Abstract
The central channel Tom40 of the preprotein translocase of outer membrane (TOM) complex is thought to be responsible for the import of virtually all preproteins synthesized outside the mitochondria. In this study, we analyze the topogenesis of the peripheral benzodiazepine receptor (PBR), which integrates into the mitochondrial outer membrane (MOM) through five hydrophobic transmembrane segments (TMSs) and functions in cholesterol import into the inner membrane. Analyses of in vitro and in vivo import into TOM component–depleted mitochondria reveal that PBR import (1) depends on the import receptor Tom70 but requires neither the Tom20 and Tom22 import receptors nor the import channel Tom40, (2) shares the post-Tom70 pathway with the C-tail–anchored proteins, and (3) requires factors of the mitochondrial intermembrane space. Furthermore, membrane integration of mitofusins and mitochondrial ubiquitin ligase, the MOM proteins with two and four TMSs, respectively, proceeds through the same initial pathway. These findings reveal a previously unidentified pathway of the membrane integration of MOM proteins with multiple TMSs.
Introduction
More than 99% of mitochondrial proteins are encoded by the nuclear genome, synthesized in the cytoplasm as precursor proteins, and imported posttranslationally into the mitochondria (Koehler, 2004; Bohnert et al., 2007). Most proteins destined for the matrix and several proteins destined for the inner membrane or intermembrane space (IMS) possess a cleavable N-terminal presequence that functions for the mitochondrial matrix targeting. Unlike these proteins, the mitochondrial outer membrane (MOM) proteins are synthesized as mature proteins, and the targeting signals are contained within the mature protein sequence (Rapaport, 2003). MOM contains three distinct protein types: proteins embedded in the membrane in a β-barrel structure such as porin and Tom40 (Paschen et al., 2005), proteins anchored to the membrane through one or several α-helical hydrophobic transmembrane segments (TMSs), and proteins peripherally associated with the MOM such as Sam35/Tom38/Tob38 (Ishikawa et al., 2004; Waizenegger et al., 2004; Bohnert et al., 2007). The preprotein import receptors Tom70 and Tom20 are anchored to the MOM through their single-spanning N-terminal TMS, whereas other proteins such as Bak and Bcl-XL are anchored to the membrane through a single-spanning C-terminal TMS (C-terminal tail anchor [C-TA] proteins; Setoguchi et al., 2006). Additionally, there are several examples of proteins with multiple TMSs. Yeast Fzo1 (or mammalian mitofusin 1 [Mfn1] and Mfn2) is anchored to the MOM through two TMSs localizing at the C-terminal segment and mediates mitochondrial fusion (Fritz et al., 2001). The peripheral benzodiazepine receptor (PBR) is anchored to the MOM through five TMSs and is involved in cholesterol import (Joseph-Liauzun et al., 1998). A mitochondrial ubiquitin ligase (MITOL; also named MARCH-V) is embedded in the MOM with four putative TMSs and regulates mitochondrial dynamics (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007).
It is generally thought that the preprotein import machinery of the outer membrane (translocase of outer membrane [TOM] complex) is responsible for the import of virtually all of the nuclear-encoded mitochondrial proteins (Rapaport, 2005). Indeed, β-barrel proteins porin and Tom40 synthesized in the cytoplasm are delivered to the import receptors Tom70 and Tom20 and are transferred through the Tom40 channel into the IMS, where they are integrated into the MOM through the sorting and assembly machinery (SAM; also named TOB [topogenesis of MOM β-barrel proteins]) complex assisted by IMS-localizing small Tim proteins (Koehler, 2004; Bohnert et al., 2007). The N-terminal signal anchor proteins, yeast Tom70 and Tom20, are integrated into the MOM independently of surface receptors but dependent on a region of Tom40 other than its pore segment (Ahting et al., 2005).
To our knowledge, the import pathway of the TMS-containing MOM proteins has been analyzed only for the components of the TOM complex, and little is known for proteins other than the TOM components as to which import receptor is responsible for the initial recognition and whether the Tom40 channel is involved in their membrane insertion.
In this study, we analyzed the membrane insertion of MOM proteins with multiple TMSs using in situ immunofluorescence microscopy and import into mitochondria isolated from TOM or SAM component–depleted semi-intact and intact cells. These experiments revealed a novel MOM insertion pathway.
Results and discussion
In vitro assay for integration of PBR into the MOM
Membrane topology of PBR as deduced from yeast mitochondria harboring human PBR revealed that it assumes a five-transmembrane topology extruding the N-terminal five amino acid residues into the IMS, whereas the C-terminal 14-residue segment remains in the cytoplasm (Joseph-Liauzun et al., 1998). We analyzed membrane orientation of the exogenously expressed rat PBR (N- or C-terminal HA-tagged versions) in cultured cells using a proteinase K–induced size-shift assay and confirmed the reported terminal orientation (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200702143/DC1). We established an in vitro assay for MOM integration of PBR, taking advantage of the proteinase K sensitivity, which removes surface-adsorbed PBR and trims the C-terminal segment of the correctly inserted form (Fig. S1, C–E).
MOM integration of PBR depends on Tom70 but not on other TOM components
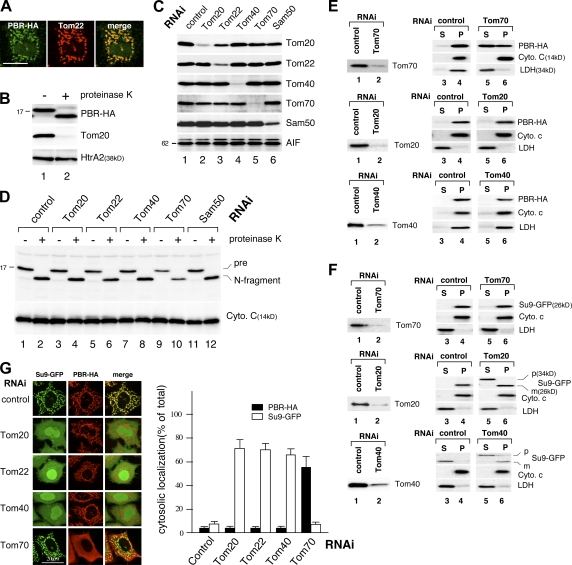
We then analyzed the import pathway of PBR using TOM component–depleted semi-intact cells. HeLa cells were treated with 25 μg/ml digitonin to selectively permeabilize the plasma membrane, cells were incubated with reticulocyte lysate–synthesized PBR-HA, and intracellular localization was analyzed by immunofluorescence microscopy. The imported PBR-HA colocalized with mitochondrial Tom22 (Fig. 1 A), exposing the C-terminal HA to the cytoplasm (Fig. 1 B). We then analyzed the effect of knockdown of TOM components on PBR import. This manipulation efficiently reduced the level of the target proteins (Fig. 1 C), and immunofluorescence microscopy (not depicted) and a proteinase K–induced size-shift assay (Fig. 1 D) revealed that PBR import was compromised by the Tom70 knockdown. Knockdown of other components, including the import receptors Tom20, Tom22, the central channel Tom40, and Sam50 had little effect on PBR import (Fig. 1 D).
Figure 1.
MOM integration of PBR in TOM component–depleted semi-intact and intact cells. (A) HeLa cells were semipermeabilized with digitonin and incubated with reticulocyte lysate–synthesized PBR-HA. After fixation and permeabilization, the cells were processed for immunofluorescence microscopy using anti-HA and anti-Tom22 antibodies. (B) Reticulocyte lysate–synthesized biotin-labeled PBR-HA was incubated with semi-intact cells as in A. The cells were treated with 50 μg/ml proteinase K at 26°C for 3 min and analyzed by SDS-PAGE and immunoblotting using HRP-conjugated streptavidin (PBR-HA), anti-Tom20, or anti-HtrA2 antibodies. (C) HeLa cells subjected to RNAi for the indicated proteins were analyzed by SDS-PAGE and subsequent immunoblotting using the indicated antibodies. AIF, loading control. (D) TOM component knockdown semi-intact cells were subjected to import assay as in B. Cytochrome c, loading control. (E and F) HeLa cells subjected to RNAi were transfected with the expression vector for Su9-GFP or PBR-HA. The cells were treated with digitonin to permeabilize the plasma membrane and centrifuged to separate the supernatant (S) and membrane (P) fractions (Otera et al., 2005), which were analyzed by SDS-PAGE and immunoblotting using the antibodies against HA or the indicated proteins. (G) Su9-GFP and PBR-HA were coexpressed in TOM component knockdown HeLa cells. The cells were analyzed by immunofluorescence microscopy. The cells (100 cells in three distinct fields) exhibiting cytosolic localization of Su9-DHFR or PBR-HA in each RNAi experiment were counted. Error bars represent SD. Bars, 20 μm.
These results were confirmed using intact cells. PBR-HA was exogenously expressed in TOM or SAM component–depleted HeLa cells, and its intracellular localization was examined by in situ immunofluorescence microscopy (unpublished data) and cell fractionation. Knockdown of Tom70 but not other TOM components significantly inhibited PBR import (Fig. 1 E). As controls, the import of both Su9-GFP and Tim23, the inner mitochondrial membrane protein with uncleavable targeting signal and four TMSs, was clearly compromised by the knockdown of Tom20, Tom22, or Tom40, whereas Tom70 knockdown had no inhibitory effect (Fig. 1 F; and not depicted for Tim23). Double expression of Su9-GFP and PBR-HA in the TOM-component knockdown cells further confirmed these results (Fig. 1 G).
Membrane integration of PBR occurs in the absence of Tom40
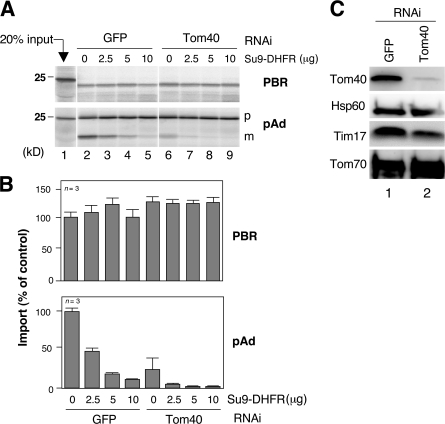
We examined whether the membrane integration of PBR depends on the import channel of Tom40. Mitochondria were isolated from Tom40 knockdown HeLa cells (Fig. 2 C), and we examined whether import of PBR would be affected by blocking the channel with tightly folded Su9–dihydrofolate reductase (DHFR). Mitochondrial import of the matrix-targeted preprotein pAd was inhibited by Tom40 knockdown, and the residual activity was further inhibited by the channel blockage (Fig. 2, A and B). As an additional control, Tom40 knockdown efficiently inhibited MOM integration of Tom40-dependent substrates VDAC2 and rat Tom5 (Setoguchi et al., 2006). In contrast, neither of these manipulations affected the import of PBR. We concluded that Tom40 is not involved in any mode in the MOM integration of PBR.
Figure 2.
Effect of Tom40 channel block on MOM integration of PBR-HA. (A) Reticulocyte lysate–synthesized 35S–PBR-HA or 35S-pAd was imported into the control or Tom40 knockdown mitochondria in the presence of recombinant Su9-DHFR. The reaction mixtures were treated with (for PBR) or without (for pAd) proteinase K and analyzed by SDS-PAGE and subsequent digital autoradiography. (B) The band intensities (proteinase K–resistant band for PBR and mature band for pAd) were quantified setting those in the absence of Su9-DHFR at 100%. Results obtained from three independent experiments are shown. (C) The mitochondria used in A were analyzed by SDS-PAGE and immunoblotting using antibodies against the indicated proteins. This procedure depleted Tom40 by ∼95%. Error bars represent SD.
Binding of PBR to Tom70
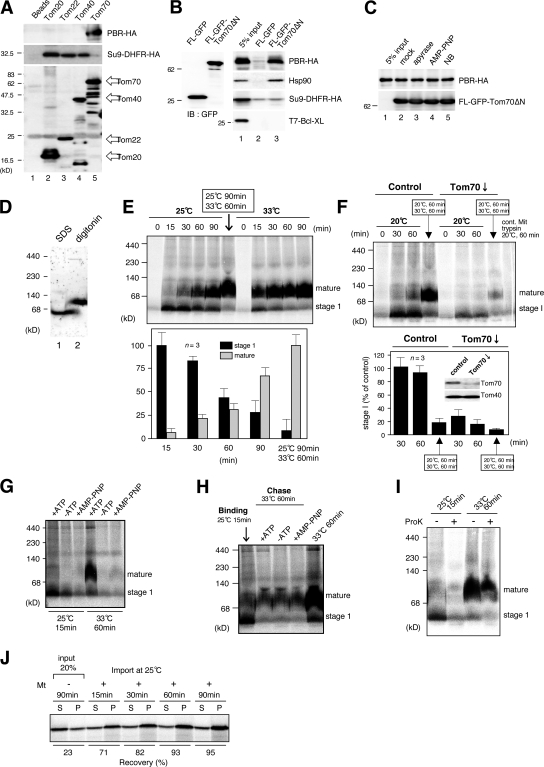
We examined binding of PBR to Tom70 by immunoprecipitation (Fig. 3, A and B). Reticulocyte lysate–synthesized PBR-HA and Su9-DHFR–HA were separately incubated with FLAG-tagged Tom20, Tom22, Tom40, or Tom70 purified from HeLa cells followed by immunoprecipitation using anti-FLAG beads. Tom70 but not other TOM components specifically bound PBR-HA. Conversely, Tom20, Tom22, and Tom40 recognized Su9-DHFR but not PBR-HA. Furthermore, FLAG-GFP-Tom70ΔN, the fusion construct between the cytoplasmic fragment of Tom70 and FLAG-GFP, specifically bound PBR-HA (Fig. 3 B). Confirming a previous study (Young et al., 2003), FLAG-GFP-Tom70ΔN also bound Hsp90 present in reticulocyte lysate. Because Hsp90 delivers phosphate carrier and peptide transporter to Tom70 before translocation through the Tom40 channel in mammalian mitochondria, we analyzed whether Hsp90 is involved in PBR binding to Tom70. Novobiocin, a coumarin antibiotic, binds to the C-terminal domain of Hsp90 to inhibit its dimerization and substrate binding and, as such, inhibits the docking of phosphate carrier and peptide transporter to Tom70 (Fan et al., 2006). However, it did not inhibit the binding of PBR to Tom70 (Fig. 3 C). Although overall MOM integration of PBR depended on ATP hydrolysis (Fig. 3, G and H), binding of PBR to Tom70 was not affected by ATP depletion or by nonhydrolyzable ATP analogue AMP-PNP (Fig. 3 C). As a control, depletion of cytoplasmic ATP completely blocked the mitochondrial import of Su9-DHFR (unpublished data). Thus, Hsp90 and ATP hydrolysis were not required for the docking of PBR onto Tom70.
Figure 3.
Analysis of import steps of PBR. (A and B) Reticulocyte lysate–synthesized PBR-HA, Su9-DHFR-HA, or T7–Bcl-XL was incubated with FLAG-tagged Tom proteins or FLAG-GFP-Tom70ΔN purified from HeLa cells. They were subjected to pull-down reaction and subsequent immunoblotting using the indicated antibodies. (C) Reticulocyte lysate–synthesized PBR-HA was incubated with 100 U/ml apyrase, 10 mM AMP-PNP, or 1 mM novobiocin (NB). The reaction mixtures were subjected to pull-down assay as in B. (D) Rat liver mitochondria were subjected to BN-PAGE followed by immunoblotting using anti-PBR antibodies. (E and F) Reticulocyte lysate–synthesized 35S–PBR-HA was incubated with control or Tom70-knockdown mitochondria under the indicated conditions. The reaction mixtures were analyzed by BN-PAGE and subsequent digital autoradiography. Band intensities of stage I and the mature form are shown, setting those of stage I (25°C for 15 min) and the mature form (25°C for 90 min plus 33°C for 60 min; E) or that of control (30 min) at 100% (F). Results obtained from three independent experiments are shown. Error bars represent SD. (G) Reticulocyte lysate–synthesized 35S–PBR-HA was passed through a spin column and subjected to import in the absence or presence of 1 mM ATP or AMP-PNP under the indicated conditions. Other conditions were set as in E. (H) Reticulocyte lysate–synthesized 35S–PBR-HA was subjected to mitochondrial import at 25°C for 15 min (binding). The mitochondria were reisolated and incubated at 33°C for 60 min (chase) in the import buffer with or without 1 mM ATP or AMP-PNP. Other conditions were set as in E. (I) Reticulocyte lysate–synthesized 35S–PBR-HA was imported into mitochondria under the indicated conditions. The reaction mixtures were treated with or without proteinase K and analyzed by BN-PAGE and digital autoradiography. (J) Reticulocyte lysate–synthesized 35S–PBR-HA was subjected to mitochondrial import. The mitochondria were treated with 100 mM Na2CO3, pH 11.5, and centrifuged to separate the supernatant (S) and membrane (P) fractions, which were analyzed by SDS-PAGE and subsequent digital autoradiography.
Analysis of the import steps of PBR
Mitochondrial PBR is organized in clusters of four to six molecules, as revealed by immunogold electron microscopy (Papadopoulos et al., 1997). Consistent with this study, digitonin-solubilized mitochondrial PBR migrated in blue-native (BN) PAGE more slowly than the SDS-solubilized monomeric form of ∼18 kD (Fig. 3 D). Therefore, we analyzed the PBR assembly process using BN-PAGE and found that PBR matured through the monomeric stage I intermediate; it was formed during import at 20–25°C and could be chased to mature form upon incubation at 30–33°C (Fig. 3, E and F). Moreover, we found that (1) the PBR assembly was strongly compromised by removing mitochondrial surface proteins by trypsin (Fig. 3 F); (2) the level of stage I intermediate was markedly decreased by Tom70 knockdown (Fig. 3 F); (3) production of stage I intermediate depended on ATP hydrolysis (Fig. 3 G); (4) maturation of stage I intermediate to the oligomeric form did not require ATP hydrolysis (Fig. 3 H); (5) PBR in stage I but not the mature form was sensitive to externally added proteinase K, indicating that stage I PBR is exposed to the mitochondrial surface (Fig. 3 I); and (6) stage I PBR as well as the mature form was resistant to alkali extraction, indicating that PBR in both stages was inserted into the membrane through hydrophobic interactions (Fig. 3 J). Together, these results suggested that PBR in stage I had left the receptor Tom70, which is associated with the surface of MOM through hydrophobic interactions and is migrated as a monomer in BN-PAGE. Considering that PBR binding to Tom70 occurred in the absence of ATP, these results suggested that the transfer of PBR from Tom70 to the following steps required ATP hydrolysis, although its function in maintaining the nascent PBR in the import-competent state was not ruled out.
PBR partly shares the MOM integration pathway with C-TA proteins
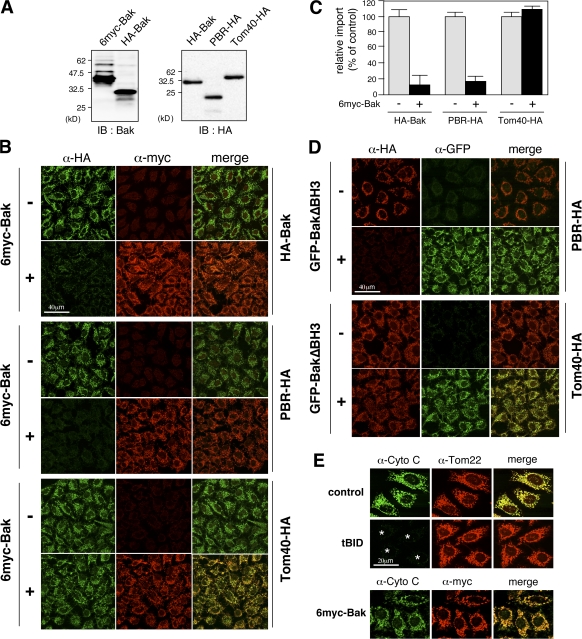
We previously demonstrated that MOM integration of C-TA proteins follows the TOM component–independent pathway (Setoguchi et al., 2006); the requirements were similar to those of PBR except for the Tom70 dependency. Therefore, we examined whether PBR and C-TA proteins have an overlapping import pathway using competition assays in semi-intact cells. Surprisingly, the import of PBR-HA, like HA-Bak, was inhibited by an excess amount of 6myc-Bak (Fig. 4, A–C). In contrast, the import of Tom40-HA (Fig. 4, B and C) and Su9-GFP (not depicted) was not affected. As a control, GFP-BakΔBH3, in which the BH3 domain of Bak was deleted, also inhibited the import, indicating that the proapoptotic activity of Bak is not involved in this inhibition (Fig. 4 D). Furthermore, proapoptotic factor tBid permeabilized MOM to release cytochrome c from mitochondria, whereas no such effect was observed with 6myc-Bak (Fig. 4 E), indicating that 6myc-Bak did not perturb MOM integrity but specifically inhibited MOM integration of PBR. We concluded that PBR shared the import pathway with the C-TA proteins after the initial docking on Tom70.
Figure 4.
Inhibition of MOM integration of PBR by an excess amount of Bak. (A) The reticulocyte lysate–synthesized proteins used (4 μl each) were analyzed by SDS-PAGE and immunoblotting (IB) using the indicated antibodies. (B) The indicated proteins were subjected to mitochondrial import in semi-intact cells in the presence or absence of an excess amount of 6myc-Bak (70 μl; import substrates, 20 μl each) and subsequent immunofluorescence microscopy. Imported proteins and 6myc-Bak are shown in green and red, respectively. (C) The extent of import was quantified using ImageJ (National Institutes of Health). Each graph indicates the mean ± SD (error bars) of three independent experiments of at least 100 cells. The fluorescence intensities of 6myc-Bak (−) cells were set at 100%. (D) Mitochondrial import of PBR-HA and Tom40-HA was performed as in B in the presence or absence of GFP-BakΔBH3. (E) Semi-intact cells were treated with tBid or 6myc-Bak, and the cells were analyzed by immunofluorescence microscopy using the antibodies against cytochrome c (green), Tom22 (red), or myc-tag (red). Asterisks indicate cytochrome c–released cells.
We noticed that PBR behaved like C-TA proteins in several additional aspects. PBR import was compromised at 4°C, very much like C-TA proteins, whereas the import of Tom40 and Su9-DHFR was less sensitive to the lower temperature (Fig. S2, A and C; available at http://www.jcb.org/cgi/content/full/jcb.200702143/DC1). Moreover, the import of PBR and C-TA proteins was significantly stimulated by the knockdown of VDAC1, the most abundant MOM protein, whereas no such stimulation was detected for the import of Tom40, VDAC2, and Su9-DHFR (Fig. S2, B and D; and not depicted for Su9-DHFR). These results might reflect the importance of the phospholipid phase as a rate-limiting step for the import of PBR and C-TA proteins, which is in contrast to import of the TOM complex–dependent substrates.
MOM integration of PBR depends on IMS components
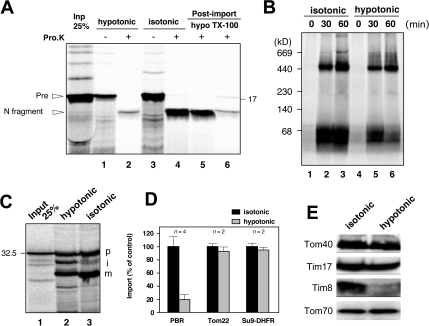
Membrane integration of mitochondrial inner membrane proteins with multi-TMSs such as ADP/ATP carrier and Tim23 and β-barrel MOM proteins depends on small Tim proteins in the IMS, which function as the chaperones to transfer incoming membrane proteins to the Tim22 complex or to the SAM complex (Koehler, 2004). Thus, we examined this point for MOM integration of PBR and found that depletion of IMS components by swelling the mitochondria strongly compromised the reaction (Fig. 5 A). MOM was not dissociated from the mitoplasts because levels of Tom40/Tom70 and Tim17 as the outer and inner membrane markers, respectively, were not changed after swelling the mitochondria (Fig. 5 E). The assembly of Tom22 to the TOM complex and matrix import of Su9-DHFR were not or were only slightly affected by this manipulation (Fig. 5, B–D), indicating that PBR import was specifically compromised by the depletion of IMS proteins.
Figure 5.
Depletion of IMS proteins inhibits PBR import. (A) Reticulocyte lysate–synthesized [35S]PBR-HA was imported into intact or hypotonic buffer–treated mitochondria. The reaction mixtures were aliquoted and incubated with or without 100 μg/ml proteinase K. The other aliquots (post-import) were treated with proteinase K under hypotonic conditions or in the presence of 1% Triton X-100. Samples were analyzed by SDS-PAGE and subsequent digital autoradiography. (B and C) Reticulocyte lysate–synthesized 35S-Tom22 (B) and 35S–Su9-DHFR (C) were imported into mitochondria and analyzed by BN-PAGE (for Tom22) or SDS-PAGE (for Su9-DHFR). p, precursor; i, intermediate; m, mature form. (D) Import efficiencies of PBR, Tom22, and Su9-DHFR were calculated setting each activity (PBR, proteinase K–resistant bands [percentage of input]; Tom22, ∼400-kD band; Su9-DHFR, m/(p + I + m]) of untreated mitochondria at 100%. Results obtained from two or four independent experiments are shown. Error bars represent SD. (E) Intact and hypotonic buffer–treated mitochondria were analyzed by SDS-PAGE and subsequent immunoblotting using the indicated antibodies.
Polytopic MOM proteins Mfn1, Mfn2, and MITOL are integrated into the membrane through a similar pathway as PBR
We addressed the integration pathway of MOM proteins with multiple TMSs other than PBR, including Mfn proteins and MITOL, and found that mitochondrial import of all of these proteins depended on Tom70 but not on any other TOM components (Fig. S3, A and B; available at http://www.jcb.org/cgi/content/full/jcb.200702143/DC1; and not depicted for Mfn1). Furthermore, MOM integration of Mfn2 as assessed by BN-PAGE was severely compromised by the depletion of IMS factors (Fig. S3 D). Thus, these polytopic MOM proteins are integrated into the MOM possibly through the PBR-like import pathway.
In conclusion, these experiments revealed a novel insertion pathway for several MOM proteins with multiple TMSs. The exact mechanism that facilitates the ATP-dependent membrane integration after targeting to Tom70 remains to be elucidated. One possibility might be spontaneous and direct insertion into the lipid bilayer without the assistance of a membrane translocation channel. However, because PBR and Mfn2 were not imported into the trypsin-treated mitochondria lacking the surface import receptors, direct insertion bypassing the Tom70 step into the lipid bilayer was ruled out. However, the possibility of Tom70-mediated direct integration into lipid bilayer remains because Tom70 has a chaperone function for several classes of membrane proteins. Similarly, IMS proteins that facilitate MOM integration of multi-TMS proteins remain to be identified. They might bind to the IMS-exposed loop and the flanking hydrophobic regions of multi-TMS proteins and function as a ratchet to promote membrane insertion, or they might be involved in releasing the insertion intermediate from the putative import conduit into the lipid bilayer.
Materials and methods
Materials
Antibodies against HA (Covance), FLAG (M2; Sigma-Aldrich), Hsp60 (Assay Designs), mHsp70 (Assay Designs), Tom20 (Santa Cruz Biotechnology, Inc.), Tom22 (Sigma-Aldrich), Tim8 (Santa Cruz Biotechnology, Inc.), cytochrome c (BD Biosciences), and rat calnexin (Assay Designs) were purchased from the indicated vendors. Antibodies against Tim17 and Tim23 (Ishihara and Mihara, 1998), Tom40 (Suzuki et al., 2000), and Tom70 (Suzuki et al., 2002) were described previously. Polyclonal antibodies against rat mitochondrial Sam50 was prepared by raising purified recombinant rat Sam50 in rabbits. Expression plasmids for Mfn1- and Mfn2-FLAG were constructed as described previously (Eura et al., 2003). MITOL-FLAG expression vector was a gift from S. Yanagi (Tokyo University of Pharmacology and Life Science, Tokyo, Japan).
Cell culture, transfection, and immunofluorescence microscopy
HeLa cells were cultured on coverslips in 35-mm dishes in 2 ml DME with 10% FCS at 37°C overnight under an atmosphere of 5% CO2 in air. Transfection was performed using FuGene 6 reagent (Roche). When mitochondria were to be stained, 40 nM MitoTracker Red CMX Ros (Invitrogen) was added to the medium and incubated for 20 min before fixation. For immunocytochemistry analysis, HeLa cells were seeded onto glass slides in mounting medium and observed by confocal fluorescence microscopy. Cells were fixed with 4% PFA, permeabilized with 1% Triton X-100 in PBS at room temperature for 5 min, and immunostained with the appropriate antibodies. Antigen–antibody complexes were detected using AlexaFluor488- or 568-conjugated goat anti–rabbit or anti–mouse IgG antibody (Invitrogen). Importantly, in these experiments, no AlexaFluor568 signal was detected in the 488-nm (green) AlexaFluor488 channel and vice versa. Immunofluorescence images were captured with the same detection sensitivity and were processed with Photoshop 8.0.1 software (Adobe). Preparation of semi-intact HeLa cells and preprotein import using semi-intact cells were conducted as described previously (Setoguchi et al., 2006).
siRNA treatment and mitochondrial protein import assay in vivo
The following RNA oligonucleotide pairs were used to create siRNA duplexes: GFP (5′-CUACAACAGCCACAACGUCdTdT-3′ and 5′-GAUGUUGUCGGUGUUGCAGdTdT-3′) and Sam50 (5′-GCUGAAAGUUAACCAGGAAdTdT-3′ and 5′-CGACUUUCAAUUGGUCCUUdAdA-3′). Validated specific siRNAs from QIAGEN (sequence not available) were used for the knockdown of Tom70, Tom40, Tom20, Tom22, Tim8B, and Tim10.
For microscopic assay of mitochondrial protein import in siRNA-treated cells, HeLa cells were transfected with the target siRNAs twice within a 24-h interval. At 72 h after the initial treatment, cells were transfected with expression plasmids for Su9-GFP, PBR-HA, Mfn2-FLAG, or MITOL-FLAG. 16 h after the DNA transfection, the cells were processed for double indirect immunofluorescence microscopy. Cell fractionation was performed as described previously (Otera et al., 2005). Fractionated samples were assessed by SDS-PAGE followed by immunoblotting using the appropriate antibodies.
cDNA cloning of rat PBR
The cDNA fragment encoding PBR was prepared by RT-PCR using rat liver poly(A)+-RNA as the template and the following oligonucleotides as the primers: 5′-CCGGAATTCATGTCTCAATCCTGGGTACCC-3 and 5′-GCGGGATCCTCACTCTGTGAGCCGGGAGCC-3′, where underlining indicates the EcoRI and BamHI sites, respectively. The PCR fragment thus obtained was digested with EcoRI and BamHI and cloned into the EcoRI–BamHI sites of p3xFLAG–cytomegalovirus (Sigma-Aldrich).
Preparation of antibodies against rat PBR
The DNA fragment encoding full-length PBR was amplified by PCR using rat PBR cDNA as the template and the following oligonucleotides as the primers: 5′-CTCGCATATGTCTCAATCCTGGGTACCC-3′ and 5′-GCGGGATCCTCACTCTGTGAGCCGGGAGCC-3′, where underlining indicates the NdeI and BamHI sites, respectively. The obtained fragment was subcloned into the pET28a vector (EMD) to create pET28a-NHis-PBR, where the (His)6-tag was attached to the N terminus of the expressed protein. His-tagged PBR was expressed in BL21 cells as inclusion bodies, which were resolved by SDS-PAGE, and the Coomassie Brilliant blue–stained band was excised from the gel and used to raise antibodies in rabbits using the Ribi Adjuvant system (Rini Immunochemical Research).
Construction of expression plasmids for rat PBR
The expression vector for the C-terminal HA-tagged PBR protein (PBR-HA) was constructed as follows. The coding region of rat PBR cDNA was amplified by PCR using rat PBR cDNA as the template and the following oligonucleotides as the primers: 5′-GCCAAGCTTCCACCATGTCTCAATCCTGGGTA-3′ and 5′-GCGGGATCCCTCTGTGAGCCAGGAGCCCCC-3′, where underlining indicates the HindIII and BamHI sites, respectively. The obtained fragment was subcloned into HindIII–BamHI-digested pcDNA3.1 to create pcDNA3.1–PBR-HA. The N-terminal HA-tagged PBR protein (HA-PBR) was constructed using rat PBR cDNA as the template and the following oligonucleotides as the primers: 5′-GCCAAGCTTCCACCATGTGCTTACCCTTACGACGTCCCTGACTACGCCTCTCTCATGTCTCAATCCT-3′ and 5′-CCGTCTAGATCACTCTGTGAGCCGGGAGCC-3′, where underlining indicates the HindIII and XbaI sites, respectively. The obtained PCR fragment was subcloned into HindIII–XbaI-digested pcDNA3.1 to create pcDNA3.1–HA-PBR. For in vitro expression, all constructs were recloned into pSP64 vectors.
Immunoblot analysis
Electroblot filters were incubated with the primary antibodies followed by peroxidase-coupled goat anti–rabbit or anti–mouse secondary antibodies (Invitrogen). Immunodetection was performed by ECL (GE Healthcare).
Preparation of rat liver mitochondria and submitochondrial fractionation
Rat liver mitochondria were prepared according to the method of Sakaguchi et al. (1992). Submitochondrial fractionation by sucrose density gradient centrifugation was performed as follows. Mitochondria were diluted into the hypotonic buffer (10 mM Hepes-KOH buffer, pH 7.4, containing 1 mM EDTA and protease inhibitor mix [5 μg/ml each of leupeptin, antipain, chymostatin, and pepstatin]) and incubated at 0°C for 30 min. The mixture was sonicated on ice five times for 30 s each time and centrifuged at 5,000 g for 10 min to obtain the supernatant. This fraction was layered over a linear gradient (0.6–1.6 M) of sucrose in the hypotonic buffer and centrifuged at 100,000 g for 15 h at 4°C.
Preparation of mitochondria from cultured COS-7 cells
Cultured cell mitochondria were prepared according to the protocol of Kanaji et al. (2000). COS-7 cells cultured in a 10-cm dish were washed with PBS and were scraped off in 1 ml PBS. Collected cells were precipitated by centrifugation at 600 g for 5 min and washed with Hepes-EDTA buffer (10 mM Hepes-KOH buffer, pH 7.5, containing 1 mM EDTA and 10% [wt/vol] sucrose). The cells were resuspended in 1 ml of the same buffer containing protease inhibitor mix, homogenized by aspirating the cells five times through a 27-gauge needle, and centrifuged at 600 g for 5 min to obtain a postnuclear supernatant. The supernatant fraction was centrifuged at 6,000 g for 15 min to obtain the mitochondria.
Mitochondrial protein import in vitro
The reaction mixtures containing 5–25 μg mitochondria and 5 μl of rabbit reticulocyte lysate–synthesized 35S-labeled pAd, Su9-DHFR, Tom22, PBR, or Mfn2 were incubated in 50–100 μl of 10 mM Hepes-KOH buffer, pH 7.4, containing 1 mM ATP, 20 mM sodium succinate, 5 mM NADPH, 0.5 mM magnesium acetate, and a protease inhibitor mixture at 30°C for 30 min. After import, the mitochondria were isolated, and the reaction mixtures were incubated with 100 μg/ml proteinase K at 10°C for 30 min and precipitated with TCA (PBR-HA) or left untreated. The mitochondria were then isolated by centrifugation and were analyzed by SDS-PAGE or BN-PAGE and subsequent immunoblotting using anti-HA antibody or digital autoradiography. The ATP requirement was determined as follows. Reticulocyte lysate–synthesized 35S–PBR-HA or 35S-pAd was passed through a spin column and used for mitochondrial import in the presence or absence of 1 mM ATP or AMP-PNP. Import of PBR-HA or pAd into the Tom40-depleted mitochondria in the presence or absence of recombinant Su9-DHFR precursor (Fig. 2) was performed as follows. ∼0–10 μg recombinant Su9-DHFR precursor was preincubated at 0°C for 10 min in the import buffer (total volume of 45 μl) containing 1 mM methotrexate, 1 mM NADPH, and 5 μg HeLa cell mitochondria, and 5 μl reticulocyte lysate–synthesized PBR-HA or pAd was added to the reaction mixture and incubated at 30°C for 30 min. The reaction mixtures were analyzed by SDS-PAGE or BN-PAGE and subsequent digital autoradiography.
BN-PAGE
50 μg mitochondria was solubilized in 50 μl of solubilization buffer, and insoluble materials were removed by centrifugation at 10,000 g for 15 min. The supernatant was mixed with 5 μl of sample buffer (5% Coomassie Brilliant blue G-250, 100 mM Bis-Tris, pH 7.0, and 500 mM 6-aminocaproic acid) and electrophoresed through a 5–16% polyacrylamide gradient gel.
Purification of recombinant Su9-DHFR-His6
5 ml of 16-h culture of Escherichia coli BL21 expressing Su9-DHFR-His6 protein (in pET28a) was diluted with 100 vol yeast extract tryptone medium. After 3 h of culture at 37°C, IPTG was added to a final concentration of 1 mM. After 3 h at 37°C, the cells were harvested, resuspended in 20 ml of ice-cold suspension buffer consisting of 50 mM Tris-HCl, pH 7.5, buffer containing 150 mM NaCl, and protease inhibitor mixture and were sonicated. The lysates were centrifuged to remove cell debris. The resulting supernatant was applied to a metal affinity resin column (TALON). The column was washed with 10 mM imidazole, and Su9-DHFR precursor was eluted with 40 mM imidazole-containing buffer. The eluted fraction was dialyzed against the homogenize buffer and concentrated by ultrafiltration to use for the general import pore block experiment.
Analysis of Tom70–PBR interaction using immunoprecipitation
HeLa cells cotransfected with PBR-HA and FLAG-GFP-Tom70ΔN were lysed with the binding assay buffer (50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, and 1 mM PMSF) and centrifuged to remove cell debris. The resulting supernatant was mixed with anti-FLAG IgG-conjugated agarose beads, and beads were washed three times with the binding assay buffer. Coimmunoprecipitated proteins were analyzed by SDS-PAGE and subsequent immunoblotting using the appropriate antibodies.
Online supplemental material
Fig. S1 shows the membrane topology of PBR and in vitro import of PBR into mitochondria. Fig. S2 shows stimulation of mitochondrial import of PBR and C-TA proteins in VDAC1-depleted semi-intact cells. Fig. S3 shows that swelling of mitochondria inhibits the mitochondrial import of Mfn2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702143/DC1.
Supplemental Material
Acknowledgments
We thank S. Yanagi for providing us with the human MITOL-FLAG expression vector. We also thank all members of the Mihara laboratory for helpful discussions.
This work was supported by grants from the Ministry of Education, Science, and Culture of Japan, the Human Frontier Science Program, and the Takeda Science Foundation.
H. Otera and Y. Taira contributed equally to this paper.
Abbreviations used in this paper: BN, blue native; C-TA, C-terminal tail anchor; DHFR, dihydrofolate reductase; IMS, intermembrane space; MITOL, mitochondrial ubiquitin ligase; MOM, mitochondrial outer membrane; PBR, peripheral benzodiazepine receptor; SAM, sorting and assembly machinery; TMS, transmembrane segment; TOM, translocase of outer membrane.
References
- Ahting, U., T. Weizenegger, W. Neupert, and D. Rapaport. 2005. Signal-anchor proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 280:48–53. [DOI] [PubMed] [Google Scholar]
- Bohnert, M., N. Pfanner, and M. van der Laan. 2007. A dynamic machinery for import of mitochondrial precursor proteins. FEBS Lett. 581:2802–2810. [DOI] [PubMed] [Google Scholar]
- Eura, Y., N. Ishihara, S. Yokota, and K. Mihara. 2003. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. (Tokyo). 134:333–344. [DOI] [PubMed] [Google Scholar]
- Fan, A.C.Y., M.K. Bhangoo, and J.C. Yound. 2006. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J. Biol. Chem. 281:33313–33324. [DOI] [PubMed] [Google Scholar]
- Fritz, S., D. Rapaport, E. Klanner, W. Neupert, and B. Westermann. 2001. Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organeller function. J. Cell Biol. 152:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara, N., and K. Mihara. 1998. Identification of the protein import components of the rat mitochondrial inner membrane, rTIM17, rTIM23, and rTIM44. J. Biochem. (Tokyo). 123:722–732. [DOI] [PubMed] [Google Scholar]
- Ishikawa, D., H. Yamamoto, Y. Tamura, K. Moritoh, and T. Endo. 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. 2004. J. Cell Biol. 166:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Liauzun, E., P. Delmas, D. Shire, and P. Ferrara. 1998. Topological analysis of the Peripheral Benzodiazepine Receptor in yeast mitochondrial membrane supports a five-transmembrane structure. J. Biol. Chem. 273:2146–2152. [DOI] [PubMed] [Google Scholar]
- Kanaji, S., J. Iwahashi, Y. Kida, M. Sakaguchi, and K. Mihara. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151:277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski, M., A. Neutzner, and R.J. Youle. 2007. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 178:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309–335. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Y. Kimura, M. Tokuda, S. Honda, and S. Hirose. 2006. MARCH-V is a novel mitofusin-2 and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 7:1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera, H., S. Ohsakaya, Z. Nagaura, N. Ishihara, and K. Mihara. 2005. Export of mitochondrial AIF in response to proapoptotic stimuli. EMBO J. 24:1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos, V., H. Amri, N. Boujrad, C. Cascio, M. Culty, M. Garnier, M. Hardwick, H. Li, B. Vidic, A.S. Brown, et al. 1997. Peripheral benzodiazepine receptor in cholesterol transport and steroidogensis. Steroids. 62:21–28. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., W. Neupert, and D. Rapaport. 2005. Biogenesis of β-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30:575–582. [DOI] [PubMed] [Google Scholar]
- Rapaport, D. 2003. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D. 2005. How does the TOM complex dediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 171:419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, M., N. Hachiya, K. Mihara, and T. Omura. 1992. Mitochondrial porin can be transported across both endoplasmic reticulum and mitochondrial membranes. J. Biochem. 112:243–248. [DOI] [PubMed] [Google Scholar]
- Setoguchi, K., H. Otera, and K. Mihara. 2006. Cytosolic factor- and TOM-independent import of C-tail-anchor mitochondrial outer membrane proteins. EMBO J. 25:5635–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Y. Okazawa, T. Komiya, K. Saeki, E. Mekada, S. Kitada, A. Ito, and K. Mihara. 2000. Characterization of rat TOM40, a central component of the proprotein translocase of the mitochondrial outer membrane. J. Biol. Chem. 275:37930–37936. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., M. Maeda, and K. Mihara. 2002. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 115:1895–1905. [DOI] [PubMed] [Google Scholar]
- Waizenegger, T., S.J. Habib, M. Lech, D. Morkanjac, S.A. Paschen, K. Hell, W. Neupert, and D. Rapaport. 2004. Tob 38, a novel essential component in the biogenesis of beta-barrel proteins of mitochondria. EMBO Rep. 5:704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro, R., S. Ishid, S. Kyo, T. Fukuda, E. Goto, Y. Matsuki, M. Ohmura-Hoshino, K. Sada, H. Hotta, H. Yamamura, et al. 2006. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 25:3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.C., N.J. Hoogenraad, and F.U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor. Cell. 112:41–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.