Abstract
Bcl-2 family member Bid is subject to autoinhibition; in the absence of stimuli, its N-terminal region sequesters the proapoptotic Bcl-2 homology 3 (BH3) domain. Upon proteolytic cleavage in its unstructured loop, Bid is activated, although structural data reveal no apparent resulting conformational change. We found that, upon Bid cleavage, the N-terminal fragment (tBid-N) is ubiquitinated and degraded, thus freeing the BH3 domain in the C-terminal fragment (tBid-C). Ubiquitination of tBid-N is unconventional because acceptor sites are neither lysines nor the N terminus. Chemical approaches implicated thioester and hydroxyester linkage of ubiquitin and mutagenesis implicated serine and possibly threonine as acceptor residues in addition to cysteine. Acceptor sites reside predominantly but not exclusively in helix 1, which is required for ubiquitination and degradation of tBid-N. Rescue of tBid-N from degradation blocked Bid's ability to induce mitochondrial outer membrane permeability but not mitochondrial translocation of the cleaved complex. We conclude that unconventional ubiquitination and proteasome-dependent degradation of tBid-N is required to unleash the proapoptotic activity of tBid-C.
Introduction
In mammalian cells, mitochondria activate the apoptotic execution machinery. Apoptotic stimuli induce mitochondrial outer membrane permeability and the release of proapoptotic proteins such as cytochrome c (Cyt c) into the cytosol. The collective action of these molecules results in effector caspase activation and apoptotic cell death (Wang, 2001). Mitochondrial permeabilization is controlled by the Bcl-2 protein family, which comprises the antiapoptotic Bcl-2 subfamily and the proapoptotic Bax/Bak and Bcl-2 homology 3 (BH3) domain–only subfamilies (Adams and Cory, 1998). Their functional activity relies on homo- and heterodimerization, which proceeds by interaction of the BH3 domain α helix with a groove formed by the BH1 and 2 domains (Sattler et al., 1997).
The BH3 domain–only subfamily has many members that specialize in responsiveness to specific apoptotic stimuli (Puthalakath and Strasser 2002). BH3-only proteins require Bax or Bak to induce cell death. They induce assembly of Bax/Bak into homomultimers in the mitochondrial outer membrane (Wei et al., 2000; Kuwana et al., 2002). These multimers are postulated to either form transmembrane pores themselves or to facilitate pore formation. Inhibitory Bcl-2 family members sequester only BH3 and/or Bax/Bak proteins, thereby preventing their participation in proapoptotic complexes (Cheng et al., 2001).
The BH3 domain–only protein Bid conveys the apoptotic signal from death receptors, such as TNF receptor 1, TNF-related apoptosis-inducing ligand (TRAIL) receptors, and Fas/CD95 to mitochondria (Li et al., 1998; Luo et al., 1998). Upon ligand binding, these death receptors induce formation of a complex between the intracellular adaptor Fas-associated death domain and procaspase-8 or -10. This results in increased local concentrations of these inducer caspases, which leads to their activation (Boatright and Salvesen, 2003). Active caspase-8/10 can cleave and activate Bid as well as effector caspases. In certain cell types, the mitochondrial route is dispensable for death receptor–induced apoptosis, whereas in others it constitutes an essential amplification loop for effector caspase activation (Yin et al., 1999).
The three-dimensional organization of full-length Bid has been resolved by nuclear magnetic resonance (NMR; Chou et al., 1999; McDonnell et al., 1999). Residues 1–12 and 43–77 form unstructured loops. The structured portion consists of eight α helices arranged in a compact fold (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200707063/DC1). Of these, helix 3 constitutes the BH3 domain, which is fixed by hydrophobic contacts with helixes 1 and 8. The sites for proteolytic cleavage by caspase-8 or granzyme B lie in the second unstructured loop (D60 and D75, respectively). In solution, N- and C-terminal fragments of Bid remain associated upon cleavage by caspase-8 and the complex undergoes no apparent conformational change (Chou et al., 1999; Kudla et al., 2000; Zha et al., 2000). Cleavage generates an N-terminal glycine in truncated C-terminal Bid (tBid-C) that can be myristoylated. This lipid modification facilitates targeting of tBid-C to mitochondria but is not required for it (Zha et al., 2000). Rather, helices 4, 5, and 6 appeared to be the critical elements for such targeting, specifically to cardiolipin-enriched domains in the mitochondrial membrane (Lutter et al., 2000).
Making the BH3 domain available for interactions with the BH1/BH2 groove of other Bcl-2 family members is essential for Bid's proapoptotic function. The solution structure of Bid indicates that the BH3 domain must be freed from the structural constraint that is exerted by the N-terminal fragment of Bid (tBid-N; Chou et al., 1999; McDonnell et al., 1999). Indeed, several biochemical studies have shown that tBid-N acts as an inhibitor of tBid-C (Tan et al., 1999; Kudla et al., 2000; Renshaw et al., 2004). Specifically, Tan et al. (1999) produced compelling evidence for autoinhibition of Bid by interactions between helix 2 and the BH3 domain. A mutant full-length Bid protein in which these interactions were abrogated was as effective as tBid-C in inducing apoptosis and binding to Bcl-xL (Tan et al., 1999). The natural occurrence of an alternative splice form of Bid that lacks the BH3 domain but possesses the inhibitory N-terminal sequences lends further support to the idea that this is an important mode of tBid-C regulation (Renshaw et al., 2004). We have addressed the question of how Bid cleavage can release tBid-C from inhibition by tBid-N. We reveal that this is accomplished by unconventional ubiquitination of the tBid-N fragment followed by its proteasomal degradation.
Results
tBid-N disappears upon its generation
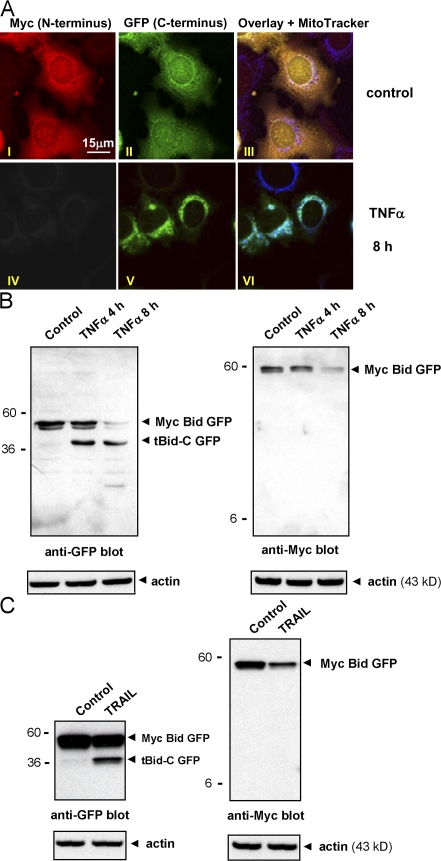
To follow the fate of tBid-N and tBid-C, Bid was N-terminally Myc tagged, C-terminally fused to GFP, and expressed in MCF-7 breast carcinoma cells, which were examined by confocal laser scanning microscopy (CLSM). In unstimulated cells, Myc and GFP signals indicated a cytoplasmic localization of full-length Myc Bid GFP (Fig. 1 A, I–III). After stimulation with TNF-α for 8 h, the GFP signal was punctate and colocalized with MitoTracker, which indicates translocation of tBid-C GFP to mitochondria (Fig. 1 A, V and VI). Surprisingly, Myc tBid-N was undetectable by CLSM (Fig. 1 A, IV).
Figure 1.
tBid-N disappears upon its generation. (A) MCF-7 cells expressing Myc Bid GFP were stimulated with TNF-α for 8 h or not (control), stained to detect mitochondria (MitoTracker) and the N-terminal Myc tag, and examined by CLSM. (B and C) MCF-7 cells expressing Myc Bid GFP were stimulated with TNF-α or TRAIL. Equal amounts of total lysate protein were separated by SDS-PAGE, blotted to nitrocellulose, probed with antibodies to the N-terminal Myc and C-terminal GFP tags, and reprobed with an anti-actin antibody as a loading control. Molecular masses (kD) of marker proteins are indicated.
Full-length Myc Bid GFP was readily detectable by both anti-Myc and -GFP immunoblotting in unstimulated MCF-7 cells and gradually decreased after stimulation with TNF-α (Fig. 1 B). Although the tBid-C GFP fragment could readily be visualized at the 4- and 8-h time points, the Myc tBid-N fragment (predicted molecular mass, 7 kD) was undetectable (Fig. 1 B). Similar results were obtained with an antibody that recognizes both N- and C-terminal epitopes on Bid (unpublished data). After stimulation with the death receptor ligand TRAIL, a similar picture emerged. At the 8-h time point, full-length Myc Bid GFP levels were decreased and a tBid-C GFP fragment had been generated. However, no Myc tBid-N was detectable (Fig. 1 C). Collectively, these results suggest that the tBid-N fragment that is generated by caspase-8/10 is rapidly degraded.
tBid-N is degraded in a proteasome-dependent manner
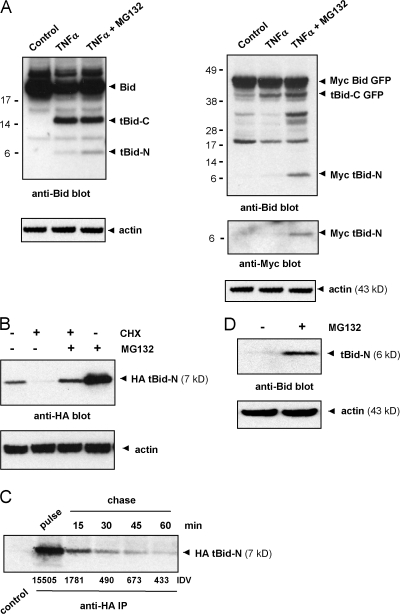
To investigate whether tBid-N was degraded by the proteasome, MCF-7 cells were stimulated with TNF-α in the presence or absence of the proteasome inhibitor MG132. Stimulation with TNF-α for 8 h resulted in the appearance of the endogenous tBid-C fragment but the tBid-N fragment was barely detectable (Fig. 2 A, left). When cells were stimulated in the presence of MG132, the amount of endogenous tBid-N was clearly increased. Similar results were obtained with transfected Myc Bid GFP; the Myc tBid-N fragment was only detectable when cells were stimulated in the presence of MG132 (Fig. 2 A, right).
Figure 2.
tBid-N is degraded by the proteasome. (A) MCF-7 cells that were not transfected (left) or transfected with Myc Bid GFP (right) were stimulated with TNF-α alone or in combination with MG132 for 8 h. Total lysates were probed for endogenous Bid (left) or transfected Myc Bid GFP (right) and cleavage fragments thereof with an anti-Bid antibody. Reprobing was performed with an anti-Myc antibody. (B) HeLa cells expressing HA-tagged tBid-N were treated for 8 h with CHX and the proteasome inhibitor MG132 as indicated. Total cell lysates were probed for HA tBid-N with an anti-HA antibody. (C) HeLa cells expressing HA tBid-N were labeled with [35S]methionine and [35S]cysteine for 2 h followed by a chase with nonradioactive amino acids for the indicated time periods. Bid protein was immunoprecipitated (IP) from detergent lysates with anti-HA mAb and resolved by SDS-PAGE. Radioactive signals were quantified by phosphorimaging. IDV, integrated density value. (D) HeLa cells transfected to express untagged tBid-N were treated or not with MG132 for 16 h. Total lysates were probed for tBid-N with an anti-Bid antibody. Blots were reprobed with an anti-actin antibody as indicated. Molecular masses (kD) of marker proteins are indicated.
To determine whether targeting of tBid-N for proteasomal degradation required a proapoptotic stimulus, the tBid-N fragment was expressed in cells. The degradation pathway was constitutively active because protein synthesis inhibition with cycloheximide (CHX) for 8 h resulted in the loss of HA-tagged tBid-N (Fig. 2 B). Proteasome inhibition with MG132 abrogated this loss and increased the HA tBid-N pool in cells that had not been treated with CHX (Fig. 2 B). HA tBid-N–expressing cells were pulse labeled for 2 h with [35S]methionine and [35S]cysteine and the fate of the labeled pool was followed during a 60-min chase period. The half-life of the labeled HA tBid-N pool was ∼5 min (Fig. 2 C). Untagged tBid-N was even more unstable than the HA-tagged version; it was only detectable when cells had been incubated with MG132 (Fig. 2 D). Collectively, these results indicate that tBid-N, upon its generation by caspase-8/10, is targeted for proteasome-dependent degradation by a pathway that is not dependent on apoptotic stimulation.
tBid-N is ubiquitinated in an unconventional manner
Because tBid-N was targeted to the proteasome, we investigated whether it was ubiquitinated. To this end, tBid-N N-terminally tagged with the HA epitope (sequence YPYDVPDYA) was expressed in HeLa cells with or without FLAG-tagged ubiquitin. An anti-HA reactive smear, characteristic of polyubiquitination, was associated with immunoprecipitated HA tBid-N (Fig. 3 A). In addition, we examined ubiquitination of tBid-N as generated from full-length Bid after death receptor stimulation. MCF-7 cells expressing Myc Bid GFP were left untreated or stimulated with TNF-α for 8 h in the presence of MG132. Lysates were immunoprecipitated with anti-GFP and immunoprecipitated sequentially with an anti-Myc antibody to recover a Myc tBid-N fragment. Immunoblotting for Bid and ubiquitin revealed that ubiquitin was uniquely associated with cleaved but not full-length Bid and specified that Myc tBid-N contained ubiquitin (Fig. 3 B). Ubiquitin was also found to be associated with tBid-C in accordance with previously published data (Breitschopf et al., 2000b).
Figure 3.
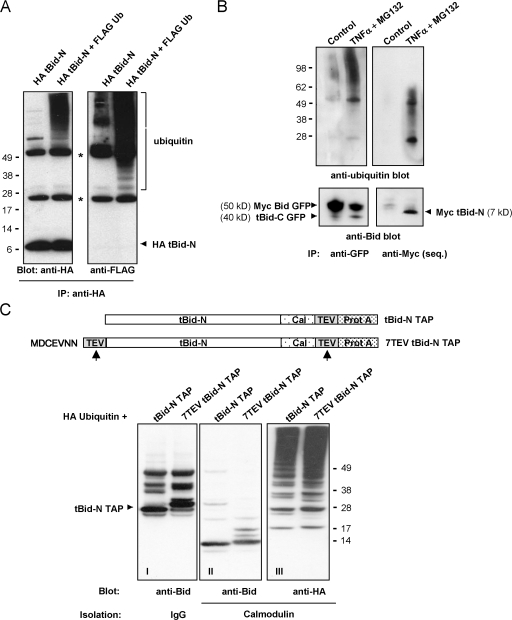
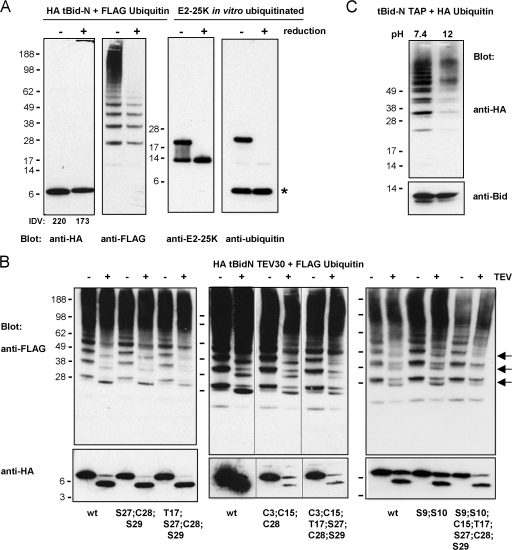
tBid-N is ubiquitinated in an unconventional manner. (A) HeLa cells were transfected to express HA tBid-N alone or in combination with FLAG-tagged ubiquitin. HA tBid-N was immunoprecipitated and probed with an anti-HA mAb to detect Bid and probed with anti-FLAG mAb to detect ubiquitin. Asterisks indicate heavy and light chains of anti-HA mAb. (B) MCF-7 cells expressing Myc Bid GFP were stimulated with TNF-α in the presence of MG132 for 8 h or not stimulated (control). Lysates were immunoprecipitated with an anti-GFP antibody and immunoprecipitated sequentially (seq) with an anti-Myc mAb, and immunoprecipitates (IP) were probed with an anti-Bid antibody and a P4D1 anti-ubiquitin mAb. (C) Assay to determine whether the N terminus is ubiquitinated. In the diagram, the tBid-N TAP proteins are assessed for ubiquitination. Cal–TEV–Prot A indicates the TAP tag with calmodulin- and IgG-binding sites separated by a cleavage site for TEV protease (arrows). HeLa cells were transfected to express HA-ubiquitin and TAP-tagged tBid-N. TAP-tagged tBid-N proteins were isolated with IgG beads, digested with TEV protease, and reisolated with calmodulin beads. Isolates were probed for ubiquitin with an anti-HA mAb and for tBid-N with an anti-Bid antibody. Note that in I, the Prot A sequence is still attached to the tBid-N TAP species and the anti-Bid antibody binds to it. Molecular masses (kD) of marker proteins are indicated.
Ubiquitination was surprising because tBid-N has no lysine residues that conventionally act as ubiquitin acceptor sites (Fig. S1 B). Typically, ubiquitin can be linked either via a peptide bond to the ɛ amino group of a lysine residue or to the α amino group of an N-terminal residue (Ciechanover and Ben-Saadon, 2004). Because linkage to lysine was excluded, we investigated whether the N terminus of tBid-N acted as a ubiquitin acceptor site. To this end, tBid-N was fused C-terminally to a tandem affinity purification (TAP) tag, which contains a calmodulin-binding domain and protein A sequence separated by a tobacco etch virus (TEV) protease-sensitive site (ENLYFQG). This allows for two-step purification; first on IgG beads and then after cleavage by TEV on calmodulin beads (Rigaut et al., 1999). In one tBid-N TAP construct (TEV tBid-N TAP 7), sequences encoding the first seven amino acids of tBid-N and a TEV protease site were cloned upstream of the tBid-N coding region (Fig. 3 C). If tBid-N would indeed be ubiquitinated at its N-terminal residue, TEV cleavage should remove the associated ubiquitin. tBid-N TAP and TEV tBid-N TAP 7 were expressed with HA-ubiquitin in HeLa cells. Fig. 3 C (I) shows the position of tBid-N TAP proteins before TEV cleavage, as isolated with IgG beads. TEV cleavage was successful, as shown by the difference in migration of Bid species recovered on calmodulin beads (Fig. 3 C, II). After TEV cleavage, ubiquitin remained attached to digestion product of TEV tBid-N TAP 7 (Fig. 3 C, III). TEV protease had efficiently cleaved at the N-terminal site because the difference in molecular mass between the two Bid species was smaller than before TEV cleavage (Fig. 3 C, compare I and II). A very small difference remained because TEV cleaves within the added protease recognition sequence. Ubiquitin was attached to Bid and not to the TAP tag because the TAP tag alone did not carry ubiquitin upon coexpression with HA-ubiquitin in HeLa cells (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200707063/DC1). This experiment argues against the possibility that the N-terminal residue of tBid-N is the ubiquitin acceptor site. Moreover, analysis of the first tryptic peptide of tBid-N TAP by MALDI-TOF/TOF mass spectrometry determined that the N terminus of Bid is acetylated and thereby not available for ubiquitin modification (unpublished data). These data indicate that tBid-N is ubiquitinated at a site that is neither a lysine nor the N terminus.
Helix 1 in Bid is critical for ubiquitination
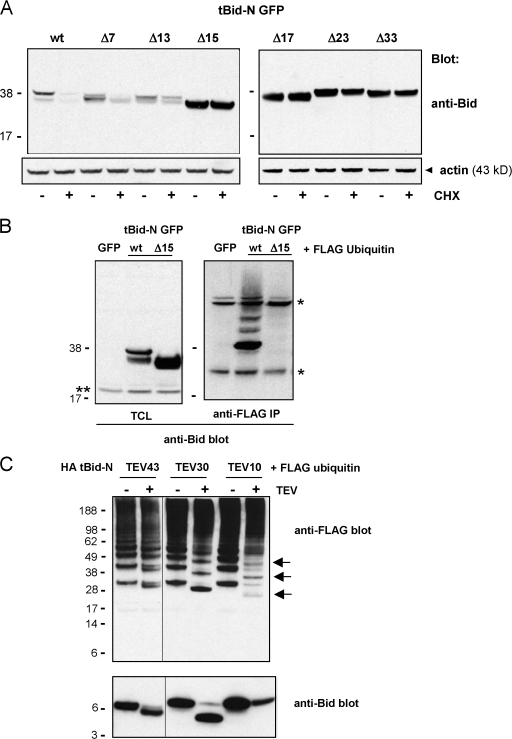
To identify regions in tBid-N that were required for ubiquitination and concomitant proteasome-mediated degradation, we made progressive N-terminal deletions. Deletion mutants of tBid-N GFP were expressed in HeLa cells, which were treated or not with CHX for 8 h and stability was assessed by immunoblotting of total cell lysates. Although tBid-N GFP remained highly unstable upon deletion of up to 13 N-terminal residues, deletion of 15 or more (17, 23, or 33) N-terminal amino acids dramatically increased its half-life (Fig. 4 A). To assess whether stabilization correlated with decreased ubiquitination, wild-type and Δ15 tBid-N GFP were expressed in HeLa cells together with FLAG-tagged ubiquitin. Anti-Bid immunoblotting of total cell lysates showed that wild-type and Δ15 tBid-N GFP were expressed to a similar extent (Fig. 4 B). Ubiquitinated tBid-N GFP species were immunoprecipitated with an anti-FLAG antibody and identified by anti-Bid immunoblotting. This analysis showed that wild-type tBid-N GFP was polyubiquitinated, whereas the Δ15 tBid-N GFP mutant bore virtually no ubiquitin (Fig. 4 B). FLAG ubiquitin expression was comparable in all transfectants (unpublished data). The combined findings in the stability and ubiquitination assays lead to the conclusion that residues 14 and/or 15 contain information that is critical for ubiquitination and destabilization of tBid-N. These are the first residues that take part in the α helix 1 of Bid (Fig. S1 B). Their deletion is expected to disrupt this structure and may therefore result in the loss of ubiquitination sites and/or the loss of a binding site for the ubiquitin ligase.
Figure 4.
Helix 1 is critical for tBid-N ubiquitination and degradation. (A) Wild-type (wt) tBid-N GFP and deletion (Δ) mutants lacking 7, 13, 15, 17, 23, or 33 N-terminal amino acids were expressed in HeLa cells, which were treated (+) or not (−) with CHX for 8 h. The tBid-N GFP proteins were detected in total lysates by immunoblotting with anti-Bid antibody. (B) GFP, wt, and Δ15 tBid-N GFP were expressed in HeLa cells together with FLAG-tagged ubiquitin. Total cell lysates (TCL) were probed with an anti-Bid antibody to assess tBid-N expression levels. Ubiquitinated protein species were isolated with an anti-FLAG mAb (IP) and probed with an anti-Bid antibody. Single asterisk indicates heavy and light chains of the anti-FLAG mAb; double asterisk indicates endogenous full-length Bid. (C) FLAG-tagged ubiquitin was coexpressed in HeLa cells with HA tBid-N versions with a TEV protease cleavage site at position 10, 30, or 43. HA tBid-N molecules were isolated, incubated with (+) or without (−) TEV protease, run on a gel, and probed by immunoblotting for Bid and ubiquitin (anti-FLAG). In the case of HA tBid-N TEV 10, the HA-tagged cleavage fragment repeatedly did not resolve on gel but the remaining undigested HA tBid-N TEV 10 and weakly ubiquitinated species of the cleavage fragment (arrows) are clearly visible. All experiments in this figure were performed three times with similar results. Molecular masses (kD) of marker proteins are indicated.
To ascertain whether helix 1 contained ubiquitination sites, we performed mapping experiments. Cleavage sites for the TEV protease were introduced in HA tBid-N after residues 10, 14, 30, or 43 and these recombinant proteins were expressed in the presence of FLAG ubiquitin. They were isolated with anti-HA antibody, digested with TEV protease, and probed for the presence of ubiquitin and Bid. In the case of the HA tBid-N TEV 43 and 30 proteins, cleavage with TEV protease resulted in a shift in molecular mass of both Bid and the ubiquitin signal. There was no apparent loss of the ubiquitin signal (Fig. 4 C). This proves that tBid-N is directly ubiquitinated and indicates that predominant ubiquitin acceptor sites are among residues 1–30 of tBid-N. The HA tBid-N TEV 14 protein did not cleave with TEV protease (unpublished data). Upon TEV digestion of HA tBid-N TEV 10, the ubiquitin signal was strongly reduced (Fig. 4 C). The HA tBid-N 1–10 fragment did not resolve on a gel, but weakly ubiquitinated species of this fragment are visible (Fig. 4 C, arrows). The remaining ubiquitin was mainly associated with the undigested HA tBid-N TEV 10. We conclude that tBid-N is predominantly ubiquitinated on sites between residues 10 and 30 but also to a certain extent on sites between residues 1 and 10. The deletion and mapping experiments collectively indicate that helix 1 (residues 14–29; Fig. S1 B) is critical for ubiquitination to occur.
tBid-N is likely ubiquitinated on cysteine, serine, and/or threonine residues
Lack of lysine residues is a common feature of tBid-N in different species (human, chimpanzee, mouse, rat, cow, and pig; Fig. S1 C). We surmised that tBid-N might be ubiquitinated on cysteine residues. Three such sites are available among residues 1–30 of human tBid-N: C3, C15, and C28. Ubiquitin linkage to cysteine is not uncommon because the E2 and E3 enzymes are charged with ubiquitin in this manner. In such case, the carboxyl group of the C-terminal glycine in ubiquitin forms a thioester bond with the sulfhydryl group in cysteine, which is easily reducible. To address this possibility, ubiquitinated HA tBid-N was isolated and subjected to reduction with DTT under denaturing conditions. HA tBid-N was reprecipitated with an anti-HA antibody to separate it from possibly dissociated ubiquitin moieties. As a positive control, we used in vitro–ubiquitinated E2-25K, an E2 that is monoubiquitin charged through a thioester bond in its active site (Pichler et al., 2005). Indeed, DTT treatment resulted in a loss of ubiquitin from E2-25K species (Fig. 5 A). Importantly, the same treatment diminished the amount of ubiquitin appended to HA tBid-N (Fig. 5 A). This experiment indicates that cysteines are among the ubiquitin acceptor sites in tBid-N. However, we found with high reproducibility that ubiquitin was partially retained on tBid-N upon reduction, which argues that other residues play roles as acceptor sites as well. Point mutation of C15 or C28 either alone or in conjunction (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200707063/DC1) did not stabilize tBid-N (Fig. S3). It also did not reproducibly reduce ubiquitination of the 1–30 fragment of HA tBid-N TEV 30 (Fig. 5 B). Additional mutation of C3 made HA tBid-N TEV 30 very unstable, such that it could only be expressed at lower levels than the wild-type protein (Figs. 5 B and S3). However, it could clearly be demonstrated that the cysteine-deficient (C3;C15;C28) mutant of HA tBid-N TEV 30 still carried ubiquitin after TEV digestion (Fig. 5 B). This relates with the partial reducibility of ubiquitination and leads to the important conclusion that residues 1–30 of tBid-N harbor residues, other than cysteine, to which ubiquitin can be conjugated.
Figure 5.
Ubiquitin is partially reducible, hydrolyzable, and affected by combined mutation of S, T, and C residues. (A) HA tBid-N was coexpressed with FLAG ubiquitin in HeLa cells. Anti-HA immunoprecipitates were reduced (+) or not (−) as described in Materials and methods and reprecipitated with an anti-HA mAb. Reprecipitates were probed for Bid (anti-HA) and ubiquitin (anti-FLAG). Recombinant E2-25K as a positive control for cysteine ubiquitination was ubiquitinated in vitro, reduced or not, and probed with mAb to E2-25K or ubiquitin. The asterisk indicates free recombinant ubiquitin. IDV, integrated density values showing comparable tBid-N loading. (B) Alanine substitution mutations were generated in HA tBid-N TEV 30. All mutants listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200707063/DC1) were tested for ubiquitination and part of the results are shown here. Wild-type and mutant proteins were expressed in HeLa cells together with FLAG ubiquitin, isolated with anti-HA mAb, and treated (+) or not (−) with TEV protease. Anti-HA immunoblotting was performed to assess Bid expression and cleavage and anti-FLAG immunoblotting was performed to examine ubiquitination. Arrows indicate positions of the ubiquitinated TEV cleavage fragment. (C) tBid-N TAP was coexpressed with HA-ubiquitin in HeLa cells. Cell lysates were treated at pH 7.4 or 12 as indicated in Materials and methods and tBid-N TAP was purified on the TAP tag. Calmodulin-bound protein was separated by SDS-PAGE and probed for Bid (anti-Bid) and ubiquitin (anti-HA). All experiments in this figure were performed three times with similar results. Molecular masses (kD) of marker proteins are indicated. Black lines indicate that intervening lanes have been spliced out.
The potential alternative ubiquitination sites in tBid-N (1–30) are serine and threonine residues because the carboxyl group of the C-terminal glycine in ubiquitin can theoretically form an ester bond with the hydroxyl group in the side chain of these amino acids. There is recent biological precedent for this type of ubiquitin conjugation by viral ubiquitin ligases (Wang et al., 2007). To test this possibility, we investigated whether the ubiquitin linkage was susceptible to alkaline hydrolysis. HeLa cells expressing tBid-N TAP and HA-ubiquitin were lysed at either pH 7.4 or 12, heated for 10 min at 70°C, diluted in buffer with neutral pH, and subjected to TAP purification. Isolates were probed for Bid and ubiquitin. Equal amounts of tBid-N TAP were recovered from both lysates but, after treatment at pH 12, the amount of ubiquitin associated with tBid-N was greatly diminished (Fig. 5 C). Thioester bonds are very easily hydrolysable but the dramatic loss of ubiquitination after hydrolysis was compatible with the notion that hydroxyl ester bonds were involved as well. To verify that the hydrolysis procedure left conventional lysine-linked ubiquitin moieties unaffected, we tested its effect on a complex of purified, in vitro–ubiquitinated Ring1b and Bmi1 (Buchwald et al., 2006). After the ubiquitination reaction, the complex was incubated at pH 7.4 or 12, exactly as was done for tBid-N TAP. In addition, it was heated in the presence of β-mercaptoethanol to test the effect of reduction. The conditions were neutralized and the complex was recovered by virtue of His tags and examined for the extent of ubiquitination by immunoblotting. None of the treatments affected the ubiquitination status of Ring1b (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200707063/DC1).
For mutation analysis of serines and threonines, we focused on the residues in helix 1. Among these, T17 is completely conserved between species (Fig. S1 C). However, alanine substitution of T17 alone did not stabilize tBid-N or affect ubiquitination of the 1–30 fragment of tBid-N (unpublished data). Additional mutation of S27, C28, and S29 or mutation of all serines, threonines, and cysteines in helix 1 did not stabilize tBid-N either (Fig. S3). It reproducibly reduced ubiquitination of the 1–30 fragment to a certain extent but did not abrogate it (Fig. 5 B). Additional mutation of C3 rendered HA tBid-N TEV 30 highly unstable (Fig. S3), but we could reveal ubiquitination on the 1–30 fragment, which supports the conclusion that residues other than cysteines are acceptor sites (Fig. 5 B). This result indicated a possible role for S9 and/or S10. However, alanine substitution of these residues either alone or in conjunction with the potential acceptor sites in helix 1 did not stabilize tBid-N (Fig. S3). Importantly, ubiquitination of the 1–30 fragment was virtually abrogated when alanine substitution of S9 and S10 was combined with alanine substitution of cysteine, threonine, and serine residues in helix 1 (Fig. 5 B). This result indicates that one or more of these residues is a ubiquitination site.
We conclude from the combined biochemical and mutation analyses that tBid-N is ubiquitinated on cysteine residues, but not exclusively so. There are ubiquitination sites other than cysteine among residues 1–30. We postulate that these are serine and/or threonine residues, given the hydrolysability of the ubiquitination and the results of mutation analysis. Unambiguously pinpointing the ubiquitination sites by mutation analysis is hampered by the effects of mutation on protein stability and by the fact that the ubiquitination machinery is promiscuous and ubiquitinates multiple sites, including alternative residues when the primary target sites are not available because of mutation.
Proteasome-dependent degradation of tBid-N is critical for its membrane-permeabilizing activity of tBid-C
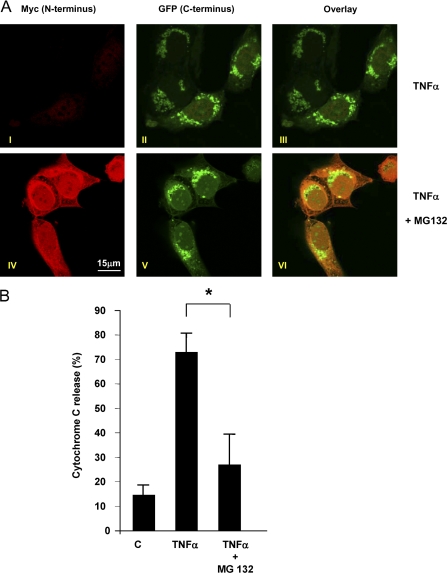
To investigate whether degradation of tBid-N was required for tBid-C function, we determined the impact of proteasome inhibition on tBid-C translocation. MCF-7 cells expressing Myc Bid GFP were stimulated with TNF-α for 8 h in the presence or absence of MG132 and examined by CLSM for the distribution of Myc tBid-N and tBid-C GFP cleavage fragments. In accordance with the biochemical data (Fig. 2 A), Myc tBid-N was rescued from degradation by MG132 (Fig. 6 A, IV). Despite this, tBid-C GFP translocated to mitochondria (Fig. 6 A, V), where it partially colocalized with Myc tBid-N (VI). This result indicates that the degradation of tBid-N is not required for tBid-C translocation. In line with a previous study (Zha et al., 2000), it also suggests that the cleaved but still associated tBid-N–tBid-C complex can translocate to mitochondria. Next, we investigated whether proteasome inhibition affected the capacity of tBid-C to permeabilize the mitochondrial outer membrane. Untransfected MCF-7 cells were stimulated with TNF-α in the presence or absence of MG132 and analyzed by flow cytometry for Cyt c content. The dramatic release of Cyt c induced by TNF-α stimulation was greatly reduced upon coincubation with MG132 (Fig. 6 B), which suggests that degradation of tBid-N is required for tBid-C function.
Figure 6.
Proteasome inhibition prohibits tBid-C function. (A) MCF-7 cells expressing Myc Bid GFP were stimulated with TNF-α with or without MG132 for 8 h. Stimulation resulted in translocation of tBid-C GFP (green) to mitochondria (II and III). In the presence of MG132, Myc tBid-N (red) remained detectable (IV), which indicates escape from proteasomal degradation. N- and C-terminal epitopes partially colocalized at mitochondria (VI). (B) TNF-α–induced Cyt c release in the presence or absence of MG132 was tested by flow cytometric analysis in untransfected MCF-7 cells after 8 h of stimulation. Results are means + SD from three independent experiments. The asterisk indicates a statistically significant difference (P < 0.05 according to a t test). C, control.
Because the proteasome may have other targets in this pathway, we aimed to test a Bid molecule harboring tBid-N that could escape from proteosomal degradation. Unfortunately, the stabilizing N-terminal deletion mutation Δ15 unleashed constitutive mitochondrial localization and permeabilizing activity in full-length Bid (unpublished data) and was therefore not useful. In the course of our study, however, we had noted that the addition of a bulky group such as GFP or a repetitive Myc tag to the N terminus of tBid-N greatly increased its stability. Therefore, we compared the functional activity of GFP Bid with a vesicular stomatitis virus (VSV) tag, which contains a stabilized GFP tBid-N, with that of Myc Bid GFP, which contains the wild-type, unstable Myc tBid-N.
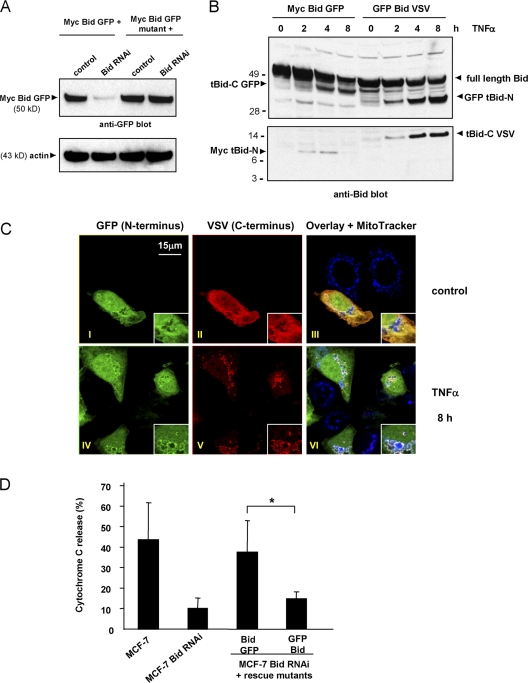
To create a cell line that exclusively expressed stabilized tBid-N, endogenous Bid was constitutively down-regulated in MCF-7 cells by RNAi (Fig. 7 A). To express stabilized GFP Bid VSV and control (unstable) Myc Bid GFP in these cells, a silent mutation was introduced into the Bid sequence that was not targeted by the siRNA. This allowed appropriate expression of the Bid constructs upon transient transfection (Fig. 7, A and B). The processing of Myc Bid GFP and GFP Bid VSV by caspase-8/10 and the half-life of the Bid fragments were followed kinetically after stimulation of the reconstituted MCF-7 cells with TNF-α. In the case of Myc Bid GFP, the tBid-C GFP cleavage product was visible from the 2-h time point onward and its signal increased in time. The Myc tBid-N fragment was detected at the 2- and 4-h time points but not at the 8-h time point (Fig. 7 B). Processing of GFP Bid VSV was also visible at 2 h, but in this case not only the tBid-C VSV fragment remained detectable in the 8-h time frame but also the GFP tBid-N fragment (Fig. 7 B). We conclude that the GFP Bid VSV protein is properly processed by caspase-8/10 and that the GFP tBid-N fragment is long-lived, whereas the Myc tBid-N fragment has a short half-life.
Figure 7.
Stabilization of tBid-N prohibits tBid-C function. (A) MCF-7 cells were depleted of endogenous Bid by stable expression of a Bid siRNA. MCF-7 Bid RNAi cells were reconstituted by transient transfection with Myc Bid GFP or GFP Bid VSV with silent mutations to allow escape from RNAi. The results for reconstitution with Myc Bid GFP wild-type and RNAi escape mutants are shown by immunoblotting for GFP in total cell lysates. Molecular masses (kD) of marker proteins are indicated. (B) Myc Bid GFP and GFP Bid VSV were expressed in MCF-7 Bid RNAi cells and cells were stimulated for the indicated time periods with TNF-α. Full-length Bid and cleavage fragments were detected by anti-Bid immunoblotting of total cell lysates. Fragments were identified additionally by immunoblotting with antibodies to Myc, GFP, and VSV (not depicted). ECL signals originate from the same blotted gel but the bottom panel represents a longer exposure than the top to allow detection of Myc tBid-N. (C) MCF-7 cells expressing GFP Bid VSV were stimulated with TNF-α for 8 h or left untreated (control). Signals diagnostic for stabilized GFP tBid-N are shown individually and in combination with the signals for tBid-C (VSV) and MitoTracker. The enlarged views allow assessment of colocalization of tBid-N and tBid-C at mitochondria. (D) TNF-α–induced Cyt c release was tested after 8 h of stimulation by flow cytometric analysis in parental MCF-7 cells, cells lacking Bid (MCF-7 Bid RNAi), and the same cells reconstituted with either Myc Bid GFP (unstable tBid-N) or GFP Bid VSV (stabilized GFP tBid-N). MCF-7 and MCF-7 Bid RNAi cells were additionally transfected to express histone-GFP. Cyt c content was monitored in cells gated for equal GFP expression to standardize for transfected Bid levels. Results are means + SD from three independent experiments. Values are corrected for Cyt c release in the absence of stimulus (15 ± 5%). The asterisk indicates statistically significant difference (P < 0.05 according to a t test).
The fate of stabilized GFP tBid-N was also followed by CLSM. In unstimulated cells, GFP and VSV signals were consistent with a cytoplasmic localization of full-length GFP Bid VSV (Fig. 7 C, I and II). At 8 h after stimulation, the VSV signal was punctate and colocalized with MitoTracker, which indicates mitochondrial localization of tBid-C VSV (Fig. 7 C, V). The GFP signal was still present at this time point (Fig. 7 C, IV) and was partially colocalized with VSV and MitoTracker signals (VI). This indicated long-term stabilization of GFP tBid-N (compare with the disappearance of Myc tBid-N; Fig. 1 A). As in the case of proteasome inhibition (Fig. 6 A), tBid-N stabilization did not interfere with mitochondrial translocation of tBid-C and both fragments partially colocalized at the mitochondria. This suggests that tBid-N and tBid-C may remain associated after cleavage of full-length Bid and translocate to mitochondria as a complex.
Next, Cyt c release upon TNF-α stimulation was compared in wild-type MCF-7 and Bid RNAi cells before and after reconstitution with unstable or stabilized Bid versions. Bid RNAi effectively blocked TNF-α–induced Cyt c release (Fig. 7 D). Importantly, introduction of GFP Bid VSV bearing stabilized tBid-N did not restore the ability of TNF-α to induce Cyt c release, whereas introduction of Myc Bid GFP bearing unstable tBid-N did (Fig. 7 D). Collectively, these results indicate that degradation of tBid-N regulates the proapoptotic function of tBid-C. Based on these data, we propose a model in which selective removal of tBid-N by proteasome-mediated degradation liberates tBid-C to induce mitochondrial membrane permeabilization.
Discussion
Sequences in Bid's N-terminal fragment sequester the proapoptotic BH3 domain. Through this autoinhibitory mechanism, full-length Bid is kept in an inactive state (Chou et al., 1999; Tan et al., 1999). Bid can be converted to its active form by cleavage in its second unstructured loop, such as at D60 by caspase-8/10 and at D75 by granzyme B (Li et al., 1998; Luo et al., 1998; Sutton et al., 2003). Mechanistically, it was unclear why cleavage should lead to Bid activation. Upon Bid cleavage in vitro, the N- and C-terminal fragments remain associated according to both NMR and biochemical analyses (Chou et al., 1999; Zha et al., 2000; Kuwana et al., 2002). Moreover, several investigators have demonstrated that the tBid-N fragment, when expressed independently, can block the proapoptotic activity of the tBid-C fragment (Tan et al., 1999; Kudla et al., 2000; Renshaw et al., 2004). Collectively, these data suggest a critical inhibitory effect of the Bid N terminus upon the C-terminal proapoptotic function both before and after Bid cleavage. Based on our results, we propose that cleavage of Bid acts as a proapoptotic activation signal by allowing ubiquitination and subsequent proteasomal destruction of the inhibitory N-terminal fragment.
Although originally recognized as a signal for the steady-state destruction of aged and misfolded proteins, ubiquitination is now known to play an essential regulatory role in cellular signaling. Various apoptosis signaling molecules are regulated by proteasomal destruction (Weissman, 2001; Jesenberger and Jentsch, 2002; Chen, 2005). These include inhibitory Bcl-2 family members Bcl-2 (Breitschopf et al., 2000a), Mcl-1 (Liu et al., 2005; Zhong et al., 2005), and Bfl-1 (Kucharczak et al., 2005); proapoptotic Bax (Li and Dou, 2000); BH3-only proteins Bim (Akiyama et al., 2003; Ley et al., 2003) and Bik (Marshansky et al., 2001); and the C-terminal fragment of Bid (Breitschopf et al., 2000b). Polyubiquitination and degradation generally negatively regulate the function of the target protein by its complete elimination. The case of tBid-N ubiquitination is different because it represents a form of positive regulation; it is required for the proapoptotic function of Bid. In the noncanonical nuclear factor κB pathway, polyubiquitination and partial protein degradation of p105/p100 also acts as an activating principle (Chen, 2005). In both the case of the nuclear factor κB precursor and Bid, a portion of the molecule escapes from degradation and is released to perform its function.
Mutation analysis specifically implicated 5 out of 13 lysines in Mcl-1 in its destruction (Zhong et al., 2005), whereas mutation of all 4 lysine residues in tBid-C was shown to increase its half-life (Breitschopf et al., 2000b). In the case of the other Bcl-2 family members, no specific target sites have been identified. In the case of Bid, ubiquitination occurred with certainty on the Bid fragment encompassing residues 1–30. Indeed, the TEV protease mapping experiments indicate that tBid-N can be ubiquitinated on sites among amino acids 11–30 as well as to a lesser extent on sites among amino acids 1–10. This classifies the ubiquitination as unconventional because it did not concern a peptide bond to an ɛ amino group of lysine or an α amino group at the N terminus of the Bid protein. After reduction, ubiquitination was partially but not completely lost. This indicates that cysteine residues are among the ubiquitin acceptor sites. A thioester linkage is not unusual for ubiquitin because the ubiquitin conjugating (E2) and ubiquitin ligase (E3) enzymes bear ubiquitin in such a manner. However, to our knowledge, cysteine ubiquitination of a target other than E2 or E3 proteins has only one precedent in the existing literature. This concerns the cytoplasmic tail of major histocompatibility complex class I that contains no lysine residues but was ubiquitinated in a reducible manner by an E3 encoded by a herpes virus (Cadwell and Coscoy, 2005). Ubiquitination of tBid-N was to a significant degree maintained upon reduction (Fig. 5 B), which is consistent with the possibility that threonine and serine residues are primary targets of the ubiquitination machinery. The fact that a tBid-N mutant lacking all three cysteine residues was still ubiquitinated on fragment 1–30 proves that residues other than cysteine are ubiquitin acceptors. This is consistent with the fact that cysteines are not fully conserved in tBid-N. In fact, rat tBid-N does not contain any cysteines (Fig. S1 C).
Our extensive mutational analysis has not pinpointed the ubiquitination sites. These experiments are hampered by the fact that ubiquitin is likely appended to other candidate sites when the original sites are not available, including residues outside region 1–30. Mutation of all cysteines, threonines, and serines in tBid-N made it much more unstable than the wild-type protein, such that it was barely detectable even in the presence of proteasome inhibitor (unpublished data). In this case, the protein likely bears little structural resemblance to the wild-type tBid-N and results are difficult to interpret. Thus far, attempts to map the ubiquitination sites by mass spectrometry have not been successful. Lability of the thioester bonds is the most likely limiting factor in this analysis, particularly after tryptic digestion, which converts the polyubiquitin chain into a reactive Gly Gly amine. Theoretically, ubiquitin can link to serine and threonine by the formation of an ester bond between the carboxyl group of the C-terminal glycine and the hydroxyl group in the side chain of these amino acids. The first evidence for such linkage in a biological system was being published when this manuscript was in the final stage of preparation. It again concerned the cytoplasmic tail of major histocompatibility complex class I, which was ubiquitinated by another herpes virus–derived E3 than the one responsible for cysteine ubiquitination (Wang et al., 2007). Our work provides additional evidence for such linkage and adds that it can occur in a mammalian system.
Although Δ13 tBid-N was as unstable as the wild-type version, the Δ15 mutant was fully stabilized. C15 is a good candidate ubiquitin acceptor site because it lies at the border of helix 1 and is accessible in the Bid structure (Chou et al., 1999; McDonnell et al., 1999). However, point mutation of C15 did not affect ubiquitination. In stark contrast, ubiquitination was almost completely lost from tBid-N GFP upon deletion of residues 1–15. This suggests that these residues, particularly residues 14 and 15 (given the instability of the Δ13 mutant), are critical for binding of the (undefined) ubiquitination machinery to tBid-N.
In our CLSM experiments, we found tBid-N together with tBid-C at the mitochondrial membrane after TNF-α stimulation when tBid-N degradation was blocked by proteasome inhibition (Fig. 6 A) or GFP fusion (Fig. 7 C). It is therefore clear that tBid-N degradation is not required for mitochondrial translocation of tBid-C. We suggest that the stabilized tBid-N fragment inhibits the function of tBid-C at the mitochondria. In time course experiments with wild-type Myc Bid GFP, we also found a degree of colocalization of Myc and GFP signals at mitochondria at 2 h after TNF-α stimulation (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200707063/DC1). These findings tie in with the in vitro observation that the cleaved complex stays intact (Chou et al., 1999) and a previous study showing the association of the cleaved Bid complex with mitochondria (Zha et al., 2000). These data suggest that tBid-N and tBid-C may remain associated during mitochondrial translocation. Further experiments are required to resolve whether tBid-N ubiquitination takes place at the mitochondrial membrane and whether this event determines its dissociation from tBid-C.
We have shown here that Bid's proapoptotic function is activated through cleavage-inducible ubiquitination and proteasomal degradation of tBid-N. Freeing of the BH3 domain by this mechanism may apply to other Bcl-2 family members as well. For instance, BimEL is cleaved by caspases at an early stage during apoptosis induction, which renders it more proapoptotic (Chen and Zhou, 2004). Moreover, Bcl-2, Bfl-1, and Bcl-xL can be converted into proapoptotic proteins after cleavage by caspases or calpain (Clem et al., 1998; Kucharczak et al., 2005). Perhaps, in these cases, a similar mechanism operates as we have shown here for Bid activation.
Materials and methods
Cells and reagents
Human cervix carcinoma line HeLa, breast carcinoma line MCF-7, and 293 embryonic kidney cell–derived packaging line ΦNX-Ampho were grown in DME medium with fetal calf serum. Human TNF-α was used at 10 ng/ml in combination with CHX at 10 μg/ml (both obtained from Sigma-Aldrich), soluble human FLAG-tagged TRAIL in combination with an enhancer (Qbiogene) at 100 ng/ml, and MG132 (EMD) at 50 μM. mAbs used were: anti-HA 12CA5, anti-Myc 9E10, anti-VSV P5D4 (purified Ig prepared from available hybridomas), anti-FLAG M2 (Sigma-Aldrich), anti-actin C4 (Chemicon), anti–Cyt c 6H2.B4 (BD Biosciences), anti-ubiquitin P4D1 (Santa Cruz Biotechnology, Inc.). Polyclonal rabbit sera used were: anti-Bid raised against a GST fusion protein of full-length human Bid (available from BD Biosciences; Werner et al., 2002), anti-GFP (Clontech Laboratories, Inc.), anti-E2-25K UG9520 (BIOMOL International, L.P.). HRP-conjugated rabbit anti–mouse Ig and pig anti–rabbit Ig were obtained from Dako, HRP-conjugated anti-HA and anti-FLAG mAb were obtained from Sigma- Aldrich, and goat anti–mouse Ig conjugated to Alexa Fluor 594 was obtained from Invitrogen.
Plasmids
Construction of cDNA encoding full-length human Bid fused C-terminally to eGFP (Bid GFP) made use of vector eGFPN1 (Clontech Laboratories, Inc.). A construct encoding full-length Bid GFP with an N-terminal Myc tag was created by placing a polylinker encoding a Myc epitope 5′ in frame to the Bid initiation codon. cDNA encoding the tBid N-terminal residues 1–60 (tBid-N) was cloned into eGFPN1, eGFPC1 (Clontech Laboratories, Inc.), pMT2SM, pMT2SM-VSV, and pMT2SM-HA (Werner et al., 2002) by PCR/restriction enzyme–based methods. The plasmid encoding histone 2B-GFP has been described previously (Tait et al., 2004). Plasmid pcDNA3.1 encoding FLAG-tagged human ubiquitin was a gift of S. Cook (Babraham Institute, Cambridge, UK). Plasmid pMT2SM encoding HA-tagged human ubiquitin has been described previously (de Melker et al., 2001). pcDNA3. 1-TAP was made by restriction enzyme–based subcloning of the TAP tag from pZomeC1 (Cellzome) into pcDNA3.1. tBid-N cDNA and deletion mutants were cloned into pcDNA3.1-TAP by restriction enzyme–based subcloning, producing constructs that were C-terminally fused to the TAP tag (Rigaut et al., 1999). Where used, polylinkers encoding TEV protease recognition sequences (encoding ENLYFQG) were inserted after the appropriate restriction digest. Point mutants and TEV insertion constructs were generated using the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene).
Western blotting, metabolic labeling, and immunoprecipitation
For immunoprecipitation, all cells (adherent and floating) were harvested, washed twice in PBS, and lysed in NP-40 buffer consisting of 50 mM Tris-HCl, pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM PMSF, and complete EDTA-free protease inhibitors (Roche). Cell lysates were clarified by centrifugation for 15 min at 13,000 g and protein content was measured by protein assay (Bio-Rad Laboratories). All SDS-PAGE analysis was done on precast 4–12% NuPAGE minigels according to the manufacturer's instructions (Invitrogen). In this procedure, samples were heated for 10 min at 70°C in reducing (100 mM DTT) sample buffer before electrophoresis. Equal amounts of protein per lane (20 μg) were separated by SDS-PAGE and transferred to nitrocellulose sheets. Immunoblotting and enhanced chemiluminescence were performed as described previously (Tait et al., 2004). For metabolic labeling, cells (106 cells in a 10-cm dish) were starved in methionine- and cysteine-free DME medium for 2 h, radiolabeled with [35S]methionine and [35S]cysteine (13.25 MBq per dish; GE Healthcare) for 2 h in the same medium. For the chase, cells were incubated for the indicated time in complete DME medium and supplemented with 2.5 mM of unlabeled methionine and cysteine. Immunoprecipitated proteins were resolved by SDS-PAGE, detected by autoradiography, and quantified by phosphorimaging (FLA-3000 and AIDA software; Fujifilm).
Tandem affinity purification
106 cells were lysed in NP-40 buffer. Lysates were clarified by centrifugation and incubated with rabbit IgG agarose (Sigma-Aldrich) for 2 h at 4°C. Agarose beads were washed in NP-40 buffer and TEV cleavage buffer (10 mM Tris, pH 8, 0.1% NP-40, 150 mM NaCl, 0.5 mM EDTA, and 1 mM DTT) and incubated with 10 U TEV protease (Invitrogen) in cleavage buffer overnight at 4°C. Next, supernatant was diluted threefold in calmodulin-binding buffer (10 mM Tris-HCl, pH 8, 0.1% NP-40, 150 mM NaCl, 1 mM Mg acetate, 1 mM imidazole, and 2 mM CaCl2) and incubated with calmodulin Sepharose beads (Stratagene) at 4°C for 1 h. Calmodulin-bound proteins were separated by SDS-PAGE and detected by Western blotting. For alkaline hydrolysis experiments, cells were lysed in PBS, pH 7.4 or 12, 1% NP-40, and complete EDTA-free protease inhibitors. Lysates were clarified by centrifugation, heated at 70°C for 10 min, diluted 100-fold in NP-40 buffer, and subjected to TAP purification.
Treatment of immunoprecipitates
For TEV protease treatment, HA Bid immunoprecipitates were resuspended in 300 μl TEV cleavage buffer and incubated with or without 2 μl TEV protease overnight at 4°C. Beads were spun down, washed three times, and taken up in sample buffer for SDS-PAGE. For reduction, HA Bid immunoprecipitates were suspended in 10 μl NP-40 buffer. To this, 15 μl of 100 mM Tris-HCl, pH 6.8, 4% SDS, 8 M urea, and 20% glycerol was added with or without 600 mM of fresh DTT. Control samples without DTT were kept on ice, whereas samples with DTT were heated at 95°C for 10 min. Next, samples were diluted out 80 times in NP-40 buffer with 10 mM iodoacetamide and 50 μg/ml RNase A (as carrier protein). Beads were spun down and supernatants were subjected to reprecipitation with anti-HA antibody. For the sequential immunoprecipitation shown in Fig. 3 B, after immunoprecipitation with anti-GFP antibody, the lysate was incubated three times with protein G–Sepharose beads followed by immunoprecipitation with anti-Myc antibody.
In vitro ubiquitination
To ubiquitinate the recombinant E2 core domain of E2-25K (Pichler et al., 2005), recombinant human ubiquitin-activating enzyme (E1; 120 ng; 0.2 μM), E2-25K1-155 (1.5 μg; 17 μM), and ubiquitin (2 μg; 47 μM) were coincubated in 20 mM Tris-HCl, pH 8, 100 mM NaCl, and 5 mM β-mercaptoethanol with 5 mM ATP and 5 mM MgCl2 for 2 min at 23°C. Reactions were terminated by 1:1 dilution in sample buffer (100 mM Tris, pH 6.8, 8 M urea, 4% SDS, and 20% glycerol with or without 600 mM DTT) and samples were kept on ice or heated at 95°C as described in the previous paragraph before electrophoresis. A complex of His-tagged Ring1b (residues 1–331) and full-length His-tagged Bmi1 were coexpressed in Sf9 insect cells by simultaneous infection with separate recombinant baculoviruses. The complex was purified on nickel-agarose (Hitrap column; GE Healthcare) and further purified as described previously (Buchwald et al., 2006). Ring1b/Bmi1 was incubated at 6 μM together with 220 nM human E1, 2 μM human UbcH5c, 20 μM ubiquitin, and 3 mM ATP in a total volume of 50 μl of ligase buffer consisting of 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM ZnCl2, and 1 mM DTT at 30°C for 1 h. For the control sample, the ubiquitination reaction was stopped by the addition of LDS sample buffer (NuPAGE; Invitrogen). Samples for immunoprecipitation were: (a) incubated with 12.5-fold excess of ligase buffer as control, (b) heated for 8 min at 95°C in the presence of 5% β-mercaptoethanol after which 12.5-fold excess ligase buffer was added, (c) incubated with 12.5-fold excess PBS, pH 7.4, and heated for 10 min at 70°C, and (d) incubated with 12.5-fold excess PBS, pH 12, and heated for 10 min at 70°C. Next, samples were diluted with NP-40 buffer (as described in the previous paragraph) to a total volume of 10 ml and subjected to immunoprecipitation with nickel-agarose beads (QIAGEN). Reaction products were separated by NuPAGE in 4–12% Bis-Tris precast gels (Invitrogen) blotted to nitrocellulose and probed independently with a P4D1 antibody directed to ubiquitin and anti-Ring1b (provided by H. Koseki, Institute of Physical and Chemical Research, Yokohama, Japan).
RNAi
A short interfering oligonucleotide targeting human Bid (nt 238–256, 5′-GAAGACATCATCCGGAATA-3′) was cloned into pSUPER and pRETRO-SUPER (Brummelkamp et al., 2002). Stable expression of Bid siRNA was achieved by retroviral transduction of cell lines of interest with the pRETRO-SUPER construct. For rescue experiments, two silent mutations were introduced in the appropriate Bid sequence (5′-GAAGACATCATAAGGAATA-3′) using the QuikChange site-directed mutagenesis kit to avoid siRNA-mediated silencing.
Transfection and retroviral transduction
Cells were transfected with FuGENE 6 according to the manufacturer's instructions (Roche). Unless otherwise indicated, cells were manipulated 20 h after transfection. Production of amphotropic retrovirus was done in ΦNX-Ampho cells as described previously (Werner et al., 2002). For transduction, virus-containing packaging cell supernatant was incubated on ice for 10 min with 10 μg/ml Dotap (Roche) and subsequently incubated with target cells overnight. Cells were selected with 1 μg/ml puromycin 48 h after transduction.
CLSM
Cells (105 per sample) were grown and transfected on glass coverslips in 6-well plates, washed in PBS, and fixed for 30 min with 4% paraformaldehyde in PBS. If applicable, cells were incubated for 30 min with 100 nM MitoTracker deep red (Invitrogen) before fixation. Fixed cells were washed in PBS, permeabilized by incubation in PBS with 1% Triton X-100 for 10 min, and blocked with 0.5% BSA in TBS-Tween for 1 h. Cells were incubated in the same buffer with the appropriate primary antibody and subsequently washed three times in TBS-Tween and incubated with Alexa Fluor 594– conjugated goat anti–mouse IgG (Invitrogen). Next, cells were washed in TBS-Tween and mounted on slides using Vectashield (Vector Laboratories). All treatments were performed at room temperature. CLSM was performed using a TCS SP2 system using a 63× 1.32 NA oil immersion objective and confocal software (all obtained from Leica). Images were processed (cropping and level adjustment) using Photoshop software (Adobe). To minimize spectral leak-through, images were obtained by sequential scanning.
Cyt c release assay
Cells were analyzed for Cyt c release essentially as described previously (Waterhouse and Trappani, 2003). All cells were collected, washed in PBS, and permeabilized with 200 μg/ml digitonin in PBS, 150 mM KCl, and 1 mM EDTA for 10 min on ice. Cells were washed once in PBS, fixed in 4% paraformaldehyde, incubated in blocking buffer (2% BSA in PBS) for 30 min at room temperature, and incubated with anti–Cyt c mAb 6H2.B4 at 1:200 dilution in blocking buffer overnight at 4°C. Cells were washed in blocking buffer and incubated with a Cy5-conjugated goat anti–mouse Ig antibody (Invitrogen) at 1:200 dilution in blocking buffer for 1 h at room temperature. After washing, cells were analyzed using a FACSCalibur and CellQuest software (BD Biosciences). Cyt c content was analyzed specifically in transfected cells by gating the GFP-positive population.
Online supplemental material
Fig. S1 shows the structure of full-length human Bid as determined by NMR (Chou et al., 1999; McDonnell et al., 1999), its amino acid sequence, and sequence homology of tBid-N between species. Fig. S2 provides the evidence that Bid, rather than the TAP tag, is ubiquitinated. Fig. S3 shows the stability assays for alanine substitutions mutants of HA tBid-N TEV 30 performed by incubating transfected HeLa cells with CHX. Fig. S4 demonstrates that reduction (β-mercaptoethanol) or pH 12 treatment does not result in loss of ubiquitin from Ring1b, which carries ubiquitin on lysine (Buchwald et al. 2006). Fig. S5 shows MCF-7 Bid RNAi cells expressing Myc Bid GFP at different time points after stimulation with TNF-α.
Supplemental Material
Acknowledgments
We are grateful to Gretel Buchwald, Puck Knipscheer, and Titia Sixma for help with the in vitro ubiquitination experiments and information on Bid structure. We thank Martina O'Flaherty, Albert J. Heck, Clive Slaughter, and Andrew High for help with mass spectrometry; Douglas R. Green for support; and Huib Ovaa for sharing his chemical expertise and commenting on the manuscript.
This work was funded by the Dutch Cancer Society.
S.W.G. Tait's present address is Department of Immunology, St. Jude Children's Research Hospital, Memphis, TN 38105.
C.S. D'Santos's present address is Proteomic Unit Bergen, University of Bergen, N-5009 Bergen, Norway.
Abbreviations used in this paper: BH, Bcl-2 homology; CHX, cycloheximide; CLSM, confocal laser scanning microscopy; Cyt c, cytochrome c; NMR, nuclear magnetic resonance; TAP, tandem affinity purification; tBid-C, C-terminal fragment of Bid; tBid-N, N-terminal fragment of Bid; TEV, tobacco etch virus; TRAIL, TNF-related apoptosis-inducing ligand; VSV, vesicular stomatitis virus.
References
- Adams, J.M., and S. Cory. 1998. The Bcl-2 protein family: arbiters of cell survival. Science. 281:1322–1326. [DOI] [PubMed] [Google Scholar]
- Akiyama, T., P. Bouillet, T. Miyazaki, Y. Kadona, H. Chikuda, U. Chung, A. Fukuda, A. Hikita, H. Seto, T. Okada, et al. 2003. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 22:6653–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright, K.M., and G.S. Salvesen. 2003. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15:725–731. [DOI] [PubMed] [Google Scholar]
- Breitschopf, K., J. Haendeler, P. Malchow, A.M. Zeiher, and S. Dimmeler. 2000. a. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: molecular characterization of the involved signaling pathway. Mol. Cell. Biol. 20:1886–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschopf, K., A.M. Zeiher, and S. Dimmerler. 2000. b. Ubiquitin-mediated degradation of the proapoptotic active form of Bid. J. Biol. Chem. 275:21648–21652. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2:243–247. [DOI] [PubMed] [Google Scholar]
- Buchwald, G., P. van der Stoop, O. Weichenrieder, A. Perrakis, M. van Lohuizen, and T.K. Sixma. 2006. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25:2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, K., and L. Coscoy. 2005. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 309:127–130. [DOI] [PubMed] [Google Scholar]
- Chen, D., and Q. Zhou. 2004. Caspase cleavage of BimEL triggers a positive feedback amplification of apoptotic signaling. Proc. Natl. Acad. Sci. USA. 101:1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J. 2005. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7:758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, E.H., M.C. Wei, S. Weiler, R.A. Flavell, T.W. Mak, T. Lindsten, and S.J. Korsmeyer. 2001. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAK- and BAK-mediated mitochondrial apoptosis. Mol. Cell. 8:705–711. [DOI] [PubMed] [Google Scholar]
- Chou, J.J., H. Li, G.S. Salvesen, J. Yuan, and G. Wagner. 1999. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 96:615–624. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103–106. [DOI] [PubMed] [Google Scholar]
- Clem, R.J., E.H. Cheng, C.L. Karp, D.G. Kirsch, K. Ueno, A. Takahashi, M.B. Kastan, D.E. Griffin, W.C. Earnshaw, M.A. Veliuona, and J.M. Hardwick. 1998. Modulation of cell death by Bcl-xL through caspase interaction. Proc. Natl. Acad. Sci. USA. 95:554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melker, A.A., G. van der Horst, and J. Borst. 2001. c-Cbl ubiquitinates the EGF receptor at the plasma membrane and remains receptor associated throughout the endocytic route. J. Cell Sci. 114:2167–2178. [DOI] [PubMed] [Google Scholar]
- Jesenberger, V., and S. Jentsch. 2002. Deadly encounter: ubiquitin meets apoptosis. Nat. Rev. Mol. Cell Biol. 3:112–121. [DOI] [PubMed] [Google Scholar]
- Kucharczak, J.F., M.J. Simmons, C.S. Duckett, and C. Gelinas. 2005. Constitutive proteasome-mediated turnover of Bfl-1/A1 and its processing in response to TNF receptor activation in FL5.12 pro-B cells convert it into a prodeath factor. Cell Death Differ. 12:1225–1239. [DOI] [PubMed] [Google Scholar]
- Kudla, G., S. Montessuit, R. Eskes, C. Berrier, J.-C. Martinou, A. Ghazi, and B. Antonsson. 2000. The destabilization of lipid membranes induced by the C-terminal fragment of caspase 8-cleaved Bid is inhibited by the N-terminal fragment. J. Biol. Chem. 275:22713–22718. [DOI] [PubMed] [Google Scholar]
- Kuwana, T., M.R. Mackey, G. Perkins, M.H. Ellisman, M. Letterich, R. Schneiter, D.R. Green, and D.D. Newmeyer. 2002. Bid, Bax and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 111:331–342. [DOI] [PubMed] [Google Scholar]
- Ley, R., K. Balmanno, K. Hadfield, C. Weston, and S.J. Cook. 2003. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 278:18811–18816. [DOI] [PubMed] [Google Scholar]
- Li, H., H. Zhu, C. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the fas pathway of apoptosis. Cell. 94:491–501. [DOI] [PubMed] [Google Scholar]
- Li, B., and Q.P. Dou. 2000. Bax degradation by the ubiquitin/proteasome- dependent pathway: involvement in tumor survival and progression. Proc. Natl. Acad. Sci. USA. 97:3850–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., H.-S. Peng, Y.-S. Cheng, H.S. Yuan, and H.-F. Yang-Yen. 2005. Stabilization and enhancement of the antiapoptotic activity of Mcl-1 by TCTP. Mol. Cell. Biol. 25:3117–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X., I. Budihardjo, H. Zou, C. Slaughter, and X. Wang. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 94:481–490. [DOI] [PubMed] [Google Scholar]
- Lutter, M., M. Fang, X. Luo, M. Nishijima, X.-S. Xie, and X. Wang. 2000. Cardiolipin provides specificity for targeting of tBID to mitochondria. Nat. Cell Biol. 2:754–756. [DOI] [PubMed] [Google Scholar]
- Marshansky, V., X. Wang, R. Bertrand, H. Luo, W. Duguid, G. Chinnandurai, N. Kanaan, M.D. Vu, and J. Wu. 2001. Proteasomes modulate balance among pro-apoptotic and anti-apoptotic Bcl-2 family members and compromise functioning of the electron transport chain in leukemic cells. J. Immunol. 166:3130–3142. [DOI] [PubMed] [Google Scholar]
- McDonnell, J.M., D. Fushman, C.L. Miliman, S.J. Korsmeyer, and D. Cowburn. 1999. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 96:625–634. [DOI] [PubMed] [Google Scholar]
- Pichler, A., P. Knipscheer, E. Oberhofer, W.J. van Dijk, R. Korner, J.V. Olsen, S. Jentsch, F. Melchior, and T. Sixma. 2005. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat. Struct. Mol. Biol. 12:264–269. [DOI] [PubMed] [Google Scholar]
- Puthalakath, H., and A. Strasser. 2002. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 9:505–512. [DOI] [PubMed] [Google Scholar]
- Renshaw, S.A., C.E. Dempsey, F.A. Barnes, S.M. Bagstaff, S.K. Dower, C.D. Bingle, and M.K.B. Whyte. 2004. Three novel Bid proteins generated by alternative splicing of the human Bid gene. J. Biol. Chem. 279:2846–2855. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Sattler, M., H. Liang, D. Nettesheim, R.P. Meadows, J.E. Harlan, M. Eberstadt, H.S. Yoon, S.B. Shuker, B.S. Chang, A.J. Minn, et al. 1997. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 275:983–986. [DOI] [PubMed] [Google Scholar]
- Sutton, V.R., M.E. Wowk, M. Cancilla, and J.A. Trappani. 2003. Caspase activation by Granzyme B is indirect and caspase autoproessing requires the release of proapoptotic mitochondrial factors. Immunity. 18:319–329. [DOI] [PubMed] [Google Scholar]
- Tait, S.W.G., A.B. Werner, E. de Vries, and J. Borst. 2004. Mechanism of action of Drosophila Reaper in human cells: Reaper globally inhibits protein synthesis and induces apoptosis independent of mitochondrial permeability. Cell Death Differ. 11:800–811. [DOI] [PubMed] [Google Scholar]
- Tan, K.O., K.M.L. Tan, and V.C. Yu. 1999. A novel BH3-like domain in BID is required for intramolecular interaction and autoinhibition of pro-apoptotic activity. J. Biol. Chem. 274:23687–23690. [DOI] [PubMed] [Google Scholar]
- Wang, X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15:2922–2933. [PubMed] [Google Scholar]
- Wang, X., R.A. Herr, W.-J. Chua, L. Lybarger, E.J.H.J. Wiertz, and T.H. Hansen. 2007. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177:613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, N.J., and J.A. Trappani. 2003. A new quantitative assay for cytochrome c release in apoptotic cells. Cell Death Differ. 10:853–855. [DOI] [PubMed] [Google Scholar]
- Wei, M.C., T. Lindsten, V.K. Mootha, S. Weiler, A. Gross, M. Ashiya, C.B. Thompson, and S.J. Korsmeyer. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Weissman, A.M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169–178. [DOI] [PubMed] [Google Scholar]
- Werner, A.B., E. de Vries, S.W.G. Tait, I. Bontjer, and J. Borst. 2002. Bcl-2 family member Bfl-1/A1 sequesters truncated Bid to inhibit its collaboration with pro-apoptotic Bak or Bax. J. Biol. Chem. 277:22781–22788. [DOI] [PubMed] [Google Scholar]
- Yin, X.-M., K. Wang, A. Gross, Y. Zhao, S. Zinkel, B. Klocke, K.A. Roth, and S.J. Korsmeyer. 1999. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 400:886–891. [DOI] [PubMed] [Google Scholar]
- Zha, J., S. Weiler, K.J. Oh, M.C. Wei, and S.J. Korsmeyer. 2000. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 290:1761–1765. [DOI] [PubMed] [Google Scholar]
- Zhong, Q., W. Gao, F. Du, and X. Wang. 2005. Mule/ARF-BP-1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 121:1085–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.