Abstract
In addition to their well-known function in apoptosis, caspases are also important in several nonapoptotic processes. How caspase activity is restrained and shut down under such nonapoptotic conditions remains unknown. Here, we show that Drosophila melanogaster inhibitor of apoptosis protein 2 (DIAP2) controls the level of caspase activity in living cells. Animals that lack DIAP2 have higher levels of drICE activity. Although diap2-deficient cells remain viable, they are sensitized to apoptosis following treatment with sublethal doses of x-ray irradiation. We find that DIAP2 regulates the effector caspase drICE through a mechanism that resembles the one of the caspase inhibitor p35. As for p35, cleavage of DIAP2 is required for caspase inhibition. Our data suggest that DIAP2 forms a covalent adduct with the catalytic machinery of drICE. In addition, DIAP2 also requires a functional RING finger domain to block cell death and target drICE for ubiquitylation. Because DIAP2 efficiently interacts with drICE, our data suggest that DIAP2 controls drICE in its apoptotic and nonapoptotic roles.
Introduction
Caspases are best known for their role in executing apoptosis, however, they also play important signaling roles in nonapoptotic processes, such as regulation of actin dynamics, innate immunity, and cell proliferation, differentiation, and survival (Lamkanfi et al., 2007). Under such conditions, caspases are activated without killing the cell.
Given the irreversible nature of caspase-mediated proteolysis, caspases' activation and activity is subject to complex regulation. After zymogen activation, caspase activity can be controlled by various cellular and viral inhibitors. Interestingly, the viral inhibitors CrmA and p35 neutralize caspases through a fundamentally different mechanism than cellular inhibitors such as the inhibitor of apoptosis (IAP) proteins (Salvesen and Duckett, 2002). CrmA and p35 function as “suicide substrates,” whereby cleavage of CrmA and p35 is required to block the caspase. Initially, CrmA and p35 are cleaved through a mechanism that resembles substrate hydrolysis. However, during the reaction, the protease's catalytic machinery is trapped via covalent linkage (Stennicke and Salvesen, 2006). This strategy is referred to as “mechanism-based” inhibition because it relies on the caspase's catalytic property. So far, cellular-derived mechanism-based caspase inhibitors have not been identified.
IAPs are defined by the presence of one to three copies of the baculovirus IAP repeat (BIR) domain, which functions as a protein interaction module. In addition, certain but not all IAPs also carry a C-terminal RING finger domain, which provides these molecules with E3 ubiquitin–protein ligase activity. In Drosophila melanogaster, DIAP1-mediated inhibition of caspases is indispensable for cell survival. Mutations that abrogate the physical association of DIAP1 with the effector caspases drICE or DCP-1 or the initiator caspase Dronc cause spontaneous caspase activation and cell death (Wang et al., 1999; Goyal et al., 2000; Lisi et al., 2000; Rodriguez et al., 2002; Zachariou et al., 2003).
The involvement of the second D. melanogaster IAP (DIAP2) in regulating caspases is less clear. Overexpression studies suggest that DIAP2 can suppress cell death induced by the IAP antagonists reaper (Rpr) and head involution defective (Hid; Hay et al., 1995) and suppress apoptosis induced by diap1-RNAi (Leulier et al., 2006b). Because diap1-RNAi–mediated death is independent of IAP antagonists, this suggests that DIAP2 can neutralize caspases. Of the seven D. melanogaster caspases, DIAP2 only binds to the effector caspase drICE and the atypical caspase Strica (Doumanis et al., 2001). In contrast, DIAP1 exhibits much broader caspase selectivity because it inhibits Dronc, drICE, and DCP-1.
Based on the domain architecture and sequence alignments, DIAP2 is the closest homologue to mammalian IAPs. In contrast, DIAP1 is more related to IAPs from insect viruses. Like the mammalian x-linked IAP (XIAP), DIAP2 carries three BIR domains and a C-terminal RING finger. XIAP blocks apoptosis by acting as a potent enzymatic inhibitor of caspase-3, -7, and -9 (Shi, 2002). Residues located immediately upstream of XIAP's BIR2 domain directly bind to the active site pockets of caspase-3 and -7, thereby obstructing substrate entry (Chai et al., 2001; Huang et al., 2001; Riedl et al., 2001a; Silke et al., 2001; Suzuki et al., 2001). Importantly, XIAP is not cleaved by caspases because it binds to the catalytic pockets of effector caspases in a reverse orientation. Thus, XIAP relies on allosteric mechanisms to block effector caspase activity. XIAP's ability to suppress effector caspases depends on the side chain of Asp148 and, to a lesser extent, Val146. It is predicted that Asp148 must be conserved in all IAPs that neutralize effector caspases. Consistently, Asp148 is conserved in cIAP1 and 2. Intriguingly, DIAP2 also shows homology to XIAP's Val146 and Asp148, as it carries equivalent residues at Val98 and Asp100 (Silke et al., 2001). In contrast, these residues are absent in DIAP1.
Of all the IAPs, XIAP appears to be the only IAP that potently inhibits caspases in vitro (Eckelman et al., 2006). Other IAPs, such as cIAP1 and 2, are poor inhibitors of caspases under these conditions. cIAP1 and 2 may rely on their E3 ligase activity to block caspases in vivo, making them caspase regulators rather than inhibitors. Although DIAP1 is a relatively good inhibitor of D. melanogaster caspases in vitro (Yan et al., 2004), DIAP1–caspase physical association alone is insufficient to neutralize caspases in the fly. In addition to binding, DIAP1 uses two independent mechanisms to regulate caspases. One relies on the E3 ubiquitin protein ligase activity of DIAP1's own RING finger, whereas the other, the “N-end” rule, functions independently of this domain. The RING finger of DIAP1 is required to target the proform of the initiator caspase Dronc for ubiquitylation and inactivation (Wilson et al., 2002). In contrast, active effector caspases seem to be neutralized through a mechanism that involves both the RING finger domain as well as the N-end rule degradation machinery that is recruited by DIAP1 (Ditzel et al., 2003).
In D. melanogaster, developmentally regulated apoptosis is induced by the IAP antagonists Rpr, Grim, and Hid (Salvesen and Abrams, 2004). Embryos lacking rpr, grim, and hid are virtually devoid of apoptosis and die at the end of embryogenesis with accumulation of supernumeral cells (White et al., 1994). The current model suggests that Rpr, Grim, and Hid induce apoptosis by binding to the BIR domains of DIAP1, thereby liberating caspases from IAP inhibition. Furthermore, IAP antagonists also deplete DIAP1 protein levels by promoting its degradation (Ryoo et al., 2002; Yoo et al., 2002) and cause mitochondrial permeability (Abdelwahid et al., 2007). Although Rpr, Grim, and Hid efficiently antagonize DIAP1, they also associate with DIAP2 (Vucic et al., 1997, 1998; Leulier et al., 2006b). This implies that DIAP2 contributes to the overall antiapoptotic threshold of a cell, a view that is supported by the notion that RNAi-mediated knockdown of DIAP2 sensitizes cultured cells to stress-induced death (Zimmermann et al., 2002). However, analyses using diap2 mutant flies have failed to expose any involvement of DIAP2 in regulating programmed cell death. Instead, they have shown that DIAP2 is required for nuclear factor κB activation during the innate immune response in D. melanogaster (Gesellchen et al., 2005; Kleino et al., 2005; Leulier et al., 2006a; Huh et al., 2007).
Here, we have used the fluorescence resonance energy transfer (FRET)–based caspase-3 indicator SCAT3 to examine DIAP2's role in regulating caspase activity in vivo. Using diap2 mutant animals, we found that these animals harbor significantly increased levels of drICE activity. Consistent with higher levels of active caspases, diap2 mutant cells are sensitized to apoptosis after exposure to sublethal doses of x-ray irradiation. Furthermore, we find that DIAP2 tightly associates with the effector caspase drICE and that DIAP2-mediated drICE inhibition requires DIAP2 cleavage at Asp100. Our data are consistent with the notion that after cleavage, the caspase forms a covalent adduct with DIAP2, which results in the stabilization of the DIAP2–caspase complex. This mode of caspase binding is similar to that of p35 or CrmA. However, in addition to the mechanism-based trapping, DIAP2 also requires a functional RING finger domain to neutralize drICE-mediated cell death. Consistently, we find that DIAP2 robustly ubiquitylates drICE in vivo.
Results
Loss of DIAP2 results in deregulated drICE activity in vivo
To investigate the role of endogenous DIAP2 in regulating drICE in vivo, we examined whether diap2 mutant animals display higher basal levels of caspase activity. In vivo caspase activity was measured using SCAT3, a FRET-based probe that detects active caspases (Fig. 1, A–H). In this system, caspase activation causes a decrease in the FRET signal, which results in a decrease of Venus/ECFP emission ratio (Takemoto et al., 2003). Hedgehog-GAL4–driven expression of the UAS-SCAT3 probe, which drives expression of the SCAT3 sensor in the posterior part of the wing disc (Fig. 1, A–D and G), revealed that caspase activity is low in wing imaginal discs of wild-type (WT) third instar larvae. In contrast, FRET analysis of diap2 loss- of-function mutants (diap27c; Leulier et al., 2006b) showed that caspase activity was significantly increased, as seen by the reduction of the FRET signal. Importantly, this increase in caspase activity was completely suppressed in the presence of a diap2 genomic rescue transgene (Fig. 1, D, P[diap2 +]), which demonstrates that it is caused by the loss of diap2. Moreover, RNAi-mediated knockdown of drICE abolished the observed increase in caspase activity (Fig. 1 C). Together, these data suggest that loss of diap2 results in an increase of drICE activity in wing discs. Similarly, salivary glands of diap2 mutant animals also displayed higher levels of endogenous caspase activity when compared with controls (Fig. 1, E, F, and H). Of note, the SCAT3 ratio of dying cells is around 1 (Fig. 1, A–F, blue; Takemoto et al., 2007). Therefore, the observed ratio in diap2 7c mutant flies is nonapoptotic. Consistent with the FRET data, we also detected increased DEVDase activity (Ac-DEVD-MCA) in adult diap2 mutant animals (Fig. 1 I). Collectively, these results indicate that the loss of diap2 leads to increased basal levels of drICE activity, albeit without causing cell death.
Figure 1.
diap2 mutant cells have increased levels of active drICE. (A–H) Spectral analysis of WT and diap2 mutant wing discs (A–D and G) and salivary glands (E, F, and H). FRET ratio images are shown. Genotypes used: +;hh-GAL4/UAS-SCAT3 (A), diap27c;hh-GAL4/UAS-SCAT3 (B), diap27c/diap27c,UAS-drICE-IR;hh-GAL4/UAS-SCAT3 (C), diap27c;hh-GAL4,UAS-SCAT3/P[diap2 + ](D), +;da-GAL4/UAS-SCAT3 (E), and diap27c;da-GAL4/UAS-SCAT3 (F). Changes in the emission ratio (530:475 nm, Venus/ECFP) of SCAT3 in the posterior region of wing discs (G) or salivary glands (H). The data shown represent an average of the FRET ratio in each tissue from >10 flies (mean ± SD). (I) Increased DEVDase activity in lysates from adult diap2 mutant animals. Caspase activity (mean ± SD) was determined using the fluorescent substrate DEVD-MCA. (J–Q) DIAP2 mutant cells are sensitized to undergo cell death, as assessed by AO staining. Genotypes used: yw1118 (J and N), diap27c/Def(2R)exel7138 (K and O), diap27c (L and P), and diap27c;P[diap2 +] (M and Q). (R) Quantification of the number of AO-positive cells (mean ± SD). Bars, 100 μm.
We next tested whether loss of DIAP2 sensitizes cells to sublethal doses of x-ray irradiation. To this end, we exposed third instar larvae to sublethal doses (1.5 Gy) of x-ray irradiation and monitored cell death after 4 h using acridine orange (AO). In nonirradiated flies, the number of AO-positive cells was low and comparable across all tested genotypes. However, after irradiation with 1.5 Gy, diap27c hemi- and homozygous flies displayed a marked increase in AO-positive cells when compared with WT flies, which displayed no significant increase in AO-positive cells (Fig. 1, J–R). Importantly, this enhanced sensitivity of diap27c mutants was rescued by a genomic construct of DIAP2. This indicates that loss of DIAP2 is responsible for the observed increase in cell death after irradiation and is consistent with a model whereby DIAP2 contributes to the apoptotic threshold (see Fig. 8 A).
Figure 8.
IAP-mediated regulation of caspases. (A) Schematic representation of the caspase cascade in D. melanogaster depicting DIAP2's epistatic position. Arrows denote activation and bars represent suppression. Bold arrows denote the predominant role of drICE downstream of apoptosome activation. (B and C) Comparison of DIAP2- and DIAP1-mediated regulation of effector caspases.
Binding of DIAP2 to drICE
We have previously shown that DIAP2 interacts with drICE (Leulier et al., 2006b). To identify the mode of caspase inhibition by DIAP2, we first established the mechanism through which DIAP2 binds to drICE. Because drICE activation results in the exposure of an evolutionarily conserved IAP-binding motif (IBM) at the neo N terminus of its p20 subunit (Tenev et al., 2005), we examined the role of this IBM for DIAP2 binding. Tandem affinity purification (TAP)–tagged DIAP2 was expressed in 293T cells (mammalian cells were chosen to avoid potential interference by DIAP1), purified (Rigaut et al., 1999), and subsequently incubated with extracts of cells expressing either drICEWT or drICEA29V lacking the Ala of the IBM (Fig. 2 B). The processed and active form of drICEWT was readily copurified by DIAP2 (Fig. 2 B). In contrast, DIAP2 failed to purify drICEA29V from cellular extracts. Note, substitution of Ala29 to Val does not affect processing or activity of drICE (Fig. 2 B, compare lanes 1 and 2; Tenev et al., 2005), which indicates that the observed differences in DIAP2 binding are caused by the lack of a functional IBM in drICEA29V.
Figure 2.
DIAP2 interacts with drICE in a BIR3- and IBM-dependent manner. (A) Schematic representation of DIAP2 proteins used in this paper. (B) The IBM mutant drICEA29V fails to associate with DIAP2. Expression of drICE and drICEA29V and their copurification with DIAP2 was determined by immunoblot analysis using the indicated antibodies. TAP-purified DIAP2 was detected by immunoblotting with an antibody recognizing the protein A portion of the TAP tag (designated as anti-TAP). (C and D) The DIAP2–drICE interaction requires the BIR3 domain, as mutation of this region abrogates drICE binding.
The Ala1 of IBMs anchors IBM-containing proteins to particular BIR domains (Wu et al., 2000, 2001). Therefore, we used DIAP2 point and truncation mutants to identify the BIR domain that is required for drICE binding (Fig. 2, C and D; and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200706027/DC1). The BIR3 domain of DIAP2 was indispensable for drICE binding because mutation of this domain abolished drICE association (Fig. 2, C and D). In contrast, deletion of the BIR1 or RING finger (Fig. S1 A) or disruption of the BIR2 through mutation of Cys149 to Gly or Asp163 to Ala did not abrogate the interaction with drICE. Although the BIR3 domain is necessary for drICE binding, it is not sufficient, as the BIR3 domain in isolation failed to associate with drICE (Fig. S1 B).
Endogenous DIAP2 is processed by a p35-sensitive caspase
We noticed that DIAP2 was readily cleaved by drICE (Fig. 2 B). To study whether DIAP2 cleavage also occurred in intact cells, we examined endogenous DIAP2 in normal D. melanogaster S2 cells. In agreement with N-terminal cleavage, we detected two distinct endogenous DIAP2 proteins with different mobilities: full-length DIAP2 and a slightly faster migrating form reflecting a smaller DIAP2 protein (Fig. 3 B). RNAi-mediated depletion of DIAP2 showed that both forms were DIAP2 derived. The presence of the smaller form was caspase dependent because it was significantly reduced in cells treated with the caspase inhibitor z-VAD-fmk or that expressed p35 (Fig. 3 B). Cleavage of DIAP2 is likely to be mediated by drICE, as drICE can directly cleave DIAP2 in vitro and RNAi-mediated depletion of drICE reduces cleavage of endogenous DIAP2 in S2 cells (Fig. 3 C and not depicted). DIAP2 carries two putative caspase cleavage sites at positions 92 and 100, N-terminal to the BIR2 domain (Fig. 3 A). Mutation of Asp100 to Glu, like z-VAD-fmk treatment, abrogated DIAP2 cleavage (Fig. 3 D). Under the same conditions, cleavage of WT DIAP2 and DIAP2D92E (unpublished data) was not affected. This indicates that Asp100, which is evolutionarily conserved, is required for caspase-mediated cleavage.
Figure 3.
DIAP2 is cleaved at Asp100 by a z-VAD– and p35-sensitive caspase. (A) Schematic representation of DIAP2. The putative caspase cleavage site is underlined and boxed. Note, XIAP's Asp148 (black circle) and Val146 (gray circle) are conserved in DIAP2 of D. melanogaster (D.mel), D. simulans (D.sim), D. yakuba (D.yak.), D. erecta (D.ere), and D. ananassae (D.ann) and in human cIAP1. DIAP1's cleavage site is indicated as a dashed box. The sequences were aligned according to ClustalX and anchored into the BIR2 of the indicated IAPs. (B) Endogenous DIAP2 is cleaved by an endogenous caspase. D. melanogaster S2 cells treated with the indicated conditions were lysed and DIAP2 was detected by immunoblotting with the indicated antibody. Actin was used as loading control. (C) In vitro cleavage assay using recombinant drICE and DIAP1 and 2. Asterisks indicate cross-reactive bands. (D) DIAP2 is cleaved at Asp100. DIAP2-GST and DIAP2D100E-GST were expressed in S2 cells in the absence or presence of z-VAD-fmk. DIAP2 was purified using GST-beads and subjected to immunoblot analysis using anti-GST. (E) Endogenous DIAP2 is cleaved in response to UV treatment. Endogenous DIAP1, DIAP2, and drICE were detected by Western blot analysis using the indicated antibodies. Black lines indicate that the intervening lanes have been spliced out.
Next, we examined the cleavage status of endogenous DIAP2 under apoptotic conditions. Exposure to UV light or expression of Rpr triggered DIAP2 cleavage (Fig. 3 E and not depicted). Under the same conditions, DIAP1 was also cleaved (Fig. 3 E; Ditzel et al., 2003). Although a small proportion of DIAP1 and 2 was processed in untreated controls, exposure to UV light converted almost all full-length DIAP1 and 2 into their cleaved forms. DIAP2 cleavage occurred predominantly 2–3 h after UV exposure and at a time point where no apoptotic morphological changes, significant drICE activation, or cleavage of Bicaudal, a bona fide caspase substrate (Martin, S., personal communication), was apparent (unpublished data). This suggests that DIAP2 cleavage, like that of DIAP1 (Ditzel et al., 2003), occurs early during apoptosis. Exposure to z-VAD-fmk restored the appearance of full-length DIAP1 and 2 (unpublished data). Consistent with previous studies, DIAP1 protein levels rapidly diminished after UV treatment (Fig. 3 E; Muro et al., 2005). Although DIAP2 was cleaved, DIAP2 levels were not depleted to the same extent as DIAP1. Collectively, these data indicate that, after induction of apoptosis, endogenous DIAP2 and 1 are cleaved by caspases with similar kinetics.
DIAP2 cleavage is required to suppress drICE-mediated cell death
To determine the functional relevance of DIAP2 cleavage, we tested the ability of WT and noncleavable mutant DIAP2 to inhibit drICE-mediated cell death. Because ectopic expression of full-length drICE does not induce cell death in D. melanogaster tissue culture cells or transgenic flies (Song et al., 2000), we used mammalian cells (NIH3T3) to study the ability of DIAP2 to suppress drICE-mediated cell death. In this system, drICE expression is sufficient to trigger apoptosis (Fig. 4 A; Tenev et al., 2005). Coexpression of DIAP1 and 2 provided protection against drICE-induced cell death. However, DIAP2D100E, which lacks the caspase cleavage site, failed to provide significant protection (Fig. 4 B). Moreover, the BIR3 mutant DIAP2C249G, which is impaired in drICE binding, also failed to inhibit this death. In contrast to drICE-induced cell death, DIAP2 was unable to suppress DCP-1–mediated apoptosis, whereas, under the same conditions, DIAP1 efficiently blocked DCP-1 killing (Fig. 4 C). This is in agreement with the notion that DIAP1 interacts with both drICE and DCP-1, whereas DIAP2 only associates with drICE (Leulier et al., 2006b).
Figure 4.
Asp100 of DIAP2 is required for drICE inhibition. (A–C) Expression of drICE and DCP-1 leads to induction of apoptosis in NIH3T3 cells. (A) DIAP2 suppresses drICE-mediated cell death. (B) The cleavage site of DIAP2 was required to block drICE killing. (C) DIAP2 failed to inhibit DCP-1– induced death. Shown are the percentages of β-galactosidase (β-Gal)–positive cells with apoptotic morphology from three independent experiments (mean ± SD). (D) Mutation of Asp100 to Glu-like deletion of the N-terminal 100 residues of DIAP2 abrogated drICE binding. The binding assay was performed as in Fig. 2.
The DIAP2 cleavage site is required for drICE binding
Next, we investigated why DIAP2 cleavage is required for drICE inhibition. Crystal structure and mutational analysis indicate that XIAP's Val146 and Asp148 play a central role in the recognition of caspase-3 and -7, serving as anchoring residues for the hydrophobic and hydrophilic contact regions, respectively (Chai et al., 2001; Riedl et al., 2001a). Because the P1 residue of DIAP2's caspase cleavage site (Asp100) corresponds to XIAP's Asp148 (Fig. 3 A; Silke et al., 2001), we determined its involvement in the binding to drICE. Although active drICE readily interacted with WT DIAP2 and DIAP278–498, its binding to the noncleavable DIAP2D100E mutant was strongly affected (Fig. 4 D). This suggests that Asp100 is required for the recognition of drICE, which provides a likely explanation for why DIAP2D100E failed to block drICE-induced killing. However, the cleavage motif on its own was not sufficient to sustain DIAP2–drICE complex formation (Fig. S1 C). Thus, drICE binding occurs through a bimodular interaction that involves the BIR3 and Asp100. Our data indicate that BIR3 binding actually precedes binding to and cleavage of Asp100 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200706027/DC1).
We also tested whether the N-terminal region of DIAP2 needs to be cleaved for DIAP2 to bind to drICE. DIAP2101–498, which lacks the N-terminal portion and hence mimics a cleaved form of DIAP2, failed to interact with drICE (Fig. 4 D, lane 5). Moreover, full-length WT DIAP2 readily bound to a fully processed drICE mutant (drICEC211A; Fig. S3, available at available at http://www.jcb.org/cgi/content/full/jcb.200706027/DC1). These observations indicate that the N-terminal portion of DIAP2 does not act as an autoinhibitory region that might interfere with drICE binding.
Collectively, our observations are consistent with a model whereby DIAP2 binds to drICE through a mechanism that relies on the BIR3 domain and Asp100. Mutation of either motif abrogates drICE binding. In contrast to XIAP, DIAP2 is effectively cleaved at Asp100, which suggests that DIAP2 and XIAP bind to the caspases' catalytic pocket differently.
DIAP2 functions as a mechanism-based regulator of drICE
DIAP2-mediated regulation of drICE strongly resembles the inhibitory mechanism of p35. p35 arrests caspase-mediated proteolysis at the thioacyl intermediate stage, resulting in a covalent p35–caspase complex in which the cleavage residue Asp87 of p35 links up with the catalytic cysteine of the caspase (Riedl et al., 2001b). We examined whether DIAP2 similarly traps the active caspase via a stable thio acyl linkage. To test this, we assessed the sensitivity of the DIAP2–drICE complex to treatment with hydroxylamine (NH2OH), a strong nucleophile often used to break ester bonds that is known to abrogate p35-mediated caspase-3 inhibition (Riedl et al., 2001b). Thus, if the DIAP2–drICE complex is indeed stabilized by a covalent thioacyl bond between Asp100 and the caspase's catalytic cysteine, it should not form in the presence of NH2OH or DTT. Accordingly, DIAP2 completely failed to bind to drICE in the presence of NH2OH (Fig. 5 A) or DTT (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200706027/DC1). Under the same conditions, formation of the p35–drICE complex was weakened but not fully abrogated. In contrast, NH2OH did not affect the interaction of XIAP to caspase-3 (Fig. 5 B) nor did it abrogate DIAP1's ability to bind drICE (not depicted). Importantly, NH2OH also did not disrupt DIAP2 binding in general because DIAP2 readily interacted with Rpr in the presence of NH2OH (Fig. 5 C). Furthermore, NH2OH and DTT did not interfere with the catalytic activity of drICE because both DIAP2 and p35 were processed in the presence of NH2OH.
Figure 5.
DIAP2 and active drICE form a stable complex that is sensitive to nucleophile attack. (A) The interaction between DIAP2 and drICE is abrogated by exposure to 1 M NH2OH. (B) NH2OH exposure had no effect on the binding of XIAP with caspase-3, whereas it weakened the p35–caspase-3 interaction. (C) NH2OH did not affect the binding of Rpr to DIAP2 or 1. (D) DIAP2's interaction with drICEC211A is insensitive to NH2OH treatment. The binding assay was performed as described in Fig. 2 except that NH2OH was added together with cell extracts containing either active drICE or caspase-3.
If DIAP2 traps the active caspase via a stable thioacyl linkage between Asp100 and the catalytic cysteine of the enzyme, such linkage should not form with a processed catalytic mutant of drICE. Thus, the observed interaction between DIAP2 and processed drICEC211A (Fig. S3) should be resistant to NH2OH because drICEC211A lacks the catalytic thiol (Cys211). Accordingly, we found that NH2OH treatment did not interfere with the binding of DIAP2 to drICEC211A, whereas it abrogated drICEWT binding (Fig. 5 D). Note, the catalytic Cys is not involved in positioning the scissile peptide bond across the catalytic apparatus. Residues of the substrate-binding pocket other than the catalytically active Cys mediate substrate binding (Timmer and Salvesen, 2007).
Collectively, our data are consistent with the notion that DIAP2 and WT drICE form a stable complex that critically depends on the P1 residue (Asp100) of DIAP2. Moreover, the stability of this complex is acutely sensitive to strong nucleophiles such as NH2OH or DTT, which is diagnostic of a stable thioester or disulfide adduct (Riedl et al., 2001b) between DIAP2 and drICE.
DIAP2 ubiquitylates drICE in a RING finger–dependent manner
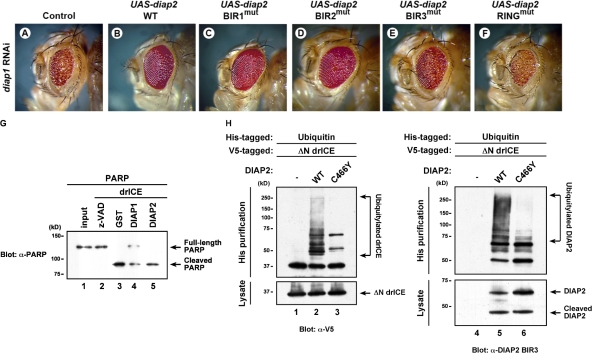
To test the contribution of individual BIR and RING finger domains in regulating cell death in vivo, we examined their ability to block diap1 RNAi–mediated cell death. RNAi-mediated depletion of DIAP1 triggers caspase-induced cell death, which is predominantly executed by drICE and occurs independently of the action of IAP antagonists (Leulier et al., 2006b). Expression of WT DIAP2 or DIAP2 mutants that carry single point mutations in either the BIR1 or 2 efficiently suppressed the eye phenotype caused by diap1 RNAi (Fig. 6; Leulier et al., 2006b). In contrast, a BIR3 mutant DIAP2 failed to rescue this phenotype, indicating that the BIR3 domain of DIAP2 is essential for apoptosis inhibition. This is consistent with our finding that the BIR3, but not the BIR1 and 2, is indispensable for drICE binding. In addition, the RING finger domain of DIAP2 was also required, as a DIAP2 RING mutant failed to suppress the diap1 RNAi phenotype (Fig. 6 F). This suggests that DIAP2 relies on its E3 ubiquitin–protein ligase activity to block cell death. This notion is further supported by the observation that, in vitro, DIAP2 is unable to block drICE-mediated poly ADP ribose polymerase (PARP) cleavage (Fig. 6 G). Consistent with the involvement of the RING, we found that DIAP2 robustly ubiquitylates drICE in a RING finger–dependent manner (Fig. 6 H). Moreover, a BIR3 mutant (C249G) of DIAP2, which fails to bind drICE, also failed to ubiquitylate drICE (unpublished data). Collectively, these results suggest that DIAP2 blocks diap1 RNAi–mediated cell death by ubiquitylating drICE.
Figure 6.
The RING finger domain of DIAP2 is required for suppression of cell death induced by diap1 RNAi. The eye phenotype caused by DIAP1 depletion (A) is rescued by expression of WT DIAP2 (B) and BIR1 (C) and BIR2 (D) mutants of DIAP2. Mutation in the BIR3 (E) or RING finger domain (F) failed to rescue this phenotype. Phenotypes were fully penetrant and representative fly heads are shown. (G) Recombinant DIAP2 is unable to block drICE-mediated PARP cleavage in vitro, as assessed by immunoblot analysis using an anti-PARP antibody. (H) DIAP2 ubiquitylates drICE in a RING-dependent manner. His-ubiquitin and ΔN-drICE-V5 were coexpressed in S2-I cells with the indicated constructs. Ubiquitylated proteins were purified and subject to Western blot analysis using the indicated antibodies. Genotypes used: w+;GMR-GAL4,UAS-diap1-IR/+ (A), w+;GMR-GAL4,UAS-diap1-IR/+;UAS-diap2WT/+ (B), w+;GMR-GAL4,UAS-diap1-IR/UAS-diap2C46G (BIR1mut) (C), w+;GMR-GAL4,UAS-diap1-IR/UAS-diap2C149G (BIR2mut) (D), w+;GMR-GAL4,UAS-diap1-IR/+; UAS-diap2C249G (BIR3mut)/+ (E), and w+;GMR-GAL4,UAS-diap1-IR/+;UAS-diap2C466Y (RINGmut)/+ (F).
IAP antagonists display differential binding to specific BIR domains of DIAP2
The IAP antagonists Rpr, Grim, and Hid interact with DIAP1 and 2 (Vucic et al., 1997; Vucic et al., 1998; Leulier et al., 2006b). When comparing the efficiency with which Rpr and Hid associate with both these IAPs, we found that DIAP1 and 2 copurified comparable amounts of Rpr and Hid (Fig. 7 A; Leulier et al., 2006b), which suggests that Rpr and Hid target both IAPs equally. Rpr and Grim preferentially bound to the BIR2 domain of DIAP2 because mutation of this domain abrogated DIAP2 association, whereas mutation of the BIR1 or 3 had no effect (Figs. 7 B and S5, available at http://www.jcb.org/cgi/content/full/jcb.200706027/DC1). In contrast, Hid binding appeared to be mediated by both BIR2 and 3 domains, as mutation of both these domains was required to disrupt Hid binding (Fig. 7 C).
Figure 7.
IAP antagonists thwart DIAP2 function. (A) DIAP2 and 1 bind to Rpr with comparable efficiencies. (B) The BIR2 domain is required for Rpr binding. The indicated constructs were coexpressed in D. melanogaster S2/p35 cells. (C) Hid binds to the BIR2 and 3 of DIAP2. (D) In the presence of Hid, DIAP2 failed to associate with drICE. DIAP2-GST was coexpressed with ΔN-drICE-V5 in D. melanogaster S2/p35 cells in the absence or presence of Hid-V5. (E and F) DIAP2-mediated inhibition of Hid-killing. (E) Hid-based killing assay in S2 cells in the presence or absence of DIAP1 and WT and mutant DIAP2. Shown are the percentages of viable, β-galactosidase–positive cells relative to controls from three independent experiments (mean ± SD). (F) Hid-binding study using mutant DIAP2.
Hid blocks DIAP2 from binding to drICE
Because both Hid and drICE bind to the BIR3 domain of DIAP2, we tested whether Hid interfered with DIAP2's ability to bind drICE. To this end, we coexpressed DIAP2-GST with AKG-drICEC211A in the presence or absence of Hid. AKG-drICEC211A is a catalytic mutant of drICE that was expressed using the ubiquitin fusion technique (Bachmair et al., 1986; Varshavsky, 2000; Tenev et al., 2005), which generates drICE with an unblocked N-terminal IBM through which it interacts with the BIR3 domain of DIAP2 (Fig. 2 B). drICE and Hid were both V5 tagged in this assay for direct comparison of protein expression levels. In the absence of Hid, DIAP2 readily copurified AKG-drICE (ΔN-drICE) from cellular extracts (Fig. 7 D). When Hid was coexpressed, however, DIAP2 did not copurify drICE and instead purified Hid, which demonstrates that Hid outcompetes drICE for free DIAP2. Of note, Hid was unable to displace drICE from a preformed DIAP2–drICE complex (unpublished data). Collectively these data indicate that Hid competes with drICE for DIAP2 binding rather than displacing it.
DIAP2 suppresses Hid killing independently of Hid binding
Overexpression of DIAP2 suppresses cell death induced by ectopic expression of IAP antagonists in the eye and cells in culture (Hay et al., 1995; Kaiser et al., 1998). Next we tested whether DIAP2 suppresses cell death by simply sequestering IAP antagonists away from DIAP1; or, alternatively, whether DIAP2 blocks apoptosis downstream of Hid. Induced expression of Hid triggered apoptosis in 84% of D. melanogaster S2 cells within 24 h, whereas coexpression of DIAP2 or 1 efficiently suppressed Hid killing (Fig. 7 E). In contrast, the DIAP2 mutants DIAP2D100E and DIAP2101–498 were severely impaired in their ability to block Hid-induced cell death. Interestingly, these DIAP2 mutants bound to Hid as efficiently as WT DIAP2 (Fig. 7 F). Because both DIAP2 mutants were fully competent in Hid binding but greatly impaired in suppressing Hid killing, these data suggest that Hid binding is not sufficient to block Hid killing.
Discussion
Our data suggest that DIAP2 is a caspase regulator that contributes to the caspase activity threshold. Several lines of evidence support this view. First, loss of diap2 causes an increase in basal levels of caspase activity in vivo. Consistent with this increase in basal levels of caspase activity, diap2 mutant cells are sensitized to sublethal doses of x-ray irradiation, and, in tissue culture cells, depletion of DIAP2 by RNAi sensitizes cells to treatment with chemotherapeutic drugs (Zimmermann et al., 2002). Second, DIAP2 overexpression in the developing fly eye efficiently suppresses cell death triggered by diap1 RNAi. Importantly, diap1 RNAi causes spontaneous and unrestrained caspase activation and cell death (Wang et al., 1999; Goyal et al., 2000; Lisi et al., 2000; Rodriguez et al., 2002) that occurs independently of the action of IAP antagonists (Leulier et al., 2006b). The efficiency with which DIAP2 rescues the diap1 RNAi phenotype is highly reminiscent of that of p35. Both DIAP2 and p35 rescue the diap1 RNAi eye size and pigmentation to an apparent normal morphology but fail to restore the formation of bristles (Leulier et al., 2006b). The notion that DIAP2 expression phenocopies the expression of p35, in the absence of any involvement of IAP antagonists, suggests that DIAP2 can function as a direct caspase inhibitor for a p35-sensitive caspase. Third, DIAP2 physically interacts with the effector caspase drICE and suppresses drICE-mediated cell death. Fourth, endogenous DIAP2 is readily cleaved by caspases, which indicates that DIAP2 encounters caspases in vivo. Moreover, after induction of apoptosis, the timing and extent of DIAP2 cleavage is highly reminiscent to that of DIAP1, which suggests that cleavage of DIAP2 is a relatively early event during apoptosis. Finally, IAP antagonists bind to DIAP1 and 2 with similar efficiencies. Consistent with the view that the caspase inhibitory activity of both IAPs is antagonized by IAP antagonists, we find that Hid blocks DIAP1 (Zachariou et al., 2003) and DIAP2 (this paper) from binding to drICE. Together, our data support a model in which DIAP1 and 2 coordinately control drICE, thereby blocking the amplification of the caspase cascade (Fig. 8 A). Accordingly, DIAP2 contributes to the apoptotic threshold by regulating drICE. However, DIAP1 controls initiation of apoptosis, as it is the sole IAP that regulates the initiator caspase Dronc and, with it, the formation of the Dronc/Dark apoptosome. Ultimately, programmed cell death is induced when IAP antagonists such as Hid synchronously bind to DIAP1 and 2, allowing apoptosome formation and unhindered amplification of the caspase cascade.
Although DIAP2 seems to be a bona fide regulator of drICE, its loss does not cause spontaneous apoptosis. This is most likely caused by its epistatic position in the caspase cascade (Fig. 8 A) and its restricted specificity for caspases. Because DIAP2 exclusively regulates drICE and does not bind other caspases (Leulier et al., 2006b; unpublished data), its loss merely causes an increase in caspase activity, presumably because of a small amount of uninhibited active drICE. Under such conditions, drICE activation remains regulated because it requires proteolytic input from Dronc, which, in turn, is under the control of DIAP1. Although loss of DIAP2 results in sublethal levels of active caspases, RNAi-mediated depletion of DIAP1 causes strong, apoptosome-driven activation of downstream effector caspases, which leads to the amplification of the proteolytic signal and death of the cell. Under diap1 RNAi conditions, levels of endogenous DIAP2 seem to be insufficient to suppress the amount of active caspases generated. But, if the levels of DIAP2 are increased, it efficiently blocks diap1 RNAi–mediated cell death. However, when programmed cell death is triggered through the activation of IAP antagonists, both IAPs are targeted equally, thereby releasing the inhibition of the cell death machinery. Thus, controlled activation of caspases and cell survival are ensured by both DIAP1 and 2.
The notion that diap2 mutant animals have increased levels of caspase activity indicates that cells can generate and tolerate a certain amount of activated caspases without undergoing apoptosis. This is consistent with the notion that caspases fulfill important signaling functions independent of executing cell death. For instance, DmIKKɛ activation was recently found to reduce DIAP1 protein levels, thereby allowing activation of sublethal levels of Dronc, which, in turn, is required for the proper development of sensory organ precursor cells (Kuranaga et al., 2006). Thus, modulating caspase activity in a selective, transient, and possibly spatially restricted manner is likely to be a widespread phenomenon that allows caspases to take part in signaling functions without jeopardizing cell viability. Concerted reduction of DIAP1 and 2 protein levels, or their controlled and selective inhibition, may therefore allow controlled activation of caspases and caspase-mediated signaling. The cellular concentration of IAPs would thereby set a cell type–specific threshold for caspase activation and activity. In DIAP2 mutants, however, cells are left in an unbalanced, sensitized state as they accumulate higher than normal levels of active caspases. Accordingly, diap2 mutant third instar larvae are sensitized to apoptosis when subjected to sublethal doses of x-ray irradiation. This sensitized state is only visualized by exposing diap2 mutant animals to sublethal insults. When strong proapoptotic stimuli are used, such as 40 Gy of x-ray irradiation (Huh et al., 2007) or overexpression of Rpr, Hid, and Grim, diap2 mutant animals display the same cell death phenotypes as WT counterparts (Huh et al., 2007; unpublished data). Under these strong proapoptotic conditions, IAP antagonists neutralize both IAPs, causing full activation and amplification of the proteolytic caspase cascade. Thus, the contribution of the sublethal amount of active caspases in diap2 mutants seems to be masked by the higher concentration of active caspases generated under such apoptotic conditions.
Our molecular characterization has uncovered an unexpected mechanism through which DIAP2 restrains the target enzyme drICE. Surprisingly, DIAP2 acts as a pseudosubstrate that, after cleavage, seems to trap the active caspase via a covalent linkage between DIAP2 and the catalytic machinery of drICE. Mutation of the DIAP2 caspase cleavage site abrogates its ability to bind drICE and suppress drICE-mediated cell death. It is unusual for proteins that regulate enzymes to use a mechanism-based strategy, and most types avoid the catalytic machinery by simply blocking the substrate cleft in a lock and key strategy. The cleavage of DIAP2 distinguishes it from all natural inhibitors with the exception of p35, CrmA, serpins, and α-macroglobulins. Serpins and p35 both require cleavage to function as protease inhibitors. p35 arrests proteolysis at the thioacyl intermediate stage, which results in a covalent adduct between p35 and the caspase. In particular, the cleavage site residue Asp87 of p35 links up with the catalytic cysteine of the caspase (Riedl et al., 2001b). Thus, cleaved p35 locks the catalytic machinery of the caspase in a nonproductive inactive configuration. The exquisite sensitivity of the DIAP2–drICE complex to strong nucleophiles such as NH2OH or DTT is diagnostic of a stable thioester adduct (Riedl et al., 2001b) between DIAP2 and drICE. Nevertheless, it remains possible that this complex could form because of a disulfide bridge between DIAP2 and the catalytic thiol of drICE. However, the requirement of DIAP2's Asp100 for drICE binding suggests that Asp100 is involved in forming a thioester covalent adduct with the caspase active site Cys. Ultimately, crystal structure or mass spectrometric analysis will be required to identify the chemical nature of this complex. However, the relatively harsh conditions required for sample preparations for mass spectrometric analysis combined with the labile nature of thiol esters will make it difficult to identify a fragment containing a covalent linkage between DIAP2 and drICE (Riedl et al., 2001b; Xu et al., 2001). Collectively, our data suggest that the DIAP2–drICE complex is stabilized by a covalent linkage (Fig. 8, B and C).
Despite the notion that p35 and DIAP2 both occupy and produce a covalent linkage with the active site of caspases, the modes of interaction and mechanism of inhibition are very different. Unlike p35, DIAP2 requires additional domains for caspase binding and inhibition. First, DIAP2 needs to associate with drICE through a bimodular interaction, whereby the BIR3 domain binds to the IBM of drICE and the 97-Ser-Val-Val-Asp-100 region of DIAP2 occupies the catalytic pocket of the caspase. Each motif on its own is insufficient for caspase binding as mutation of either the BIR3 or cleavage site interferes with complex formation. Second, physical interaction with drICE is not sufficient to block caspase activity. In addition, DIAP2 requires a functional RING finger domain to regulate drICE-mediated cell death. This is evident because a DIAP2 RING finger mutant fails to suppress cell death induced by RNAi-mediated depletion of DIAP1. Moreover, DIAP2 fails to suppress drICE in vitro. In this respect, DIAP2 functions as mechanism-based regulator (relying on the enzyme's catalytic property to trap it) rather than inhibitor. After capture, DIAP2 targets drICE for ubiquitylation (Fig. 8).
An important question is why DIAP2 does not function as an inhibitor like p35 if it also establishes a covalent linkage with the catalytic active site cysteine of the caspase. One possibility is that DIAP2 and p35 differ in their ability to protect the highly labile thioester bond from hydrolysis. This view is supported by the notion that the DIAP2–drICE interaction is significantly more sensitive to nucleophile attack than p35–drICE. Thus, in the situation of DIAP2, the thioester linkage with drICE may simply help to stabilize the caspase interaction, whereas in p35–drICE, it is used to irreversibly inhibit the protease. The more labile nature of the DIAP2–drICE complex may allow regulation, which cannot occur with p35. Moreover, p35 inhibits all effector caspases, whereas DIAP2 only regulates drICE. The observation that the nontarget caspase DCP-1 can also cleave DIAP2 but is not inhibited raises the possibility that DCP-1 controls the level of functional DIAP2 through proteolytic inactivation. Examples of such regulation are manifold and include the proteolytic inactivation of serpins by nontarget proteases (Gettins, 2002). Because DIAP2 tightly interacts with drICE, it is likely to control its apoptotic and nonapoptotic functions.
Materials and methods
Generation of constructs and antibodies
DIAP2 constructs were cloned into pcDNA3 and C-terminally tagged with TAP (provided by B. Seraphin, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France; Rigaut et al., 1999). Point mutants were generated by site-directed mutagenesis (Stratagene) according to the manufacturer's instructions. WT (SVVDS) and mutant (SVVES) DIAP2 cleavage site motifs were N-terminally fused to TAP. For ubiquitylation assays, untagged DIAP2, DIAP2C466Y, or DIAP2C249G were cloned in pAcV5/His. For recombinant assays, DIAP1 and 2 were cloned into pGEX-6P-1 and N-terminally tagged with GST and C-terminally tagged with V5 His, whereas drICE was cloned into pGEX-5X-3 and C-terminally tagged with V5 His. drICE-V5, drICEA29V-V5, ΔN–drICE29-339/C211A-V5, Rpr-V5, Grim-V5, Hid-V5, DEP1-TAP, and anti-drICE and anti-BIR3 DIAP2 antibodies were described previously (Tenev et al., 2005; Leulier et al., 2006b). Constructs were generated by PCR and verified by DNA sequencing. Anti-V5 (AbD Serotec), anti-GST (GE Healthcare), anti-actin (Santa Cruz Biotechnology, Inc.), and biotin-calmodulin (EMD) were used according to the manufacturers' instructions.
Tissue culture and cell death assays
S2, S2-I, HEK293T, and NIH3T3 cells were cultured as described previously (Tenev et al., 2005) and transfected with either calcium phosphate (BD Biosciences), Effectene (QIAGEN), or Cellfectin (Invitrogen). Apoptosis assays were performed as described previously (Tenev et al., 2002). In brief, 0.09 × 106 NIH3T3 cells were transfected with the following DNA ratios: drICE, 26%; DIAP1/DIAP2, 54%; and LacZ, 20%; 1 μg total. Cells were assayed for β-galactosidase (β-Gal) activity after 24 h and cell viability was determined based on morphology of at least 900 β-Gal–positive cells. Live blue cells were scored and expressed as a percentage of total positively transfected blue cells, which was established by treating cells with z-VAD-fmk (BIOMOL International, L.P.). Note, z-VAD-fmk treatment allows normalization of differences in transfection efficiencies. In Fig. 7 (E and F), S2 and S2/p35 cells stably expressing p35 were used. In brief, S2 cells were seeded into a 24-well plate and transfected with the following DNA ratios: Hid, 58%; DIAP1/DIAP2, 40%; and LacZ, 2%; 0.5 μg total. 24 h after transfection, cells were assayed for β-Gal activity.
Ubiquitylation assay
S2 cells with constitutively knocked-down endogenous DIAP2 (S2-I cells) were used. S2-I cells were derived from S2 cells by constitutively expressing diap2-IR, which consists of diap2's 3′ untranslated region sequence separated by the intronic sequence of the Ret gene (Pili-Floury et al., 2004). S2-I cells were cotransfected with His-ubiquitin and ΔN-drICE-V5 together with either vector control, WT DIAP2, DIAP2C466Y, or DIAP2C249G. The following DNA ratios were used: ubiquitin, 60%; drICE, 20%; and DIAP2, 20%; 2.5 μg total. Cells were harvested and washed once in PBS. 10% of the cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and 1 mM EDTA lysis buffer and extracts, whereas the rest was lysed in guanidinium lysis buffer I (6 M guanidinium-HCl, 100 mM NaH2PO4, 10 mM Tris-HCl, pH 8, 5 mM imidazole, and 10 mM β-mercaptoethanol). Ubiquitylated proteins were purified using Ni-NTA agarose beads (QIAGEN) as described previously (Rodriguez et al., 1999) and eluted using 200 mM imidazole, 150 mM Tris, pH 6.8, 30% glycerol, 720 mM β-mercaptoethanol, and 5% SDS.
Chemicals and UV treatment
S2 cells were UV irradiated at 150 kμJ and lysed at the indicated time points. z-VAD-fmk was used at a final concentration of 40 μM. In recombinant assays, a final concentration of 50 μM was used. 1M NH2OH and 16 mM DTT (Sigma-Aldrich) was added to DIAP2 together with drICE- expressing lysate.
Cleavage assay
TAP-purified DIAP2 was incubated with lysates of 293T cells expressing drICE or drICEA29V. The in vitro cleavage reaction was performed in 10 mM Tris, pH 7.5, 150 mM NaCl, 2 mM DTT, and 0.1% Triton X-100 at room temperature for the indicated time points.
Immunoprecipitation and immunoblot analysis
Immunoprecipitation assays were performed as described previously (Tenev et al., 2005). In brief, cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and 1 mM EDTA and extracts were spun at 14,000 rpm for 10 min at 4°C. GST- or TAP-tagged proteins were purified using glutathione- or IgG-coupled Sepharose 4B beads (GE Healthcare), respectively. After 1-h incubation, beads were washed three times in 10 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and 5% glycerol. To assess DIAP2 binding, purified and immobilized DIAP2 was exposed to extracts from 293T cells expressing drICE. Processed active-site mutant drICEC211A was generated by incubating drICEC211A-expressing cells with 50 μM MG132 (Sigma-Aldrich). DIAP2-bound samples were washed and examined by immunoblot analysis using either chemiluminescence (GE Healthcare) or Odyssey technology (LI-COR Biosciences). GST alone or GST-tagged DIAP1 or 2 were coexpressed with RprV5 in D. melanogaster S2/p35 cells.
In vitro assays
Recombinant active drICE was purified from bacterial lysates using Ni-NTA agarose beads (QIAGEN) according to the manufacturer's instructions. Recombinant DIAP1 and 2 were obtained by sequential use of Ni-NTA agarose (His purification) and glutathione-Sepharose beads (GST purification). For drICE-mediated PARP cleavage, reactions were performed with 4 μg of recombinant PARP (R&D Systems), 0.5 μg of DIAP1, DIAP2, or GST, and 160 ng of in vitro purified active drICE. PARP cleavage reactions were performed at room temperature in 100 μl of in vitro cleavage buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 5% glycerol, and 1 mM DTT). Note that drICE was incubated for 30 min with IAPs before the addition of PARP for 30 min. For drICE-mediated cleavage of IAPs, 25 ng of in vitro purified active drICE were incubated with 0.25 μg of recombinant GST, DIAP1, or DIAP2. Reactions were performed in 50 μl of in vitro cleavage buffer for 1 h at room temperature. In both assays, z-VAD-fmk was used in a final concentration of 50 μM.
Fly strains, crosses, and x-ray irradiation
diap2 mutant flies were described previously (Leulier et al., 2006a). Crosses were performed at 25°C. Third instar larvae were x-ray irradiated with 1.5 Gy. After 4 h of recovery, wing imaginal discs were isolated and stained with AO (Abrams et al., 1993). Quantification of AO-stained cells was performed with Photoshop CS2 (Adobe) with a threshold setting of 100 (scoring only strongly stained cells). Untagged DIAP2, DIAP2C46G, DIAP2C149G, DIAP2C249G, and DIAP2C466Y were cloned into pUAST and transgenic flies were generated (Leulier et al., 2006a). The diap2 rescue construct was generated by cloning a WT genomic fragment of 3.3 kb covering the diap2 locus in pCasper4.
Image acquisition
Images of adult heads were acquired using a stereo microscope (MZ APO; Leica) equipped with a Plan Apo 1.0× lens using a digital camera (D1; Nikon) and software (Capture; Nikon). Images of AO-stained wing discs were taken using an Axiovert 100 (Carl Zeiss, Inc.) with a 20×/0.3 lens using the aforementioned mentioned digital camera and software. Quantification of AO-positive cells was done using Photoshop CS2 software by applying a threshold filter of 100 to all images collected, allowing the scoring of only strongly stained cells. All images were taken at room temperature and processed with Photoshop CS2 software. Images were linearly rescaled to optimize brightness and contrast uniformly without altering, masking, or eliminating data. Confocal FRET images were acquired on the Aquacosmos/Ashura system (Hamamatsu Photonics) using an IX-71 equipped with a UPlan Apo10×/040 NA objective (Olympus), a spinning disk-type confocal unit (CSU10; Yokogawa), a diode-pumped solid-state laser (430 nm; Melles Griot), and a three charge-coupled device color camera (C7780-22; Hamamatsu Photonics). Image acquisition and analysis were performed using Aquacosmos/Ashura software.
FRET assay
The confocal imaging analysis of FRET-based caspase activation was performed as described previously (Takemoto et al., 2003). FRET ratio data were collected from >10 flies. After the acquisition of each image (intensity of CFP-CFP and CFP-Venus), the background signal was subtracted. The ratio image (Venus intensity/CFP intensity) was made for each individual tissue and the average ratio was calculated in all pixels of the area in which SCAT3 was expressed.
Online supplemental material
Fig. S1 shows additional experiments for DIAP2–drICE interaction. Fig. S2 shows kinetics studies of DIAP2 cleavage. Fig. S3 shows interaction of DIAP2 with drICEC211A. Fig. S4 shows the influence of DTT in DIAP2–drICE binding. Fig. S5 shows the interactions of DIAP2 with Rpr and Grim. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706027/DC1.
Supplemental Material
Acknowledgments
We would like to thank B. Seraphin for reagents. We are grateful to A. Tonoki, M. Ditzel, and M. Gyrd-Hansen for technical support. We thank members of the Meier laboratory for critical reading of the manuscript and helpful discussions.
P.S. Ribeiro is supported by a PhD fellowship (SFRH/BD/15219/2004) from the Fundação para a Ciência e Tecnologia and F. Leulier is supported by a Human Frontier Science Program fellowship (LT-0177/2004). E. Kuranaga and M. Miura are supported in part by grants from the Japanese Ministry of Education, Science, Sports, Culture and Technology (19109003, 17025010, 19040006, 19041026, and 19044013) and the Takeda Science Foundation.
Abbreviations used in this paper: AO, acridine orange; BIR, baculovirus IAP repeat; DIAP2, Drosophila melanogaster IAP 2; FRET, fluorescence resonance energy transfer; Hid, head involution defective; IAP, inhibitor of apoptosis protein; IBM, IAP-binding motif; PARP, poly ADP ribose polymerase; Rpr, reaper; TAP, tandem affinity purification; WT, wild type; XIAP, x-linked IAP.
References
- Abdelwahid, E., T. Yokokura, R.J. Krieser, S. Balasundaram, W.H. Fowle, and K. White. 2007. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell. 12:793–806. [DOI] [PubMed] [Google Scholar]
- Abrams, J.M., K. White, L.I. Fessler, and H. Steller. 1993. Programmed cell death during Drosophila embryogenesis. Development. 117:29–43. [DOI] [PubMed] [Google Scholar]
- Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 234:179–186. [DOI] [PubMed] [Google Scholar]
- Chai, J., E. Shiozaki, S.M. Srinivasula, Q. Wu, P. Datta, E.S. Alnemri, Y. Shi, and P. Dataa. 2001. Structural basis of caspase-7 inhibition by XIAP. Cell. 104:769–780. [DOI] [PubMed] [Google Scholar]
- Ditzel, M., R. Wilson, T. Tenev, A. Zachariou, A. Paul, E. Deas, and P. Meier. 2003. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 5:467–473. [DOI] [PubMed] [Google Scholar]
- Doumanis, J., L. Quinn, H. Richardson, and S. Kumar. 2001. STRICA, a novel Drosophila melanogaster caspase with an unusual serine/threonine-rich prodomain, interacts with DIAP1 and DIAP2. Cell Death Differ. 8:387–394. [DOI] [PubMed] [Google Scholar]
- Eckelman, B.P., G.S. Salvesen, and F.L. Scott. 2006. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 7:988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesellchen, V., D. Kuttenkeuler, M. Steckel, N. Pelte, and M. Boutros. 2005. An RNA interference screen identifies inhibitor of apoptosis protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 6:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettins, P.G. 2002. Serpin structure, mechanism, and function. Chem. Rev. 102:4751–4804. [DOI] [PubMed] [Google Scholar]
- Goyal, L., K. McCall, J. Agapite, E. Hartwieg, and H. Steller. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, B.A., D.A. Wassarman, and G.M. Rubin. 1995. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 83:1253–1262. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Y.C. Park, R.L. Rich, D. Segal, D.G. Myszka, and H. Wu. 2001. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell. 104:781–790. [PubMed] [Google Scholar]
- Huh, J.R., I. Foe, I. Muro, C.H. Chen, J.H. Seol, S.J. Yoo, M. Guo, J.M. Park, and B.A. Hay. 2007. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J. Biol. Chem. 282:2056–2068. [DOI] [PubMed] [Google Scholar]
- Kaiser, W.J., D. Vucic, and L.K. Miller. 1998. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 440:243–248. [DOI] [PubMed] [Google Scholar]
- Kleino, A., S. Valanne, J. Ulvila, J. Kallio, H. Myllymäki, H. Enwald, S. Stöven, M. Poidevin, R. Ueda, D. Hultmark, et al. 2005. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 24:3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga, E., H. Kanuka, A. Tonoki, K. Takemoto, T. Tomioka, M. Kobayashi, S. Hayashi, and M. Miura. 2006. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 126:583–596. [DOI] [PubMed] [Google Scholar]
- Lamkanfi, M., N. Festjens, W. Declercq, T. Vanden Berghe, and P. Vandenabeele. 2007. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 14:44–55. [DOI] [PubMed] [Google Scholar]
- Leulier, F., N. Lhocine, B. Lemaitre, and P. Meier. 2006. a. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol. Cell. Biol. 26:7821–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier, F., P.S. Ribeiro, E. Palmer, T. Tenev, K. Takahashi, D. Robertson, A. Zachariou, F. Pichaud, R. Ueda, and P. Meier. 2006. b. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 13:1663–1674. [DOI] [PubMed] [Google Scholar]
- Lisi, S., I. Mazzon, and K. White. 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 154:669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro, I., J.C. Means, and R.J. Clem. 2005. Cleavage of the apoptosis inhibitor DIAP1 by the apical caspase DRONC in both normal and apoptotic Drosophila cells. J. Biol. Chem. 280:18683–18688. [DOI] [PubMed] [Google Scholar]
- Pili-Floury, S., F. Leulier, K. Takahashi, K. Saigo, E. Samain, R. Ueda, and B. Lemaitre. 2004. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 279:12848–12853. [DOI] [PubMed] [Google Scholar]
- Riedl, S.J., M. Renatus, R. Schwarzenbacher, Q. Zhou, C. Sun, S.W. Fesik, R.C. Liddington, and G.S. Salvesen. 2001. a. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 104:791–800. [DOI] [PubMed] [Google Scholar]
- Riedl, S.J., M. Renatus, S.J. Snipas, and G.S. Salvesen. 2001. b. Mechanism-based inactivation of caspases by the apoptotic suppressor p35. Biochemistry. 40:13274–13280. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A., P. Chen, H. Oliver, and J.M. Abrams. 2002. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J. 21:2189–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, M.S., J.M. Desterro, S. Lain, C.A. Midgley, D.P. Lane, and R.T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo, H.D., A. Bergmann, H. Gonen, A. Ciechanover, and H. Steller. 2002. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4:432–438. [DOI] [PubMed] [Google Scholar]
- Salvesen, G.S., and J.M. Abrams. 2004. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 23:2774–2784. [DOI] [PubMed] [Google Scholar]
- Salvesen, G.S., and C.S. Duckett. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3:401–410. [DOI] [PubMed] [Google Scholar]
- Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 9:459–470. [DOI] [PubMed] [Google Scholar]
- Silke, J., P.G. Ekert, C.L. Day, C.J. Hawkins, M. Baca, J. Chew, M. Pakusch, A.M. Verhagen, and D.L. Vaux. 2001. Direct inhibition of caspase 3 is dispensable for the anti-apoptotic activity of XIAP. EMBO J. 20:3114–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z., B. Guan, A. Bergman, D.W. Nicholson, N.A. Thornberry, E.P. Peterson, and H. Steller. 2000. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol. Cell. Biol. 20:2907–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke, H.R., and G.S. Salvesen. 2006. Chemical ligation–an unusual paradigm in protease inhibition. Mol. Cell. 21:727–728. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Y. Nakabayashi, K. Nakata, J.C. Reed, and R. Takahashi. 2001. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes. J. Biol. Chem. 276:27058–27063. [DOI] [PubMed] [Google Scholar]
- Takemoto, K., T. Nagai, A. Miyawaki, and M. Miura. 2003. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160:235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, K., E. Kuranaga, A. Tonoki, T. Nagai, A. Miyawaki, and M. Miura. 2007. Local initiation of caspase activation in Drosophila salivary gland programmed cell death in vivo. Proc. Natl. Acad. Sci. USA. 104:13367–13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev, T., A. Zachariou, R. Wilson, A. Paul, and P. Meier. 2002. Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 21:5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev, T., A. Zachariou, R. Wilson, M. Ditzel, and P. Meier. 2005. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat. Cell Biol. 7:70–77. [DOI] [PubMed] [Google Scholar]
- Timmer, J.C., and G.S. Salvesen. 2007. Caspase substrates. Cell Death Differ. 14:66–72. [DOI] [PubMed] [Google Scholar]
- Varshavsky, A. 2000. Ubiquitin fusion technique and its descendants. Methods Enzymol. 327:578–593. [DOI] [PubMed] [Google Scholar]
- Vucic, D., W.J. Kaiser, A.J. Harvey, and L.K. Miller. 1997. Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl. Acad. Sci. USA. 94:10183–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic, D., W.J. Kaiser, and L.K. Miller. 1998. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol. Cell. Biol. 18:3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.L., C.J. Hawkins, S.J. Yoo, H.A. Müller, and B.A. Hay. 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 98:453–463. [DOI] [PubMed] [Google Scholar]
- White, K., M.E. Grether, J.M. Abrams, L. Young, K. Farrell, and H. Steller. 1994. Genetic control of programmed cell death in Drosophila. Science. 264:677–683. [DOI] [PubMed] [Google Scholar]
- Wilson, R., L. Goyal, M. Ditzel, A. Zachariou, D.A. Baker, J. Agapite, H. Steller, and P. Meier. 2002. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4:445–450. [DOI] [PubMed] [Google Scholar]
- Wu, G., J. Chai, T.L. Suber, J.W. Wu, C. Du, X. Wang, and Y. Shi. 2000. Structural basis of IAP recognition by Smac/DIABLO. Nature. 408:1008–1012. [DOI] [PubMed] [Google Scholar]
- Wu, J.W., A.E. Cocina, J. Chai, B.A. Hay, and Y. Shi. 2001. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell. 8:95–104. [DOI] [PubMed] [Google Scholar]
- Xu, G., M. Cirilli, Y. Huang, R.L. Rich, D.G. Myszka, and H. Wu. 2001. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 410:494–497. [DOI] [PubMed] [Google Scholar]
- Yan, N., J.W. Wu, J. Chai, W. Li, and Y. Shi. 2004. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat. Struct. Mol. Biol. 11:420–428. [DOI] [PubMed] [Google Scholar]
- Yoo, S.J., J.R. Huh, I. Muro, H. Yu, L. Wang, S.L. Wang, R.M. Feldman, R.J. Clem, H.A. Müller, and B.A. Hay. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4:416–424. [DOI] [PubMed] [Google Scholar]
- Zachariou, A., T. Tenev, L. Goyal, J. Agapite, H. Steller, and P. Meier. 2003. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 22:6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, K.C., J.E. Ricci, N.M. Droin, and D.R. Green. 2002. The role of ARK in stress-induced apoptosis in Drosophila cells. J. Cell Biol. 156:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.