Abstract
The retinoblastoma tumor suppressor protein (pRb) is involved in mitotic exit, promoting the arrest of myoblasts, and myogenic differentiation. However, it is unclear how permanent cell cycle exit is maintained in differentiated muscle. Using RNA interference, expression profiling, and chromatin immunoprecipitations, we show that pRb is essential for cell cycle exit and the differentiation of myoblasts and is also uniquely required to maintain this arrest in myotubes. Remarkably, we also uncover a function for the pRb-related proteins p107 and p130 as enforcers of a G2/M phase checkpoint that prevents progression into mitosis in cells that have lost pRb. We further demonstrate that pRb effects permanent cell cycle exit in part by maintaining trimethylation of histone H3 lysine 27 (H3K27) on cell cycle genes. H3K27 trimethylation silences other genes, including Cyclin D1, in a pRb-independent but polycomb-dependent manner. Thus, our data distinguish two distinct chromatin-based regulatory mechanisms that lead to terminal differentiation.
Introduction
The mammalian cell cycle is controlled at the transcriptional level by the E2F family of transcription factors. E2F activity is repressed through binding to pocket proteins, a family that includes the retinoblastoma tumor suppressor protein (pRb) as well as the related p107 and p130 proteins (Macaluso et al., 2006). The cell implements a signaling cascade whereby growth factors override the repressive action of pocket proteins by activating Cdks. This leads to the activation of E2F target genes and allows progression through the G1–S phase transition. Under growth-repressive conditions, such as those occurring in the absence of mitogens, repressor E2Fs replace activator E2Fs on the promoters of target genes, in some cases recruiting pocket proteins and associated corepressors, which silences expression of these genes and promotes quiescence (Takahashi et al., 2000; Cam et al., 2004; Balciunaite et al., 2005).
Pocket proteins repress transcription by recruiting enzymatic complexes that modify the chromatin environment. The list of repressor proteins recruited by the pRb family includes histone deacetylases and polycomb group (PcG) repressor complex 2, although direct evidence for a specific role for PcG repressor complex 2 (PRC2) in pRb-mediated transcriptional repression is lacking (Huang et al., 1991; Qian et al., 1993; Kuzmichev et al., 2002; Benevolenskaya et al., 2005; Bracken et al., 2006). Multisubunit PcG complexes are essential for maintaining proper expression of homeotic genes in Drosophila melanogaster and generally function as transcriptional repressors (Orlando, 2003; Ringrose and Paro, 2004). The repressive action of PRC2 requires its Ezh2 component, a histone methyltransferase (HMTase) that methylates lysine 27 of histone H3 (H3K27), which has been shown to facilitate the recruitment of the PRC1 complex and subsequent repression. In higher eukaryotes, the function of multiple subunits of the PRC2 complex, including Suz12 and Ezh2, is essential for the establishment of H3K27 methylation, embryonic development, and cellular proliferation (Faust et al., 1998; O'Carroll et al., 2001; Erhardt et al., 2003; Pasini et al., 2004). Recently, it has been shown by chromatin immunoprecipitation (ChIP)–on-chip that PRC2 promotes transcriptional silencing of many regulators of cell fate, development, and differentiation, and that this repression is progressively lifted as differentiation proceeds (Bracken et al., 2006; Lee et al., 2006). This regulatory mechanism is also relevant to muscle differentiation, wherein Ezh2 activity and the presence of the H3K27Me3 indicate a correlation with repression of muscle differentiation genes in myoblasts (Caretti et al., 2004). Although repression by PcG complexes plays an essential role in proliferation and development, the mechanisms underlying their recruitment and eventual dissociation from specific target genes remain largely unknown.
A hallmark of vertebrate differentiation is irreversible cell cycle exit, wherein cells become refractory to subsequent mitogen stimulation and differentiated skeletal muscle cells never resume proliferation (Molkentin and Olson, 1996). Although reversible cell cycle exit has been studied extensively and the role of pocket proteins is rather well understood (Zhang et al., 2000; Sage et al., 2003; Cam et al., 2004; Korenjak et al., 2004; Balciunaite et al., 2005), how an irreversibly arrested state is established and maintained is less clear. The pocket proteins have been the focus of intense scrutiny and many studies have implicated pRb in the cell cycle blockade of muscle cells. pRb knockout mice die in utero before the development of skeletal muscle. However, the skeletal muscles of animals rescued from embryonic death by hypomorphic expression of an RB minigene or by tetraploid aggregation show signs of apoptosis, improper cell cycle exit, and endoreduplication (Zacksenhaus et al., 1996; Wu et al., 2003). Mice deficient in either p107 or p130 do not exhibit detectable muscle phenotypes and, although mice deficient in both p107 and p130 die at birth, they also lack a discernible muscle phenotype, which suggests that pRb is the sole pocket protein involved in the control of myogenic differentiation (Cobrinik et al., 1996). However, in spite of the information obtained from germ-line knockout studies, a definitive model for the role of each pocket protein in regulating myogenesis has not emerged, largely owing to the fact that compensatory mechanisms confound the analysis of contributions from individual pRb family members.
Here, we investigate the mechanisms underlying maintenance of cell cycle arrest in differentiated skeletal muscle cells. To circumvent complications arising from cell cycle alterations, we suppressed pRb expression in fully differentiated myotubes using RNAi. We have made the notable finding that pRb is not only important for initial cell cycle exit at the onset of myogenesis but also for the maintenance of this arrest in mature myotubes. Depletion of pRb causes both C2C12 and primary myotubes to resume proliferation and complete S phase. Importantly, we also discovered a role for p107 and p130 in preventing progression of pRb-depleted myotubes into M phase. Our studies show that H3K27Me3 marks promoters of cell cycle genes, which strongly coincides with pRb-mediated repression, cell cycle exit, and maintenance of terminal differentiation. However, we also identified a distinct mechanism for the cyclin D1 (Ccnd1) gene, which acquires the H3K27Me3 mark in a pRb-independent but PcG-dependent manner. We describe a transcriptional regulatory framework that explains, at least in part, the irreversibility of cell cycle exit in muscle cells.
Results
Cell cycle reentry after impairment of pRb function in differentiated muscle
The role of pRb in skeletal muscle differentiation and cell cycle exit has been studied using several models that include functional inactivation with viral oncoproteins, germ line ablation, and conditional excision. It is likely that functional compensation by p107 and p130 and the pocket protein–independent effects of oncoproteins have obscured the individual contribution of each pocket protein to myogenesis and cell cycle arrest. To address this issue, we performed acute, RNAi-mediated suppression of pocket proteins in growing and terminally differentiated C2C12 mouse myotubes. The C2C12 system represents a well-established in vitro differentiation model, and our work previously indicated that the gene expression program activated during C2C12 myogenic differentiation closely resembles the one observed in primary myoblasts undergoing differentiation (Blais et al., 2005).
First, we tested the efficacy of siRNAs that target each individual pocket protein. We found that pRb is required for skeletal myogenesis and initiation of cell cycle exit because its ablation in myoblasts impaired their differentiation and was accompanied by their failure to cease proliferation (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1). These results are consistent with previous studies using knockout cells. Importantly, the block to differentiation specifically resulted from pRb knockdown and did not occur after ablation of the other two pocket proteins. This finding strongly supports the notion that pRb is uniquely required to promote cell cycle arrest and myogenic differentiation.
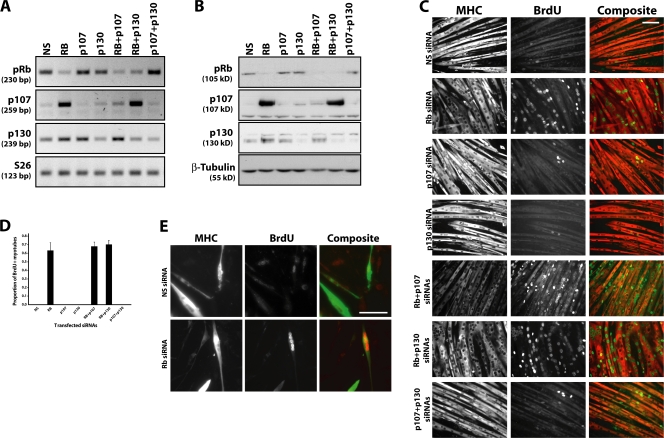
Next, we acutely depleted each pocket protein in fully differentiated cells by transfecting siRNAs into myotubes. Our experiments indicated that myotubes were fully differentiated after 4 d in differentiation medium (Fig. S1 B). Delivery of each siRNA specifically suppressed expression of its target and we achieved equally dramatic reductions in the level of each protein, which could be detected as early as 18–24 h after transfection (Fig. 1, A and B; Fig. S1 C; and not depicted). Importantly, we tested additional siRNAs targeting pRb and p130 and showed that each siRNA behaved with comparable specificity and efficiency (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1). We determined whether pRb ablation would affect the quiescent state of myotubes. Cell cycle exit is a prerequisite for myogenic differentiation and is thought to be irreversible. To this end, we examined BrdU incorporation after transfection of siRNAs targeting each pocket protein and determined whether cell cycle reentry occurred in differentiated muscle cells. Remarkably, myotubes treated with siRNAs targeting pRb reentered S phase, in sharp contrast with nonspecific control duplexes (Fig. 1 C). On average, >60% of fully differentiated muscle cells reentered the cell cycle, which attests to the robustness of the phenotype (Fig. 1 D). S phase reentry was not enhanced by concomitant growth factor stimulation (unpublished data), presumably because pRb is itself the ultimate target of growth factors through Cdk-mediated phosphorylation. To ensure that this effect was specific for pRb and to rule out off-target effects, we tested multiple distinct siRNAs (Fig. S2 B) and obtained identical results. To test the possibility that cell cycle reentry after pRb knockdown was restricted to C2C12 cells, an established cell line, we isolated primary myoblasts from the skeletal muscle of newborn mice, subjected them to a differentiation protocol, and transfected the resulting myotubes. Although the transfection efficiency of primary myotubes was lower than C2C12 myotubes (∼30 vs. 80%), the results were qualitatively similar (Fig. 1 E); primary muscle cells also require functional pRb to remain terminally arrested. Importantly, this result further confirms the validity of using the C2C12 model in our experiments.
Figure 1.
pRb depletion leads to cell cycle reentry in terminally differentiated muscle cells. (A) RT-PCR detection of pocket protein mRNA expression 48 h after transfection of C2C12 myotubes with siRNA duplexes. (B) Western blot detection of each pocket protein indicates the extent of knockdown achieved in a typical experiment. (C) Immunofluorescent detection of BrdU incorporation (green) and MHC expression (red) in differentiated C2C12 myotubes after transfection with a nonspecific siRNA duplex or duplexes targeting various pocket protein combinations. (D) Histogram representing the proportion of C2C12 myotubes with at least one BrdU+ nucleus after transfection with the indicated siRNA. Error bars indicate SD of three independent experiments. (E) Immunofluorescent detection of BrdU incorporation (red) and MHC expression (green) in primary myotubes after transfection with a nonspecific siRNA duplex or a duplex targeting pRb. Bars, 100 μm.
We tested the possibility that suppression of p107 or p130 expression could similarly promote cell cycle reentry in differentiated myotubes. In striking contrast to cells depleted of pRb, we observed no increase in BrdU-positive myotubes lacking either p107 or p130 (Fig. 1, C and D). Moreover, the combined loss of both p107 and p130 also failed to induce cell cycle reentry. We note that p107 is not expressed in myotubes or differentiated muscle (Fig. S1 A) but it is significantly induced after knockdown of pRb (Fig. 1, A and B), a phenomenon that has also been observed by others (Schneider et al., 1994; Novitch et al., 1996) presumably because it is directly repressed by pRb (Hurford et al., 1997). p130 levels also increased moderately after pRb knockdown (Fig. 1, A and B). However, the dramatic elevation in p107 and p130 levels is not sufficient to bypass a requirement for pRb in suppressing cell cycle reentry and is consistent with the absence of inappropriate proliferation in myotubes lacking p107 and p130.
Because p107 and p130 also repress E2F activity and potentially compensate for pRb loss, we simultaneously suppressed both pRb and p107 or p130 in differentiated muscle cells. We reasoned that if p107 and/or p130 could compensate for diminished levels of pRb, then cell cycle reentry would be enhanced by depleting a second pocket protein in addition to pRb. However, we did not observe a further increase in the proportion of BrdU-positive cells, irrespective of which proteins were depleted (Fig. 1, C and D). Furthermore, depleting all three pocket proteins did not further expand the proportion of cells in S phase (unpublished data). These data strongly suggest that the maintenance of cell cycle arrest, and thus the differentiated state, critically depends on pRb but not the other two pocket proteins.
Interestingly, however, we noticed that myotubes depleted of both pRb and a second pocket protein frequently exhibit pinching between nuclei, in some cases causing entry into mitosis and apparent myotube fragmentation (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1). These data suggest that p107 and p130 play a fail-safe role of blocking cells at the G2/M boundary that have progressed through S phase. Loss of this G2/M “checkpoint” results in further progression of differentiated muscle cells into mitosis. This function is only uncovered in cells that have simultaneously lost pRb, which further emphasizes the utility of our acute knockdown approach.
Cells depleted of pocket proteins exhibit alterations in genome-wide expression profiles
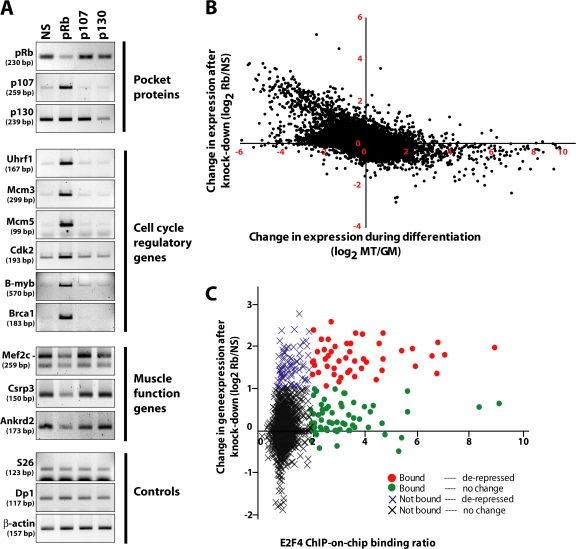
We have conclusively shown that myotubes reenter the cell cycle after pRb loss, prompting us to determine whether this event is associated with changes at the gene expression level. We performed genome-wide expression profiling on myotubes treated with pRb-specific siRNA and observed widespread derepression of cell cycle genes (Fig. 2 A and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1). As expected, because its expression is barely detectable in myotubes, p107 suppression had very mild effects on the gene expression profile. In addition, we observed remarkably few differences after suppressing p130 or p107 and p130 (Fig. 2 A; Table S1, and not depicted), despite the fact that p130 is expressed at high levels in myotubes and was suppressed to an extent comparable to pRb (Fig. 1, A and B).
Figure 2.
Loss of pRb leads to derepression of cell cycle control genes. (A) RT-PCR detection of pocket protein, cell cycle, and muscle function gene expression 48 h after suppression of pocket protein expression in C2C12 myotubes. (B) Scatter plot representing the change in gene expression in C2C12 myotubes upon knockdown of pRb as a function of the gene expression changes during the normal course of myogenic differentiation. Each spot represents one measurement with one microarray probe. MT, untransfected myotubes; GM, untransfected growing myoblasts; NS, myotubes transfected with a nonspecific siRNA duplex; pRb, myotubes transfected with the pRb-specific duplex. All data points represent the combination of three independent experiments. (C) Scatter plot representing the change in gene expression in myotubes after pRb knockdown as a function of the degree of binding of E2F4 in a ChIP-on-chip microarray experiment performed in C2C12 myotubes. The binding ratio serves as an indirect measure of the strength of binding of E2F4 to its target genes; a cut-off corresponding to a twofold enrichment was used.
We used gene ontology to analyze the changes associated with pRb loss and found that the majority of derepressed genes are involved in cell cycle control, DNA replication, segregation, cytokinesis, and the response to DNA damage. The vast majority (74%) of these pRb-repressed genes exhibit diminished expression in differentiated cells as compared with proliferating myoblasts (Fig. 2 B, top left quadrant). Finally, expression profiling of cells transfected with a second siRNA against pRb showed nearly interchangeable profiles with the first dataset (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1), which suggests that the derepression of cell cycle genes we observed is not caused by off-target effects.
We inspected the proximal promoters of derepressed genes and found that a large portion are known or predicted E2F targets as was expected (unpublished data). We then performed ChIP-on-chip with an antibody against E2F4 because it is the predominant “repressor E2F” family member expressed in myotubes (unpublished data). We confirmed that many genes derepressed after the loss of pRb are bound by E2F4 in differentiated muscle cells. This is associated with a p-value of 9.4 × 10−26 by hypergeometric distribution (Fig. 2 C), which indicates that there is a very strong association between derepression after pRb ablation and regulation by E2F4. Furthermore, E2F4 target genes are induced 3.1-fold after pRb knockdown on average, whereas genes that are not bound by E2F4 are not significantly induced (1.2-fold; P = 4 × 10−47 for t test; Fig. 2 C). We have so far been unable to knockdown the expression of E2F4 and thus have not been able to further investigate the involvement of E2F4 in pRb-mediated repression. Together, these data suggest that depletion of pRb causes cell cycle reentry of terminally arrested myotubes through widespread deregulation of E2F target gene expression.
The expression of 62 genes diminished significantly upon loss of pRb. Interestingly, these genes, which include Mef2c, Ankrd2, Csrp3, and various myosin isoforms, are strongly associated with muscle development and function, and many of them are known or predicted targets of the muscle regulatory factors MyoD, myogenin, or Mef2c (Fig. 2, A and B, bottom right quadrant; Blais et al., 2005). These genes also have a strong tendency to be induced during muscle differentiation (6.5-fold induction during myogenesis on average as compared with no change for the mean of all other genes; P = 4 × 10−95 for t test; Fig. 2 B).
Perhaps our most striking observation was that expression of myogenic differentiation genes decreases subsequently to the establishment of a terminally differentiated state in the absence of pRb. That is, because our experiments were performed with isolated myotubes that had already differentiated before pRb depletion, our conclusions are not confounded by the block to differentiation that is encountered by myoblasts that have lost pRb (Fig. S1). These studies strongly suggest that loss of pRb promotes “dedifferentiation” of myotubes, returning them to a proliferative, myoblast-like state (Cobrinik et al., 1996; Zacksenhaus et al., 1996; Wu et al., 2003). Thus, pRb function is essential not only for the establishment of cell cycle arrest during myogenic differentiation but also for its maintenance.
A mechanism for permanent cell cycle exit: histone H3K27 methylation
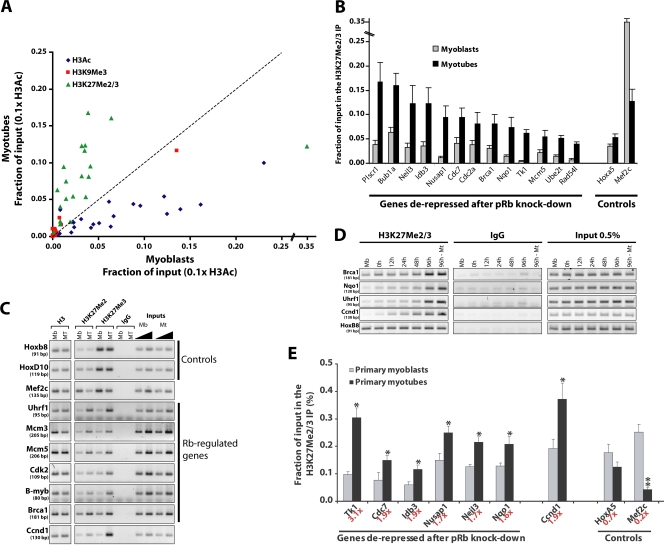
We postulated that pRb-mediated recruitment of chromatin-modifying activities and subsequent modification of chromatin could establish a stable, repressive mark on such promoters and lead to the heterochromatinization of its target genes. To test this hypothesis, we performed a systematic screen for 10 different histone tail modifications associated with both transcriptionally active and repressed chromatin in growing and differentiated muscle cells. Our goal was to determine which chromatin modifications are most robustly associated with irreversible cell cycle arrest during myogenic differentiation. A complete analysis of the results of this screen will be described elsewhere (unpublished data). Here, we focused exclusively on those genes that were transcriptionally derepressed in myotubes depleted of pRb. We found that one particular mark, di- and trimethylation of histone H3K27 (here we do not distinguish between the di- and trimethylated state and refer to this modification as H3K27Me2/3), increased most significantly on a large number of genes that were both transcriptionally silenced during myogenic differentiation and derepressed after pRb ablation (Fig. 3 A and Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1). H3K27Me2/3 levels were elevated 2–13-fold in myotubes when compared with growing myoblasts as assessed by quantitative real-time PCR (qChIP; Fig. 3 B). In contrast, other marks known to be associated with repressed chromatin, including trimethylation of histone H3K9 and histone H4K20, exhibited little or no difference on 20 genes that we surveyed during the transition from myoblasts to myotubes. The Mef2c promoter displayed the opposite pattern: H3K27Me2/3 levels dramatically decreased as cells differentiated into myotubes (Fig. 3, A and B). These Mef2c methylation data are consistent with previous results, which indicates that certain muscle-specific genes are repressed via trimethylation of H3K27 specifically in myoblasts (Caretti et al., 2004). Other genes, including stably silenced homeotic control genes, exhibited no change in H3K27Me2/3 levels (Fig. 3, A and C; and not depicted), which indicates that changes in H3K27 methylation levels were highly promoter specific.
Figure 3.
H3K27 trimethylation of cell cycle gene promoters during myogenic differentiation. (A) qChIP on chromatin from either C2C12 myoblasts or myotubes using antibodies that recognize H3K27Me2/3, H3K9Me3, and H3Ac. A scatter plot indicates the fraction of input DNA immunoprecipitated from each state. Values obtained with the H3Ac antibody were divided by 10 to fit on the same plot as the other two datasets. Results represent the mean of at least three independent experiments. (B) Histogram representing the H3K27Me3 signal from A. The Hoxa5 and Mef2c genes served as specificity controls. Error bars indicate SD of three independent experiments. (C) ChIP assay performed in C2C12 myoblasts and myotubes using antibodies against histone H3, H3K27Me2 or H3K27Me3, or rabbit IgG. (D) ChIP assay performed on cells at different times after induction of differentiation. The 96-h Mt sample represents pure myotubes, whereas the samples labeled 96 h and earlier time points represent the mixed cell population before separation of myotubes from reserve cells. (E) Quantitative ChIP assays for H3K27Me2/3 performed in primary myoblasts and matched myotubes. The mean of four ChIP assays and standard errors are shown. The ratio of H3K27Me2/3 signal in myotubes over that in myoblasts is indicated in red below each gene name. Asterisks indicate an H3K27Me2/3 signal significantly higher in myotubes than in myoblasts; P < 0.03 by t test. The double asterisk indicates an H3K27Me2/3 signal significantly lower in myotubes than in myoblasts; P < 0.001 by t test.
We also determined whether the increase in signal detected in Fig. 3 (A and B) was caused by an increase in di- and/or trimethylation of H3K27 in ChIP assays using antibodies specific for both forms of H3K27. As shown in Figs. 3 C and S5 A (available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1), the majority of pRb target genes display marked increases in both di- and trimethyl H3K27. We note that although these histone marks increase or decrease in abundance during myogenic differentiation, the global levels of nucleosome occupancy at the tested loci remain relatively constant (Fig. 3 C), which suggests that histone modification rather than nucleosome loading or eviction is the primary mechanism explaining these fluctuations.
We examined the temporal appearance of the H3K27Me2/3 mark during the course of differentiation and found that it remains stable or gradually rises during the early stages of myogenesis. Interestingly, the mark dramatically appears between 2 and 4 d after induction of differentiation, reaching maximal levels after 4 d (Fig. 3 D). These data suggest that the acquisition of this mark is a relatively late event in the differentiation process.
We verified that the appearance of the H3K27Me2/3 mark during differentiation is not exclusive to C2C12 myoblasts because the mark is also acquired as primary myoblasts differentiate (Fig. 3 E). Here, quantitative ChIP assays were performed using primary myoblasts that were obtained from mouse muscle tissue and maintained in the proliferative state or induced to differentiate. Fig. 3 E clearly demonstrates that the H3K27Me2/3 ChIP signal significantly and reproducibly increases (P < 0.03 by t test) at several pRb target genes during myogenic differentiation of primary myoblasts, whereas control genes are not significantly affected. The mean effect is twofold for pRb-regulated genes. This increase in H3K27Me2/3 was somewhat lower than in C2C12 cells, most likely because primary myoblasts tend to spontaneously exit the cell cycle and differentiate even when maintained in growth medium at relatively low confluence, as we have done here (unpublished data).
The role of pRb in establishing the H3K27Me3 mark
We sought to establish a causal relationship between pRb function and H3K27 methylation at cell cycle control genes. If methylation of H3K27 is responsible for maintaining these genes in an irreversibly repressed state and pRb is specifically required to promote and maintain this mark, then pRb depletion but not stimulation of myotubes with mitogens should result in the disappearance of this mark and thereby lead to their reexpression. We tested this idea by performing ChIP assays on fully differentiated myotubes treated with serum or transfected with pRb-specific or control siRNAs.
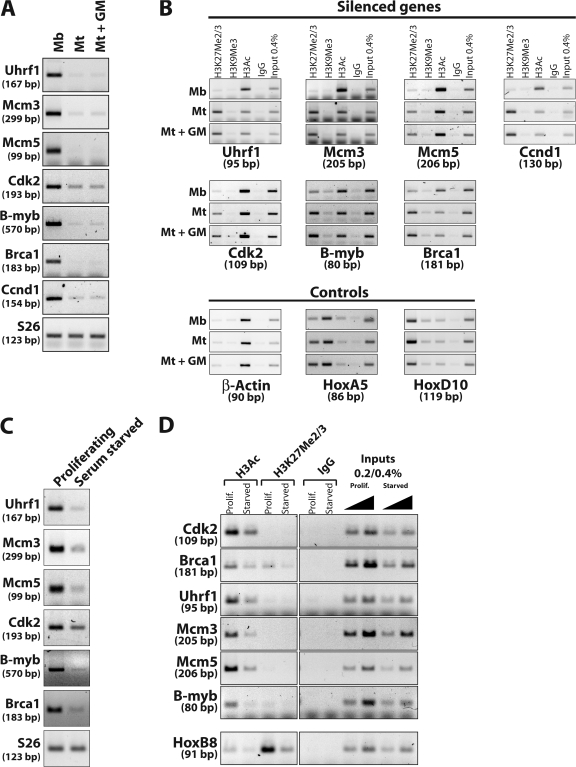
Differentiated muscle cells are refractory to mitogenic stimulation, and cell cycle control genes remain silent upon such stimulation (Fig. 4 A). As shown in Fig. 4 B, the extent of H3K27 methylation of cell cycle regulatory genes is unaffected by treatment of mature myotubes with high concentrations of mitogens, which indicates that once established this histone mark is stable and may thus contribute to the permanent nature of the cell cycle exit. In addition, the degree of histone H3 acetylation is also unchanged after mitogen treatment, which is consistent with the observation that these genes are not reexpressed.
Figure 4.
Trimethylation of H3K27 is insensitive to mitogenic stimulation and does not occur during reversible, mitogen starvation–induced quiescence. (A) RT-PCR assay of gene expression in C2C12 myoblasts and myotubes or myotubes treated with high serum concentrations for 24 h (Mt + GM). (B) ChIP assay performed on samples from cells analyzed in A. The input control corresponds to 0.4% of the amount used per immunoprecipitation. The Hoxa5 gene is shown as a control for efficiency of immunoprecipitation by the anti-H3K9Me3 antibody. (C) MEFs were harvested during asynchronous growth or after 3 d of starvation in serum-free medium. RNA was extracted and cell cycle gene expression was assessed by RT-PCR. (D) ChIP assays performed on cross-linked cells treated and processed in parallel with those indicated in C.
Furthermore, if H3K27 methylation is involved in terminal transcriptional silencing of pRb target genes, then this histone mark should be specific to instances of irreversible cell cycle arrest. We tested this idea by performing analogous experiments using mouse embryonic fibroblasts (MEFs) that had been brought to quiescence through mitogen deprivation. It is known that this state is fully reversed upon mitogen stimulation (Pardee, 1974). As expected, the cell cycle control genes we examined previously are repressed when MEFs are rendered quiescent (Fig. 4 C), but this silencing is not accompanied by the appearance of the H3K27 di- or trimethylation marks, despite the fact that histone H3 acetylation levels are markedly decreased (Fig. 4 D). These results indicate that trimethylation of H3K27 at pRb target genes is not a generic mark of nonproliferative cells. Instead, methylation of these targets is context dependent and specific to permanently arrested cells.
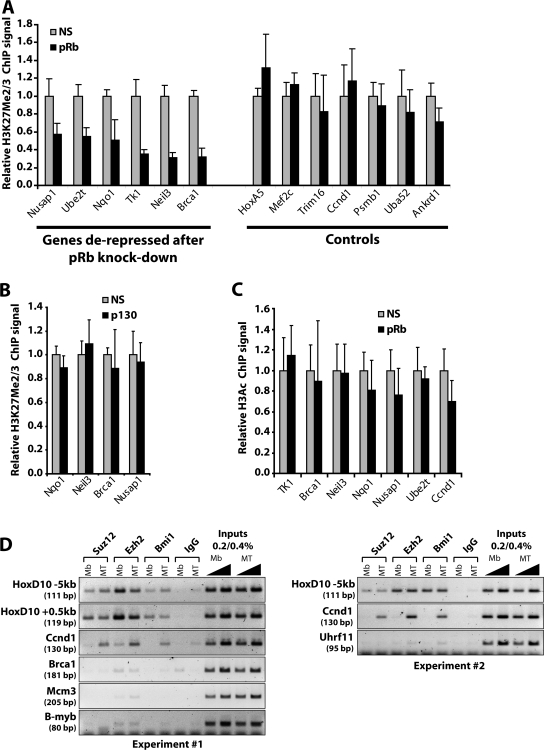
Next, we examined the impact of pRb depletion on H3K27 methylation by performing ChIP on chromatin from myotubes depleted of pRb. In striking contrast with serum addition, pRb knockdown resulted in the marked diminution of the H3K27Me2/3 signal at several genes compared with controls (Fig. 5 A and Table S2). This coincides with derepression of these genes under the same conditions (Fig. 2 A and Table S1). Notably, although p130 could also be depleted with comparable efficiency (Fig. 1), its loss did not lead to a reduction in H3K27Me2/3 signal at these genes (Fig. 5 B). Further, other genes, including Ccnd1, whose expression was not affected by ablation of pRb, did not exhibit any significant differences in H3K27Me2/3 levels after pRb knockdown (Fig. 5 A, controls; P = 0.001 by one-tailed t test). Importantly, these findings rule out the possibility that histone replacement during DNA replication (induced by pRb depletion) is responsible for the decrease in the H3K27Me2/3 signal. Interestingly, depletion of pRb did not affect histone H3 acetylation levels at the promoters of derepressed genes (Fig. 5 C).
Figure 5.
pRb-dependent trimethylation of H3K27 at cell cycle gene promoters. (A) Histogram indicating H3K27Me2/3 levels in C2C12 myotubes transfected with a nonspecific control or pRb-silencing siRNA. The ChIP signal intensity is reported as a fraction of the signal obtained in cells transfected with the nonspecific siRNA. (B and C) Histograms depicting H3K27Me2/3 (B) and H3Ac (C) levels in C2C12 myotubes transfected with siRNAs targeting p130 or pRb or with a nonspecific control siRNA as indicated in A. Error bars represent SD of three independent experiments. (D) ChIP assay performed in C2C12 myoblasts and myotubes using antibodies directed against Suz12, Ezh2, and Bmi-1. Two regions of the Hoxd10 gene were surveyed to evaluate PRC binding. The results of two independent experiments are shown.
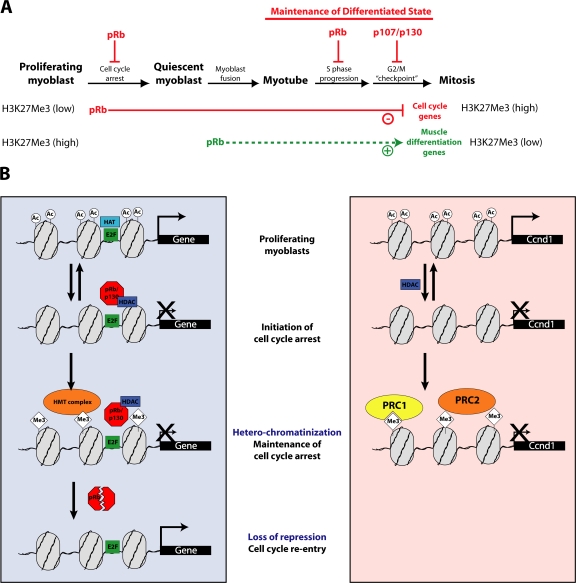
We have thus shown that pRb is essential for the maintenance of the H3K27Me2/3 mark at the promoter of cell cycle regulatory genes in terminally arrested muscle cells. Ablation of pRb causes the loss of trimethylation at H3K27 and ultimately leads to reexpression, causing the cells to reenter the cell cycle (Fig. 6 B).
Figure 6.
Model detailing pRb function in differentiated muscle. See text for details.
Role of PcG repressor complex
We sought to determine if pRb-regulated genes recruit PcG complexes known to be involved in H3K27 methylation and subsequent gene silencing. ChIP assays using antibodies directed toward a PRC1 component, Bmi-1, and two core components of PRC2, Suz12 and Ezh2, failed to reveal convincing binding to pRb target genes during differentiation, although we surveyed up to ∼3 kb upstream of several genes and showed that Ezh2 is expressed in myotubes (Figs. 5 D and S5 B; and not depicted). In contrast, recruitment of these proteins at homeotic control genes (Hoxd10 and Hoxb8) was readily detected irrespective of differentiation status (Fig. 5 D and not depicted).
In addition to pRb target genes, we also examined the Ccnd1 gene because it plays a key role in regulating cell cycle progression in response to growth factors. Its silencing during myogenesis is accompanied by trimethylation of H3K27 but is pRb independent (Fig. 5 A). Indeed, Ccnd1 exhibits the most dramatic increase in methylation during differentiation and is among the most strongly repressed cell cycle genes (Fig. 3, C–E; Fig. 4, A and B; and Table S1). Interestingly, we found that the PRC2 and PRC1 complexes are recruited to the Ccnd1 promoter specifically in myotubes (Fig. 5 D). Additionally, several other genes implicated in growth control or response to mitogens were also found to be silenced during myogenesis in a H3K27Me3-associated, pRb-independent manner (Fig. S5 A and Table S1).
These data therefore suggest the existence of at least two distinct pathways toward repression of the cell cycle machinery in differentiated muscle cells: one group of genes is trimethylated on H3K27 and repressed via pRb, whereas a second group is repressed independently of pRb through a mechanism that likely involves PcG complexes (Fig. 6 B).
Discussion
Acute ablation of pocket proteins in differentiated myotubes
Our findings regarding a specific requirement for pRb in cell cycle exit and myogenic differentiation are consistent with previous knockout studies as well as recent studies using conditional ablation of RB in myoblasts in mice and cultured cells (Camarda et al., 2004; Huh et al., 2004). However, our acute depletion experiments allowed us to show definitively that the combined loss of p107 and p130 has no effect on myogenic differentiation. Our experiments are less likely to be complicated by the issue of compensation by pRb, which may have occurred in earlier germline knockout studies and confounded the analysis (Cobrinik et al., 1996). In contrast to earlier studies (Gu et al., 1993; Endo and Nadal-Ginard, 1998), our use of RNAi has allowed us to avoid the use of viral oncogenes, which are known to deregulate all three pocket proteins as well as a plethora of other cellular proteins, and has enabled us to perform more precise gene ablation experiments.
In contrast with a demonstrated role for pRb in differentiation, the role for this protein in maintenance of terminal differentiation is less clear. We discovered that pRb is also involved in maintaining the postmitotic state of differentiated muscle cells by suppressing the expression of cell cycle control genes. Importantly, these findings were obtained using both well-characterized C2C12 myoblasts and myotubes prepared from primary myoblasts, which rules out the immortal nature of the cell line as the cause of cell cycle reentry. However, Camarda et al. (2004) and Huh et al. (2004) showed that conditional ablation of Rb in differentiated myotubes using viral transduction or muscle creatine kinase–driven Cre expression produced no obvious cell cycle reentry and suggested that pRb is involved in the establishment but not maintenance of the differentiated state. Although we cannot currently explain the differences between our results and those published recently, several important technical differences are noteworthy. First, differences may stem from the kinetics of knockdown; i.e., we acutely ablated pRb from differentiated myotubes and knockdown occurred typically within 18 h after siRNA delivery. In contrast, other studies required a longer duration to achieve suppression of pRb. A second possibility is that Cre-mediated excision triggers or contributes to a checkpoint because Cre-mediated recombination has been shown to suppress the growth of proliferating cells and elicits a DNA damage response in mammalian cells (Loonstra et al., 2001; Pfeifer et al., 2001; Silver and Livingston, 2001; Baba et al., 2005). This could conceivably prevent cell cycle reentry or survival of pRb-deleted myotubes. A third explanation derives from the extent to which pRb expression was suppressed. We note that our RNAi approach does not lead to a complete loss of pocket proteins but rather a strong decrease in their expression, possibly generating a hypomorphic phenotype. In such a scenario, pRb levels are lowered below the threshold necessary for blocking cell cycle progression but remain high enough to execute certain functions such as the prevention of apoptosis. In this regard, it is interesting to note that conditional excision of RB alleles in muscle creatine kinase–Cre–expressing myoblasts promotes apoptosis to some extent during the differentiation time course, and it is possible that this is a consequence of deregulated E2F1 function after pRb loss, which may not occur with our approach. In fact, massive induction of apoptosis after excision of RB in myotubes was also reported by Camarda et al. (2004), prompting them to overexpress the antiapoptotic protein Bcl-2 to counteract this effect in at least some of their experiments. We note that Bcl-2 has been shown to exert antiproliferative effects in certain cell types and thus its enforced expression could have precluded cell cycle reentry in pRb-depleted myotubes (Vairo et al., 1996, 2000). Thus, the fact that apoptosis was provoked by depletion of pRb from myotubes confirms our conclusion regarding an essential role for pRb in maintaining their postmitotic state.
Remarkably, our depletion of pRb from myotubes not only led to reexpression of genes associated with proliferation but also dramatically reduced expression of genes involved in muscle development and function. Because we performed these experiments with fully differentiated cells, it is clear that reduced expression of these genes is not simply a consequence of defective differentiation. Instead, the cells depleted of pRb exhibit the hallmarks of dedifferentiation. One model that could explain our data posits that pRb acts as a coactivator for the expression of muscle differentiation genes by cooperating with other transcription factors that promote differentiation (Singh et al., 1995; Chen et al., 1996; Thomas et al., 2001). Moreover, our findings may also be consistent with a second model in which pRb enhances the activation of MyoD target genes by sequestering the histone deacetylase 1 repressor away from MyoD (Puri et al., 2001). Our genome-wide studies have vastly extended the notion that pRb acts positively to maintain expression of muscle-specific genes after terminal differentiation.
Reevaluating the role of pocket proteins: a model for irreversible cell cycle exit and maintenance of the differentiated state
Although p107 and p130 are highly related to pRb and clearly display highly overlapping biochemical activities, neither p107 nor p130 mutations have been widely implicated in human cancer (Paggi et al., 1996; Dannenberg and te Riele, 2006; Wikenheiser-Brokamp, 2006). This finding has been ascribed to redundancy between pocket proteins as well as tissue-specific requirements (Lee et al., 1996; Robanus-Maandag et al., 1998; Lee et al., 2002; Dannenberg et al., 2004). We found that the function of p107 and p130 is not required for the establishment or the maintenance of cell cycle exit and that their absence does not lead to derepression of cell cycle control genes. This situation is unlike the one prevailing in fibroblasts, where these pocket proteins are essential for the silencing of cell cycle genes during quiescence (Rayman et al., 2002; Balciunaite et al., 2005), and suggests that cell cycle arrest in differentiated muscle is mechanistically very different from that of other cell types. However, our data reveal a role for p107 and p130 in control over a G2/M checkpoint that is uncovered only in the absence of pRb and provide a novel explanation for the role of p107 and p130 in the context of differentiation. A role for p130 in G2/M had been suggested previously (Mayol et al., 1995), which is a hypothesis we have experimentally substantiated here. Moreover, because this function is uncovered exclusively in cells that have simultaneously lost pRb, our findings could explain a long-standing conundrum in the field; namely, why pRb is a tumor suppressor with strong connections to human cancer, whereas mutations in the p107 and p130 genes are not widely associated with tumors.
We propose that pRb acts first to uniquely promote myogenic differentiation and, subsequently, block cell cycle reentry and DNA replication in myotubes (Fig. 6 A). Cells that escape this barrier (if and only if pRb function is compromised) are likely to arrest in late S or G2 phase because a fail-safe mechanism that requires p107 or p130 blocks further cell cycle progression. Loss of this G2/M checkpoint results in further progression of differentiated muscle cells into mitosis.
A role for H3K27 methylation in maintenance of the differentiated state
We have found that trimethylation of H3K27 plays an unanticipated and widespread role in repressing expression of cell cycle genes during differentiation of C2C12 and primary myoblasts. Four characteristics indicate that it constitutes a chromatin mark essential for mediating the effect of pRb in the repression of these genes. First, the mark appears at the promoter of pRb-repressed genes specifically during the differentiation of myotubes, which coincides with gene silencing. Second, the mark is erased after the loss of pRb in myotubes, which coincides with gene derepression. Third, trimethylation of H3K27 is not erased after treatment of myotubes with growth factors, paralleling the serum-insensitive repression of these genes. Fourth, the H3K27Me3 mark appears to be specific to irreversibly arrested myotubes and is not detected when genes are temporarily silenced during cellular quiescence.
Previous studies have implicated methylation of H3K9 in repressing certain cell cycle genes during myogenesis or quiescence (Nielsen et al., 2001; Ait-Si-Ali et al., 2004). Here, H3K27 and H3K9 trimethylation have been more extensively examined during differentiation. We have confirmed that certain promoters (Bub1 and Mcm5) exhibit enhanced trimethylation of H3K9 during myogenic differentiation but that the absolute levels of H3K9 methylation at these loci are substantially lower than those generally observed for H3K27 methylation of the promoters we have examined (unpublished data). One explanation for this difference could relate to differences in antibody specificity; it is possible that the anti–trimethyl H3K9 (“branched”) antibodies used in previous experiments also recognized H4K20Me3 and H3K27Me3 (Nielsen et al., 2001; Ait-Si-Ali et al., 2004; Sarma et al., 2004). In contrast, the anti-H3K27 antibody used in our study is specific for di- and trimethylated forms of H3K27 and does not bind to other histone residues (Sarma et al., 2004). In addition, we note that our studies examined pure myotubes rather than a mixed population of quiescent/undifferentiated and differentiated cells.
Despite numerous attempts to identify the HMTase responsible for silencing pRb targets, we have failed to detect PRC2 binding (Fig. 5 D and not depicted). Preliminary ChIP-on-chip experiments using antibodies against Suz12 have reliably identified several fate determination genes silenced by PRC2 (notably, Hox gene clusters) but did not reveal a clear association between PRC2- and pRb-regulated genes. The interaction of PRC2 with these loci may be outside the range surveyed by our microarrays, be short-lived, or occur at an earlier stage of differentiation. However, a most intriguing possibility is that enzymes distinct from PRC2 may possess H3K27 HMTase activity and target pRb-regulated genes; G9a and Whistle (Tachibana et al., 2001; Kim et al., 2006) are candidates. Both enzymes are expressed in C2C12 cells (unpublished data) and it is therefore possible that they are involved in the trimethylation of H3K27 at pRb-regulated genes during myogenic differentiation. It is thought that PRC2 is responsible for most H3K27 trimethylation in the nucleus (Erhardt et al., 2003; Pasini et al., 2004; Schoeftner et al., 2006) but, because these observations have been made using low-resolution techniques at early developmental stages, a role for other HMTases cannot currently be ruled out.
It is also not presently known how pRb promotes H3K27 methylation of its targets. Our limited ChIP analyses have failed to detect pRb recruitment to relevant promoters, which is reminiscent of recent studies of the Cdkn2a gene, which is also silenced by H3K27 methylation in a pRb-dependent manner without detectable pRb recruitment (Bracken et al., 2007; Kotake et al., 2007). It is possible that a pRb complex transiently establishes a specific chromatin environment at target genes that promotes the subsequent methylation of H3K27 by a distinct enzyme. In such a scenario, stepwise modification of histone tail residues could occur, with deacetylation of H3Ac representing an early, reversible event (Fig. 6 B). H3K27Me3 could constitute an intermediate or final mark, although it appears at genes relatively late in the differentiation process (Figs. 3 D and 6 B). The H3K27Me3 mark is lost and cell cycle genes are reexpressed upon knockdown of pRb, which suggests that there would be constant turnover of these histone modifications in the absence of pRb and that one role of pRb in the establishment of irreversible cell cycle arrest is to tip the scale toward maintenance of this repressive modification.
In contrast with the aforementioned group of pRb targets, we found that Ccnd1 expression was silenced via pRb-independent H3K27 trimethylation and PcG complex binding (Fig. 6 B). It is worth noting that Ccnd1 is at the core of the cell's mitogen-sensing machinery, as it mediates (with Cdk4) the initial pRb inactivation steps in late G1 phase (Blagosklonny and Pardee, 2002). Permanent silencing of its expression could therefore complement the repressive action of pRb on cell cycle effector genes (Fig. 6 B). Additional experiments must be performed to determine the mode of recruitment of PRC complexes at the Ccnd1 promoter.
Materials and methods
Cell culture
The C2C12 murine myoblast cell line (obtained from the American Type Culture Collection) was cultured in growth medium consisting of DME supplemented with 10% fetal bovine serum, 2 mM glutamine, and antibiotics. In all experiments, care was taken to harvest myoblasts in proliferation at a point well before confluence (<60% confluence) to avoid cell cycle exit and spontaneous differentiation. To induce differentiation, myoblasts were grown to confluence, rinsed twice with PBS, and grown in differentiation medium consisting of DME supplemented with 2% horse serum. To harvest myotubes and separate them from the undifferentiated cells (so-called reserve cells), the differentiated cultures were harvested using diluted trypsin, which results in the selective detachment of myotubes (Blais et al., 2005). Primary mouse myoblasts were obtained from the lab of M. Rudnicki (Ottawa Health Research Institute, Ottawa, Canada) or prepared as described previously (Rando and Blau, 1994). In both cases, the cells were grown on Matrigel-coated dishes (Becton Dickinson). Some experiments were also repeated using mouse satellite cells isolated from single soleus muscle fiber cultures as described previously (Rosenblatt et al., 1995). Once enough satellites cells had migrated out of the fibers, the cultures were trypsinized, expanded for one passage, and plated on 8-well slides (Thermo Fisher Scientific) coated with Matrigel. After 4–5 d, the cells had started to differentiate and were transferred to differentiation-promoting medium. 1 wk later, the resulting primary myotubes were transfected with siRNA duplexes as described in RNAi assays. Primary MEFs from mixed backgrounds were provided by G. David and B. Grandinetti (New York University School of Medicine, New York, NY) and grown in growth medium for a maximum of six passages to avoid replicative senescence. For mitogen starvation, they were cultured for 3 d in medium devoid of serum.
Antibodies
The MF20 antibody against myosin heavy chain (MHC; Bader et al., 1982) and the G3G4 antibody against BrdU (George-Weinstein et al., 1993) were obtained from the Developmental Studies Hybridoma Bank. The FITC-coupled anti-BrdU antibody was obtained from Roche. Anti–phospho-Ser10-histone H3 was obtained from Millipore and anti–γ-tubulin and –α-tubulin antibodies were obtained from Sigma-Aldrich. The anti–cyclin B1 monoclonal (GNS1) was a gift of S. Shiff (Robert Wood Johnson University Hospital, New Brunswick, NJ). Anti-H3K27Me2/3 (clone 7B11; a gift of D. Reinberg, New York University School of Medicine) has been characterized previously (Sarma et al., 2004). Anti-H3K27Me2 (ab24684), anti-H3K27Me3 (ab6002), and anti-H3K9Me3 (ab8898) were obtained from Abcam. Anti–pan-acetylated H3 (K9Ac + K14Ac, 06-599) was obtained from Millipore. The anti-Bmi1 mAb was provided by K. Helin (Biotech Research and Innovation Center, Copenhagen, Denmark)
Immunofluorescence and image acquisition
Cells were plated on slide flasks (Thermo Fisher Scientific) and grown and differentiated as described in Cell culture. They were fixed using formalin (Sigma-Aldrich), permeabilized with 1% Triton X-100 in PBS, and blocked in PBS containing 3% BSA. Antibodies were diluted in blocking buffer and FITC- or Cy3-coupled secondary antibodies were used (Jackson ImmunoResearch Laboratories). For BrdU incorporation, myotubes were pulsed for 12 h with 10 μM BrdU (Roche) and then processed for immunofluorescence. After staining with primary antibodies against MHC and secondary Cy3-coupled anti–mouse antibody, the samples were fixed a second time with formalin and denatured for 10 min in 4 N HCl. After neutralization, the samples were incubated with FITC-coupled anti-BrdU (Roche). After incubation with biotin-labeled anti-BrdU antibodies, the samples were further incubated with FITC-labeled streptavidin (Jackson ImmunoResearch Laboratories). For H3S10P staining, the cells were first extracted sequentially for 5 min in CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM piperazine-N,N′-bis[2-ethane-sulfonic acid], Pipes, pH 6.8, and 3 mM MgCl2) and 5 min in CSK buffer containing 0.5% Triton X-100 (Martini et al., 1998) before being fixed with formalin as before. All samples were mounted in Prolong anti-fade medium (Invitrogen). Photomicrography was performed at room temperature using a fluorescence microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a camera (Retiga 2000R; QImaging) and controlled with Metamorph 7.0 software (MDS Analytical Technologies). All photomicrographs were taken using 20× Plan Apo 0.75 differential interference contrast M infinity/0.17 WD 1.0 (for most figures) or 100× Plan Apo 1.40 oil differential interference contrast H infinity/0.17 WD 0.13 (for Fig. S3) objectives (both obtained from Nikon). For all photographic data, images were acquired as 8-bit-per-channel TIF files. In some instances where the overall signal was low, intensity levels were adjusted using the Levels tool of Photoshop one color channel at a time but gamma adjustments were never made. For gel photographs, all lanes for a given gene/protein were subjected to identical adjustments so that the band intensities can be directly compared. Care was also taken to avoid pixel clipping.
RNAi assays
Cells were grown to confluence, induced to differentiate for 4–5 d, and transfected with siRNA duplexes (Thermo Fisher Scientific) using the siMPORTER reagent (Millipore), according to the manufacturer's recommendations. The sequences are listed in Table S2. The amount of RNA transfected was kept constant in each transfection experiment. For experiments performed in myoblasts, cells were transfected at 50% confluence and induced to differentiate for 2 d after they reached confluence.
RT-PCR and expression profiling
RT-PCR was performed using 250 ng of total RNA per reverse transcriptase reaction. 1% of the resulting cDNA was then used as a template for semiquantitative PCR. In each case, PCR reactions were determined to be in the linear phase of the amplification. The sequence of the oligonucleotide primers used is given in Table S3 (available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1) along with the expected size of the amplicons. Expression profiling with microarrays was performed in triplicate (except when noted) as described previously (Blais et al., 2005) using MOE430A 2.0 chips (Affymetrix). The data were analyzed using the program dChip (Li and Hung Wong, 2001) with the PM-MM model and averaging of replicate samples. Additional filtering was performed manually, removing microarray probes for which the maximum mean value across samples was <75 and those for which the number of “present” calls among all samples is <2.
ChIP assays
ChIP assays were performed essentially as described previously (Takahashi et al., 2000). For histone marks, less chromatin was used per immunoprecipitation to maintain the antibody in excess. 1 μg chromatin was used for H3, H3Ac, and H3K27Me2. 7.5 μg chromatin was used for H3K27Me3. In some cases, real-time PCR quantitation (qChIP) of the immunoprecipitated material was performed using the SYBR green method. The value obtained with a nonspecific antibody (normal rabbit IgG) was subtracted from the value obtained with the test antibodies and the overall efficiency was expressed as a fraction of the material used in the ChIP reaction. Values were adjusted to zero when the ChIP signal was lower than that obtained with the irrelevant antibody control. The raw numerical data is presented in Table S2 and the sequences of the oligos used are listed in Table S3. ChIP assays on primary myoblasts and myotubes were done similarly using two sources of primary myoblasts: with fresh cultures prepared according to a previously published method (Rando and Blau, 1994) and with cultures obtained from the lab of M. Rudnicki. In both cases, chromatin was prepared from myoblasts and myotubes and duplicate ChIP assays were performed for each sample for a total of four ChIP assays. The signal of H3K27Me2/3 was then quantitated using real-time PCR.
ChIP-on-chip experiments
A mouse proximal promoter microarray was used to identify the target genes of E2F4 and p130 in C2C12 myotubes using ChIP-on-chip (Blais et al., 2005). The antibodies used were rabbit anti-E2F4 (Santa Cruz Biotechnology, Inc.) and mouse anti-p130 (Becton Dickinson).
Online supplemental material
Fig. S1 shows the expression profiles of pocket proteins during C2C12 differentiation. It also demonstrates the failure of myogenesis and improper cell cycle exit in the absence of pRb. Fig. S2 demonstrates that distinct siRNA duplexes targeting the pocket proteins generate essentially the same effect at the level of target gene expression. Fig. S3 documents mitotic entry after the knockdown of pRb together with either p107 or p130 and suggests the existence of a G2/M checkpoint in which these proteins participate. Fig. S4 further demonstrates that two distinct pRb-targeting siRNA duplexes generate essentially the same effect at the level of gene expression. Fig. S5 shows the di- and trimethylation of H3K27 of additional genes silenced during differentiation in a pRb-dependent and -independent manners. It also demonstrates the expression profile of the Ezh2 HMTase during C2C12 differentiation. Table S1 represents the summary of our expression profiling data. Table S2 gives the raw quantitative ChIP data represented in the main text. Table S3 gives the sequence of all DNA and RNA oligonucleotides used in this paper. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705051/DC1.
Supplemental Material
Acknowledgments
We thank all members of the Dynlacht laboratory; P. Asp for technical help; D. Reinberg, A. Peschiaroli, G. David, and K. Helin for the gift of antibodies and advice; M. Rudnicki for the gift of primary mouse myoblasts and helpful comments; the New York University School of Medicine Genomics Core for invaluable help; B. Grandinetti for the gift of MEFs; and V. d'Escamard, M. Finnerty, and N. Simpson for help with isolation of primary cells from mice.
This work was funded by postdoctoral fellowships from the Canadian Institutes of Health Research to A. Blais and the Susan G. Komen Breast Cancer Foundation (PDF 0504345) to C.J.C. van Oevelen, a grant from the Department of Defense (PC050535) to R. Margueron, and funds from the National Institutes of Health (GM067132 and CA77245) to B.D. Dynlacht.
A. Blais and C.J.C. van Oevelen contributed equally to this paper.
Abbreviations used in this paper: Ccnd1, cyclin D1; ChIP, chromatin immunoprecipitation; HMTase, histone methyltransferase; MEF, mouse embryonic fibroblast; MHC, myosin heavy chain; PcG, polycomb group; pRb, retinoblastoma tumor suppressor protein; PRC, PcG repressor complex.
References
- Ait-Si-Ali, S., V. Guasconi, L. Fritsch, H. Yahi, R. Sekhri, I. Naguibneva, P. Robin, F. Cabon, A. Polesskaya, and A. Harel-Bellan. 2004. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 23:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, Y., M. Nakano, Y. Yamada, I. Saito, and Y. Kanegae. 2005. Practical range of effective dose for Cre recombinase-expressing recombinant adenovirus without cell toxicity in mammalian cells. Microbiol. Immunol. 49:559–570. [DOI] [PubMed] [Google Scholar]
- Bader, D., T. Masaki, and D.A. Fischman. 1982. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J. Cell Biol. 95:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunaite, E., A. Spektor, N.H. Lents, H. Cam, H. Te Riele, A. Scime, M.A. Rudnicki, R. Young, and B.D. Dynlacht. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25:8166–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya, E.V., H.L. Murray, P. Branton, R.A. Young, and W.G. Kaelin Jr. 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell. 18:623–635. [DOI] [PubMed] [Google Scholar]
- Blagosklonny, M.V., and A.B. Pardee. 2002. The restriction point of the cell cycle. Cell Cycle. 1:103–110. [PubMed] [Google Scholar]
- Blais, A., M. Tsikitis, D. Acosta-Alvear, R. Sharan, Y. Kluger, and B.D. Dynlacht. 2005. An initial blueprint for myogenic differentiation. Genes Dev. 19:553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken, A.P., N. Dietrich, D. Pasini, K.H. Hansen, and K. Helin. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken, A.P., D. Kleine-Kohlbrecher, N. Dietrich, D. Pasini, G. Gargiulo, C. Beekman, K. Theilgaard-Monch, S. Minucci, B.T. Porse, J.C. Marine, et al. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam, H., E. Balciunaite, A. Blais, A. Spektor, R.C. Scarpulla, R. Young, Y. Kluger, and B.D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 16:399–411. [DOI] [PubMed] [Google Scholar]
- Camarda, G., F. Siepi, D. Pajalunga, C. Bernardini, R. Rossi, A. Montecucco, E. Meccia, and M. Crescenzi. 2004. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. J. Cell Biol. 167:417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti, G., M. Di Padova, B. Micales, G.E. Lyons, and V. Sartorelli. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18:2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.L., D.J. Riley, Y. Chen, and W.H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794–2804. [DOI] [PubMed] [Google Scholar]
- Cobrinik, D., M.H. Lee, G. Hannon, G. Mulligan, R.T. Bronson, N. Dyson, E. Harlow, D. Beach, R.A. Weinberg, and T. Jacks. 1996. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 10:1633–1644. [DOI] [PubMed] [Google Scholar]
- Dannenberg, J.H., L. Schuijff, M. Dekker, M. van der Valk, and H. te Riele. 2004. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 18:2952–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg, J.H., and H.P. te Riele. 2006. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl. Cell Differ. 42:183–225. [DOI] [PubMed] [Google Scholar]
- Endo, T., and B. Nadal-Ginard. 1998. Reversal of myogenic terminal differentiation by SV40 large T antigen results in mitosis and apoptosis. J. Cell Sci. 111:1081–1093. [DOI] [PubMed] [Google Scholar]
- Erhardt, S., I.H. Su, R. Schneider, S. Barton, A.J. Bannister, L. Perez-Burgos, T. Jenuwein, T. Kouzarides, A. Tarakhovsky, and M.A. Surani. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 130:4235–4248. [DOI] [PubMed] [Google Scholar]
- Faust, C., K.A. Lawson, N.J. Schork, B. Thiel, and T. Magnuson. 1998. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 125:4495–4506. [DOI] [PubMed] [Google Scholar]
- George-Weinstein, M., R.F. Foster, J.V. Gerhart, and S.J. Kaufman. 1993. In vitro and in vivo expression of alpha 7 integrin and desmin define the primary and secondary myogenic lineages. Dev. Biol. 156:209–229. [DOI] [PubMed] [Google Scholar]
- Gu, W., J.W. Schneider, G. Condorelli, S. Kaushal, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 72:309–324. [DOI] [PubMed] [Google Scholar]
- Huang, S., W.H. Lee, and E.Y. Lee. 1991. A cellular protein that competes with SV40 T antigen for binding to the retinoblastoma gene product. Nature. 350:160–162. [DOI] [PubMed] [Google Scholar]
- Huh, M.S., M.H. Parker, A. Scime, R. Parks, and M.A. Rudnicki. 2004. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 166:865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurford, R.K., Jr., D. Cobrinik, M.H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447–1463. [DOI] [PubMed] [Google Scholar]
- Kim, S.M., H.J. Kee, G.H. Eom, N.W. Choe, J.Y. Kim, Y.S. Kim, S.K. Kim, H. Kook, H. Kook, and S.B. Seo. 2006. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem. Biophys. Res. Commun. 345:318–323. [DOI] [PubMed] [Google Scholar]
- Korenjak, M., B. Taylor-Harding, U.K. Binne, J.S. Satterlee, O. Stevaux, R. Aasland, H. White-Cooper, N. Dyson, and A. Brehm. 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 119:181–193. [DOI] [PubMed] [Google Scholar]
- Kotake, Y., R. Cao, P. Viatour, J. Sage, Y. Zhang, and Y. Xiong. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 21:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev. 16:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E.Y., H. Cam, U. Ziebold, J.B. Rayman, J.A. Lees, and B.D. Dynlacht. 2002. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell. 2:463–472. [DOI] [PubMed] [Google Scholar]
- Lee, M.H., B.O. Williams, G. Mulligan, S. Mukai, R.T. Bronson, N. Dyson, E. Harlow, and T. Jacks. 1996. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 10:1621–1632. [DOI] [PubMed] [Google Scholar]
- Lee, T.I., R.G. Jenner, L.A. Boyer, M.G. Guenther, S.S. Levine, R.M. Kumar, B. Chevalier, S.E. Johnstone, M.F. Cole, K. Isono, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 125:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., and W. Hung Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra, A., M. Vooijs, H.B. Beverloo, B.A. Allak, E. van Drunen, R. Kanaar, A. Berns, and J. Jonkers. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 98:9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso, M., M. Montanari, and A. Giordano. 2006. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene. 25:5263–5267. [DOI] [PubMed] [Google Scholar]
- Martini, E., D.M. Roche, K. Marheineke, A. Verreault, and G. Almouzni. 1998. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 143:563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayol, X., J. Garriga, and X. Grana. 1995. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene. 11:801–808. [PubMed] [Google Scholar]
- Molkentin, J.D., and E.N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445–453. [DOI] [PubMed] [Google Scholar]
- Nielsen, S.J., R. Schneider, U.M. Bauer, A.J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R.E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature. 412:561–565. [DOI] [PubMed] [Google Scholar]
- Novitch, B.G., G.J. Mulligan, T. Jacks, and A.B. Lassar. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135:441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll, D., S. Erhardt, M. Pagani, S.C. Barton, M.A. Surani, and T. Jenuwein. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V. 2003. Polycomb, epigenomes, and control of cell identity. Cell. 112:599–606. [DOI] [PubMed] [Google Scholar]
- Paggi, M.G., A. Baldi, F. Bonetto, and A. Giordano. 1996. Retinoblastoma protein family in cell cycle and cancer: a review. J. Cell. Biochem. 62:418–430. [DOI] [PubMed] [Google Scholar]
- Pardee, A.B. 1974. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA. 71:1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini, D., A.P. Bracken, M.R. Jensen, E. Lazzerini Denchi, and K. Helin. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23:4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer, A., E.P. Brandon, N. Kootstra, F.H. Gage, and I.M. Verma. 2001. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc. Natl. Acad. Sci. USA. 98:11450–11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, P.L., S. Iezzi, P. Stiegler, T.T. Chen, R.L. Schiltz, G.E. Muscat, A. Giordano, L. Kedes, J.Y. Wang, and V. Sartorelli. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell. 8:885–897. [DOI] [PubMed] [Google Scholar]
- Qian, Y.W., Y.C. Wang, R.E. Hollingsworth Jr., D. Jones, N. Ling, and E.Y. Lee. 1993. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 364:648–652. [DOI] [PubMed] [Google Scholar]
- Rando, T.A., and H.M. Blau. 1994. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125:1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman, J.B., Y. Takahashi, V.B. Indjeian, J.H. Dannenberg, S. Catchpole, R.J. Watson, H. te Riele, and B.D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose, L., and R. Paro. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38:413–443. [DOI] [PubMed] [Google Scholar]
- Robanus-Maandag, E., M. Dekker, M. van der Valk, M.L. Carrozza, J.C. Jeanny, J.H. Dannenberg, A. Berns, and H. te Riele. 1998. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12:1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt, J.D., A.I. Lunt, D.J. Parry, and T.A. Partridge. 1995. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. Anim. 31:773–779. [DOI] [PubMed] [Google Scholar]
- Sage, J., A.L. Miller, P.A. Perez-Mancera, J.M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 424:223–228. [DOI] [PubMed] [Google Scholar]
- Sarma, K., K. Nishioka, and D. Reinberg. 2004. Tips in analyzing antibodies directed against specific histone tail modifications. Methods Enzymol. 376:255–269. [DOI] [PubMed] [Google Scholar]
- Schneider, J.W., W. Gu, L. Zhu, V. Mahdavi, and B. Nadal-Ginard. 1994. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 264:1467–1471. [DOI] [PubMed] [Google Scholar]
- Schoeftner, S., A.K. Sengupta, S. Kubicek, K. Mechtler, L. Spahn, H. Koseki, T. Jenuwein, and A. Wutz. 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25:3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, D.P., and D.M. Livingston. 2001. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 8:233–243. [DOI] [PubMed] [Google Scholar]
- Singh, P., J. Coe, and W. Hong. 1995. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 374:562–565. [DOI] [PubMed] [Google Scholar]
- Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309–25317. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., J.B. Rayman, and B.D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Thomas, D.M., S.A. Carty, D.M. Piscopo, J.S. Lee, W.F. Wang, W.C. Forrester, and P.W. Hinds. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell. 8:303–316. [DOI] [PubMed] [Google Scholar]
- Vairo, G., K.M. Innes, and J.M. Adams. 1996. Bcl-2 has a cell cycle inhibitory function separable from its enhancement of cell survival. Oncogene. 13:1511–1519. [PubMed] [Google Scholar]
- Vairo, G., T.J. Soos, T.M. Upton, J. Zalvide, J.A. DeCaprio, M.E. Ewen, A. Koff, and J.M. Adams. 2000. Bcl-2 retards cell cycle entry through p27(Kip1), pRB relative p130, and altered E2F regulation. Mol. Cell. Biol. 20:4745–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp, K.A. 2006. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell. Mol. Life Sci. 63:767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., A. de Bruin, H.I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J.C. Thompson, M.C. Ostrowski, et al. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 421:942–947. [DOI] [PubMed] [Google Scholar]
- Zacksenhaus, E., Z. Jiang, D. Chung, J.D. Marth, R.A. Phillips, and B.L. Gallie. 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 10:3051–3064. [DOI] [PubMed] [Google Scholar]
- Zhang, H.S., M. Gavin, A. Dahiya, A.A. Postigo, D. Ma, R.X. Luo, J.W. Harbour, and D.C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 101:79–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.