic side of integrins.
We suggest that Zasp regulates or strengthens the link of integrins to the actin cytoskeleton after the initial attachment of integrins to actin via talin. Two lines of evidence support this proposal. First, the zasp muscle-attachment defect is weaker than a complete loss of function of βPS integrin or talin and is most similar to mutants in factors linking integrin to the cytoskeleton, like integrin-linked kinase (Zervas et al., 2001). Second, we observed muscle detachment after muscle contractions began in late–stage 17 embryos and larvae. The strongest evidence that Zasp plays a crucial role in the assembly of functional integrin adhesion sites is the strong genetic interaction between αPS2 integrin and Zasp.
Related to Zasp's function at integrin adhesion sites, Zasp also organizes the Z line. There is ample evidence of α-actinin binding and cross-linking actin, and it has been established that Alp/Enigma proteins directly bind α-actinin (Faulkner et al., 1999; Zhou et al., 1999; Pashmforoush et al., 2001). We also observed a physical interaction of Zasp and α-actinin in D. melanogaster, but our data imply a considerably larger role for Zasp in establishing Z line structure and function than was previously appreciated. Our data indicate that Zasp acts upstream and recruits α-actinin because in the absence of Zasp, α-actinin no longer localizes to Z lines. In contrast, Zasp still localizes to Z lines in α-actinin–null mutants. Titin, which is anchored to Z lines by directly binding α-actinin (Ohtsuka et al., 1997; Sorimachi et al., 1997; Young et al., 1998), is not present at Z lines in Actn mutants, confirming the previously published biochemical data. Molecular modeling recently predicted a ternary complex of Zasp, α-actinin, and titin, with Zasp and titin binding to different surfaces of α-actinin (Au et al., 2004). Our data are in agreement with such a complex and indicate that Zasp is the most upstream component of the complex. Recruiting α-actinin to the Z line cannot be Zasp's only function because a well-characterized α-actinin–null mutant still shows Z lines (Fyrberg et al., 1998; Fig. 7 E), which are disrupted in zaspΔ mutant larvae. We suggest that this additional function of Zasp is again dependent on integrins. Integrins connect Z lines laterally to the ECM surrounding muscle fibers and are required for sarcomere assembly (Volk et al., 1990; Schwander et al., 2003; Lecroisey et al., 2007).
Mutations in PDZ-LIM family members cause myopathies. Our work suggests that other mutations in members of the PDZ-LIM family should also be involved in integrin-related diseases in vertebrates.
In conclusion, we have identified a novel regulator of integrin function that plays a crucial role in assembling integrin adhesion sites.
Materials and methods
Tissue culture and RNAi
D. melanogaster S2 and S2R+ cells were treated with dsRNAs as previously described (Kiger et al., 2003; Rogers et al., 2003). For dsRNA synthesis, T7-flanked PCR products (provided by N. Perrimon, Harvard Medical School, Boston, MA) were used as templates for in vitro transcription with T7 RNA polymerase using the MEGAscript kit (Ambion). For zasp, the following additional primers flanked by the T7 promoter (TAATACGACTACTATAGGGAGA) were used for PCR followed by dsRNA synthesis: exon 3 (263 bp) CGCTGCACTGTGATGACAAA and GCCCAACCACAGCTGCTGCAA; and exon 5 (153 bp) CCGAGCACACCGCCAGCCAA and CAACGCGGCCCGTCCCTTCTC. Cells were incubated with dsRNA for 5–7 d before being harvested for microscopy analysis, RT-PCR, and Western blotting. Control experiments were performed in parallel without the addition of dsRNA.
Before fixation, dsRNA-treated S2R+ cells were replated on glass slides (VWR) and allowed to spread for 4 h, whereas dsRNA-treated S2 cells were replated on glass slides coated with concanavalin A and were allowed to spread for 2 h. Glass slides were coated for 30 min with 0.5 mg/ml concanavalin A, and then air dried for another 30 min before the addition of S2 cells. Cells were then fixed in 4% formaldehyde in PBS for 20 min.
After being permeabilized in 1× PBS containing 0.1% Triton X-100 for 3 min, cells were washed twice in PBT (PBS containing 0.05% Tween 20), followed by incubation in blocking solution containing 1% BSA in 1× PBS for at least 30 min at room temperature, and then stained with the primary antibody overnight at 4°C. After washing with PBT with several changes over 30 min, cells were incubated with the secondary antibody for 45 min at room temperature. After several washes, the glass slides were mounted in Prolong Gold antifade (Invitrogen). For RT-PCR, mRNA was isolated using Trizol (Invitrogen) according to the manufacturer's instructions. Gene-specific cDNAs were reverse transcribed and amplified with the same set of primers used previously for dsRNA synthesis.
Generation of anti-Zasp antibody
We prepared a rabbit polyclonal antibody using a 6× His-tagged fusion protein corresponding to ZaspEnigma. cDNA was amplified from EST RH03424 as a template with CACCATGGCCCAACCACAGCTGCTG and GCGCGCGTGATTCTTGCAG as primers and cloned into the Gateway pENTR/D-TOPO vector (Invitrogen). Recombination between the entry clone and the Gateway pDEST17 destination vector generated expression clones, which we transformed into TOP10 competent cells. Expression was induced in the presence of 0.2% l-arabinose. We purified the recombinant protein under denaturing conditions on Ni2+-affinity columns (QIAGEN) according to the manufacturer's instructions. We tested antibody specificity by Western blotting and immunofluorescence detection comparing wild-type, G00189, and zaspΔ embryos.
Immunoprecipitation assay
The indirect immunoprecipitation strategy was used by covalently binding the anti-Zasp antibody to AffiPrep protein A–Sepharose beads (Bio-Rad Laboratories) using dimethylpimelimidate as a cross-linker. 10 μl of anti- Zasp serum or rabbit preimmune serum were incubated with 110 μl of beads for 2 h at room temperature. Previous to treatment with dimethylpimelimidate, the beads were washed in 0.2 M sodium borate, pH 9. The cross-linking reaction was stopped by washing the beads with 0.2 M ethanolamine/0.2 M NaCl, pH 8.5, for 1 h at room temperature. First-instar larvae extracts were incubated with 50 μl of conjugated beads for 4 h at 4°C. They were rinsed, followed by three washes with the lysis buffer (50 mM Hepes, pH 7.4, 1 mM EGTA, 1 mM MgCl2, 300 mM KCl, 0.05% NP-40, 0.5 mM DTT, 10% glycerol, and one tablet of EDTA-free complete protease inhibitor cocktail). Beads were eluted by heating in 2× sample elution buffer (2% SDS, 62.5 mM Tris, pH 6.8, and 10% glycerol) without DTT for 10 min at 50°C to avoid IgG contamination. 1/10 of the supernatant was resolved by 8% SDS-PAGE and blotted on Hybond-C extra nitrocellulose membrane (GE Healthcare) for detection with anti–α-actinin monoclonal antibody (1:20). Anti–mouse IgG horseradish peroxidase–linked secondary antibody (1:2,500) was used together with the ECL detection kit for visualization (GE Healthcare).
Histochemistry and microscopy
RNA in situ hybridization of embryos was performed with digoxigenin-labeled RNA probes, made by in vitro transcription of RH03424 with T3 and T7 RNA polymerase yielding full-length antisense and sense probes, respectively. Embryos mutant for zasp were identified by the absence of staining with the antisense probe in ∼25% of an unsorted collection. We also sorted homozygous zaspΔ mutant embryos by the absence of the GFP balancer CyO, twi-Gal4 UAS-2xEGFP. In this case we hybridized green and nongreen embryos in parallel and observed a signal only in green embryos. Images were obtained on a stereomicroscope (MZ16-FA; Leica) using a Plan Apo 2.0× objective with a digital camera (Qicam) and OpenLab software (Improvision).
Embryos and larvae were fixed using heat fixation (Tepass, 1996). In brief, embryos were dechorionated in 50% bleach for 90 s, rinsed in water, immersed in boiling 1× embryonic wash buffer (70 mM NaCl and 0.05% Triton X-100) for 10 s, immediately cooled by adding 3 vol of ice-cold embryonic wash buffer, and placed on ice for 30 min. Embryos were devitellinized in methanol/heptane.
Primary antibodies used were the following: mouse anti–α-actinin (1:10; provided by J. Saide, Boston University School of Medicine, Boston, MA; Saide et al., 1989), rat anti-αPS2 integrin (1:10; 7A10; provided by N.H. Brown and G. Tanentzapf, Gurdon Institute, University of Cambridge, Cambridge, UK; Bogaert et al., 1987), mouse anti–βPS integrin (1:10; CF.6G11; obtained from Developmental Studies Hybridoma Bank; Brower et al., 1984), mouse anti-paxillin (1:50; 165; obtained from BD Biosciences), rabbit anti-talin (1:100; provided by N.H. Brown; Brown et al., 2002), rat anti–D-titin-KZ (1:500; provided by D. Andrew, Johns Hopkins University School of Medicine, Baltimore, MD; Machado et al., 1998), rabbit antimuscle myosin heavy chain (1:400; provided by D. Kiehart, Duke University, Durham, NC; Kiehart and Feghali, 1986), mouse antiphosphotyrosine (1:200; 4G10; obtained from Millipore), mouse anti–C. elegans vinculin (1:10; MH24; obtained from Developmental Studies Hybridoma Bank; Francis and Waterston, 1985), and rabbit anti-Zasp (1:300 for cells; 1:400 for embryos). Fluorescently labeled secondary antibodies of the Alexa series (Invitrogen) were used at a 1:300 dilution.
Filamentous actin was visualized with Alexa 488– or Alexa 594–labeled phalloidin (1:50 for tissue culture and 1:200 for embryos; Invitrogen). Embryos were devitellinized by hand.
After washing for 1 h in PBT, embryos were preincubated for 1 h in PBT containing 5% normal goat serum (PBTN), followed by an overnight incubation at 4°C with primary antibody, which was diluted in PBTN containing 0.1% BSA. After a 1-h wash in PBT and preincubation in PBTN for 1 h, embryos were incubated in secondary antibody for 2 h at room temperature and embedded in Prolong Gold antifade solution after several washes in PBT.
Tissue culture images were obtained on an upright microscope (DM6000B; Leica) using a 63× 1.4 NA oil objective (HCX PL APO CS) with a digital camera (Orca-ER; Hamamatsu) and OpenLab software. Embryo images were obtained on a confocal microscope (LSM510 Meta; Carl Zeiss, Inc.) using a 40× 1.3 NA (Plan-Neofluar) or a 63× 1.4 NA (Plan Apo) oil objective and processed using ImageJ (National Institutes of Health) and Photoshop (Adobe). Live imaging was done as previously described (Schöck and Perrimon, 2003). All images were obtained at room temperature.
Electron microscopy
First-instar larvae were fixed in 5% glutaraldehyde and 0.1 M cacodylate buffer, pH 7.4, for 1 h at room temperature. After cutting off the extremities, the specimens were transferred into fresh 2.5% glutaraldehyde and 0.1 M cacodylate buffer and fixed for another 5 h at 4°C. Larvae were postfixed in 1% osmium tetroxide for 2 h at 4°C. The samples were washed with several changes in 0.1 M cacodylate buffer for 40 min at 4°C, followed by staining with 2% tannic acid in 0.1 M cacodylate buffer for 1 h at 4°C. After several washes in distilled water, the specimens were restained with 2% uranyl acetate for 1 h at 4°C and washed in distilled water for 45 min at 4°C, followed by dehydration in acetone at room temperature. Samples were embedded in epoxy resin (EPON-815; Electron Microscopy Sciences) and cut with an Ultracut AV (Reichert). Sections were then stained in 7% uranyl acetate in absolute methanol for 4 min followed by incubation in Reynolds' lead citrate for 4 min and examined on a transmission electron microscope (Tecnai 12; FEI).
Fly stocks and genetics
We used the following stocks: mysXG43 and ifSEF, obtained from N.H. Brown; if3, Actn8, Actn14, G00189, and Df(2R)Jp1, obtained from the Bloomington D. melanogaster stock center; and f04847 and f04784, obtained from the Exelixis D. melanogaster stock collection at Harvard Medical School.
We generated a zasp mutant by recombining two flippase recognition target–bearing piggyBac elements as previously described (Parks et al., 2004). To verify the presence and location of piggyBac elements, we conducted genomic PCRs using genome-specific primers (CAATCTGCGTCACAAGGATG for f04847 and TTTGCGTTTTCATTTCATCG for f04784) and piggyBac-specific primers (RB(WH+)forward, TCCAAGCGGCGACTGAGATG, and RB(WH+)reverse, CCTCGATATACAGACCGATAAAAC). We used a GFP balancer (CyO, twi-Gal4 UAS-2xEGFP) to identify homozygous zaspΔ mutant embryos.
To image Z lines in Actn mutant larvae, we crossed Actn14/FM7c, twi-Gal4 UAS-2xEGFP females to FM7c, twi-Gal4 UAS-2xEGFP; G00189/+ males. To test for genetic interaction between αPS2 integrin and Zasp, we crossed y w ifSEF/FM7, eve-lacZ females to zaspΔ/CyO, twiGal4 UAS2EGFP males. Embryos were distinguished by detecting β-galactosidase activity with X-Gal staining. In brief, nongreen embryos were fixed for 7 min in n-heptane saturated with 2.5% glutaraldehyde in PBS at room temperature. After washing and permeabilization for another 2 h with several changes of PBT, we stained the embryos with staining solution (0.2 M Na2HPO4, 0.2 M NaH2PO4, 5 M NaCl, 1 M MgCl2, 50 mM K3[Fe(CN)6], and 50 mM K4[Fe(CN)6]) containing 1/50 vol of 10% X-gal solution in dimethyl sulfoxide for 1 h at 37°C. After stopping the reaction with several washes in PBT, we proceeded with the standard antibody staining procedure.
Online supplemental material
Fig. S1 shows cell shape changes in S2 cells spread on concanavalin A and in S2R+ cells upon depletion of BRWD3, CG3799, and if. Fig. S2 shows verification and expression of G00189, an endogenous GFP-Zasp fusion. Table S1 lists dsRNAs showing cell shape changes from the screen of 72 candidate genes. Table S2 lists the PCR amplicons used to make dsRNAs. Table S3 lists zasp splice variants derived from EST analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707045/DC1.
Supplemental Material
Acknowledgments
We thank E.C. Davis and J. Mui for help with electron microscopy and N. Perrimon for providing dsDNAs with attached T7 promoter sequence for the candidate genes tested in the pilot screen. We thank N.H. Brown for flies, D.J. Andrew, N.H. Brown, D.P. Kiehart, J. Saide, G. Tanentzapf, and the Developmental Studies Hybridoma Bank for antibodies, and F. Fagotto, L. Nilson, J. Vogel, C. Gamberi, and J.-M. Kugler for comments on the manuscript.
This work was supported by the New Opportunities grant 9607 (F. Schöck) from the Canada Foundation for Innovation and by operating grants IC1-70768 and MOP-74716 (F. Schöck) from the Canadian Institutes of Health Research. F. Schöck is a Canadian Institutes of Health Research New Investigator (MSH-76596).
Abbreviations used in this paper: dsRNA, double-stranded RNA; Zasp, Z band alternatively spliced PDZ-motif protein.
References
- Alatortsev, V.E., I.A. Kramerova, M.V. Frolov, S.A. Lavrov, and E.D. Westphal. 1997. Vinculin gene is non-essential in Drosophila melanogaster. FEBS Lett. 413:197–201. [DOI] [PubMed] [Google Scholar]
- Arimura, T., T. Hayashi, H. Terada, S.Y. Lee, Q. Zhou, M. Takahashi, K. Ueda, T. Nouchi, S. Hohda, M. Shibutani, et al. 2004. A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J. Biol. Chem. 279:6746–6752. [DOI] [PubMed] [Google Scholar]
- Artero, R., A. Prokop, N. Paricio, G. Begemann, I. Pueyo, M. Mlodzik, M. Perez-Alonso, and M.K. Baylies. 1998. The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev. Biol. 195:131–143. [DOI] [PubMed] [Google Scholar]
- Au, Y., R.A. Atkinson, R. Guerrini, G. Kelly, C. Joseph, S.R. Martin, F.W. Muskett, A. Pallavicini, G. Faulkner, and A. Pastore. 2004. Solution structure of ZASP PDZ domain; implications for sarcomere ultrastructure and enigma family redundancy. Structure. 12:611–622. [DOI] [PubMed] [Google Scholar]
- Bauer, K., M. Kratzer, M. Otte, K.L. de Quintana, J. Hagmann, G.J. Arnold, C. Eckerskorn, F. Lottspeich, and W. Siess. 2000. Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood. 96:4236–4245. [PubMed] [Google Scholar]
- Bloor, J.W., and N.H. Brown. 1998. Genetic analysis of the Drosophila alphaPS2 integrin subunit reveals discrete adhesive, morphogenetic and sarcomeric functions. Genetics. 148:1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert, T., N. Brown, and M. Wilcox. 1987. The Drosophila PS2 antigen is an invertebrate integrin that, like the fibronectin receptor, becomes localized to muscle attachments. Cell. 51:929–940. [DOI] [PubMed] [Google Scholar]
- Brabant, M.C., and D.L. Brower. 1993. PS2 integrin requirements in Drosophila embryo and wing morphogenesis. Dev. Biol. 157:49–59. [DOI] [PubMed] [Google Scholar]
- Brower, D.L., M. Wilcox, M. Piovant, R.J. Smith, and L.A. Reger. 1984. Related cell-surface antigens expressed with positional specificity in Drosophila imaginal discs. Proc. Natl. Acad. Sci. USA. 81:7485–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N.H. 1994. Null mutations in the alphaPS2 and betaPS integrin subunit genes have distinct phenotypes. Development. 120:1221–1231. [DOI] [PubMed] [Google Scholar]
- Brown, N.H., S.L. Gregory, W.L. Rickoll, L.I. Fessler, M. Prout, R.A. White, and J.W. Fristrom. 2002. Talin is essential for integrin function in Drosophila. Dev. Cell. 3:569–579. [DOI] [PubMed] [Google Scholar]
- Burridge, K., K. Fath, T. Kelly, G. Nuckolls, and C. Turner. 1988. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu. Rev. Cell Biol. 4:487–525. [DOI] [PubMed] [Google Scholar]
- Bökel, C., and N.H. Brown. 2002. Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell. 3:311–321. [DOI] [PubMed] [Google Scholar]
- Clark, K.A., A.S. McElhinny, M.C. Beckerle, and C.C. Gregorio. 2002. Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 18:637–706. [DOI] [PubMed] [Google Scholar]
- Devenport, D., and N.H. Brown. 2004. Morphogenesis in the absence of integrins: mutation of both Drosophila beta subunits prevents midgut migration. Development. 131:5405–5415. [DOI] [PubMed] [Google Scholar]
- Devenport, D., T.A. Bunch, J.W. Bloor, D.L. Brower, and N.H. Brown. 2007. Mutations in the Drosophila alphaPS2 integrin subunit uncover new features of adhesion site assembly. Dev. Biol. 308:294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil, R.R., and P. Wang. 2000. Genetic analysis of the requirements for alpha-actinin function. J. Muscle Res. Cell Motil. 21:705–713. [DOI] [PubMed] [Google Scholar]
- Ervasti, J.M. 2003. Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 278:13591–13594. [DOI] [PubMed] [Google Scholar]
- Faulkner, G., A. Pallavicini, E. Formentin, A. Comelli, C. Ievolella, S. Trevisan, G. Bortoletto, P. Scannapieco, M. Salamon, V. Mouly, et al. 1999. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell Biol. 146:465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, G.R., and R.H. Waterston. 1985. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101:1532–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg, E., M. Kelly, E. Ball, C. Fyrberg, and M.C. Reedy. 1990. Molecular genetics of Drosophila alpha-actinin: mutant alleles disrupt Z disc integrity and muscle insertions. J. Cell Biol. 110:1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg, C., A. Ketchum, E. Ball, and E. Fyrberg. 1998. Characterization of lethal Drosophila melanogaster alpha-actinin mutants. Biochem. Genet. 36:299–310. [DOI] [PubMed] [Google Scholar]
- Garamvölgyi, N. 1965. Inter-Z bridges in the flight muscle of the bee. J. Ultrastruct. Res. 13:435–443. [DOI] [PubMed] [Google Scholar]
- Giancotti, F.G., and G. Tarone. 2003. Positional control of cell fate through joint integrin/receptor protein kinase signaling. Annu. Rev. Cell Dev. Biol. 19:173–206. [DOI] [PubMed] [Google Scholar]
- Ginsberg, M.H., A. Partridge, and S.J. Shattil. 2005. Integrin regulation. Curr. Opin. Cell Biol. 17:509–516. [DOI] [PubMed] [Google Scholar]
- Gotwals, P.J., L.I. Fessler, M. Wehrli, and R.O. Hynes. 1994. Drosophila PS1 integrin is a laminin receptor and differs in ligand specificity from PS2. Proc. Natl. Acad. Sci. USA. 91:11447–11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe, C., C.G. Zervas, T. Hunter, N.H. Brown, and R.H. Palmer. 2004. Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development. 131:5795–5805. [DOI] [PubMed] [Google Scholar]
- Graner, M.W., T.A. Bunch, S. Baumgartner, A. Kerschen, and D.L. Brower. 1998. Splice variants of the Drosophila PS2 integrins differentially interact with RGD-containing fragments of the extracellular proteins tiggrin, ten-m, and D-laminin 2. J. Biol. Chem. 273:18235–18241. [DOI] [PubMed] [Google Scholar]
- Grinnell, F., and M.K. Feld. 1979. Initial adhesion of human fibroblasts in serum-free medium: possible role of secreted fibronectin. Cell. 17:117–129. [DOI] [PubMed] [Google Scholar]
- Henderson, J.R., P. Pomies, C. Auffray, and M.C. Beckerle. 2003. ALP and MLP distribution during myofibrillogenesis in cultured cardiomyocytes. Cell Motil. Cytoskeleton. 54:254–265. [DOI] [PubMed] [Google Scholar]
- Huelsmann, S., C. Hepper, D. Marchese, C. Knoll, and R. Reuter. 2006. The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development. 133:2915–2924. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. 2001. Evolution of the integrin alpha and beta protein families. J. Mol. Evol. 52:63–72. [DOI] [PubMed] [Google Scholar]
- Hutson, M.S., Y. Tokutake, M.S. Chang, J.W. Bloor, S. Venakides, D.P. Kiehart, and G.S. Edwards. 2003. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 300:145–149. [DOI] [PubMed] [Google Scholar]
- Kadrmas, J.L., and M.C. Beckerle. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5:920–931. [DOI] [PubMed] [Google Scholar]
- Kiehart, D.P., and R. Feghali. 1986. Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 103:1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger, A., B. Baum, S. Jones, M. Jones, A. Coulson, C. Echeverri, and N. Perrimon. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaavuniemi, T., A. Kelloniemi, and J. Ylanne. 2004. The ZASP-like motif in actinin-associated LIM protein is required for interaction with the alpha-actinin rod and for targeting to the muscle Z-line. J. Biol. Chem. 279:26402–26410. [DOI] [PubMed] [Google Scholar]
- Kunda, P., G. Craig, V. Dominguez, and B. Baum. 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13:1867–1875. [DOI] [PubMed] [Google Scholar]
- Lecroisey, C., L. Segalat, and K. Gieseler. 2007. The C. elegans dense body: anchoring and signaling structure of the muscle. J. Muscle Res. Cell Motil. 28:79–87. [DOI] [PubMed] [Google Scholar]
- Leptin, M., T. Bogaert, R. Lehmann, and M. Wilcox. 1989. The function of PS integrins during Drosophila embryogenesis. Cell. 56:401–408. [DOI] [PubMed] [Google Scholar]
- Machado, C., C.E. Sunkel, and D.J. Andrew. 1998. Human autoantibodies reveal titin as a chromosomal protein. J. Cell Biol. 141:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown, C.R., H.F. Han, and M.C. Beckerle. 2006. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev. Dyn. 235:530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, X., R. Daneman, M. Zavortink, and W. Chia. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 98:15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha, M., and N.H. Brown. 2004. Novel functions for integrins in epithelial morphogenesis. Curr. Biol. 14:381–385. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H., H. Yajima, K. Maruyama, and S. Kimura. 1997. The N-terminal Z repeat 5 of connectin/titin binds to the C-terminal region of alpha-actinin. Biochem. Biophys. Res. Commun. 235:1–3. [DOI] [PubMed] [Google Scholar]
- Pardo, J.V., J.D. Siliciano, and S.W. Craig. 1983. A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc. Natl. Acad. Sci. USA. 80:1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, A.L., K.R. Cook, M. Belvin, N.A. Dompe, R. Fawcett, K. Huppert, L.R. Tan, C.G. Winter, K.P. Bogart, J.E. Deal, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. [DOI] [PubMed] [Google Scholar]
- Pashmforoush, M., P. Pomies, K.L. Peterson, S. Kubalak, J. Ross Jr., A. Hefti, U. Aebi, M.C. Beckerle, and K.R. Chien. 2001. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat. Med. 7:591–597. [DOI] [PubMed] [Google Scholar]
- Perrimon, N., L. Engstrom, and A.P. Mahowald. 1985. Developmental genetics of the 2C-D region of the Drosophila X chromosome. Genetics. 111:23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomies, P., T. Macalma, and M.C. Beckerle. 1999. Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J. Biol. Chem. 274:29242–29250. [DOI] [PubMed] [Google Scholar]
- Reedy, M.C., and C. Beall. 1993. Ultrastructure of developing flight muscle in Drosophila. II. Formation of the myotendon junction. Dev. Biol. 160:466–479. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, N. Stuurman, and R.D. Vale. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saide, J.D., S. Chin-Bow, J. Hogan-Sheldon, L. Busquets-Turner, J.O. Vigoreaux, K. Valgeirsdottir, and M.L. Pardue. 1989. Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster. J. Cell Biol. 109:2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353–365. [PubMed] [Google Scholar]
- Schwander, M., M. Leu, M. Stumm, O.M. Dorchies, U.T. Ruegg, J. Schittny, and U. Müller. 2003. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev. Cell. 4:673–685. [DOI] [PubMed] [Google Scholar]
- Schöck, F., and N. Perrimon. 2003. Retraction of the Drosophila germ band requires cell-matrix interaction. Genes Dev. 17:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi, H., A. Freiburg, B. Kolmerer, S. Ishiura, G. Stier, C.C. Gregorio, D. Labeit, W.A. Linke, K. Suzuki, and S. Labeit. 1997. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J. Mol. Biol. 270:688–695. [DOI] [PubMed] [Google Scholar]
- te Velthuis, A.J., T. Isogai, L. Gerrits, and C.P. Bagowski. 2007. Insights into the molecular evolution of the PDZ/LIM family and identification of a novel conserved protein motif. PLoS ONE. 2:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass, U. 1996. Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila. Dev. Biol. 177:217–225. [DOI] [PubMed] [Google Scholar]
- Vatta, M., B. Mohapatra, S. Jimenez, X. Sanchez, G. Faulkner, Z. Perles, G. Sinagra, J.H. Lin, T.M. Vu, Q. Zhou, et al. 2003. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 42:2014–2027. [DOI] [PubMed] [Google Scholar]
- Volk, T., L.I. Fessler, and J.H. Fessler. 1990. A role for integrin in the formation of sarcomeric cytoarchitecture. Cell. 63:525–536. [DOI] [PubMed] [Google Scholar]
- Xia, H., S.T. Winokur, W.L. Kuo, M.R. Altherr, and D.S. Bredt. 1997. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 139:507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa, S., J.S. Lee, and A. Ishimoto. 1998. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J. Biol. Chem. 273:32353–32359. [DOI] [PubMed] [Google Scholar]
- Young, P., C. Ferguson, S. Banuelos, and M. Gautel. 1998. Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of alpha-actinin. EMBO J. 17:1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar, R., S. Itzkovitz, A. Ma'ayan, R. Iyengar, and B. Geiger. 2007. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas, C.G., S.L. Gregory, and N.H. Brown. 2001. Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J. Cell Biol. 152:1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., P. Ruiz-Lozano, M.E. Martone, and J. Chen. 1999. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 274:19807–19813. [DOI] [PubMed] [Google Scholar]
- Zhou, Q., P.H. Chu, C. Huang, C.F. Cheng, M.E. Martone, G. Knoll, G.D. Shelton, S. Evans, and J. Chen. 2001. Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol. 155:605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.