Abstract
We have used reverse-transcription-polymerase chain reaction (RT-PCR) and DNA sequencing techniques to confirm the transcription of seven (six α and one non-α) novel candidate nicotinic acetylcholine receptor (nAChR) subunit-encoding genes identified in the genome sequence of the nematode Caenorhabditis elegans. Compared to vertebrate nAChR subunits, they most closely resemble the homomer-forming, neuronal α7 subunit. Comparison of the predicted amino acid sequences of the new nAChR subunits with those described previously in C. elegans reveals five subunits (four α and one non-α) which resemble the DEG-3-like group of subunits. To date, this highly divergent nAChR subunit group is unique to C. elegans. ACR-22 is the first non-α member of the DEG-3-like group of subunits to be identified. Two new members of the related ACR-16-like nAChR group of subunits have also been shown to be transcribed, making the ACR-16-like subunit group the largest in C. elegans. Residues in the α subunit second transmembrane region (M2) which contribute to the channel lining show variations with implications for channel function. For example, in ACR-22, the highly conserved 0` lysine of M2 is replaced by histidine. Restrained molecular dynamics simulations have been used to generate molecular models of homo-pentameric M2 helix bundles for the novel subunits, enabling identification and display of pore-lining and protein interface residues. The number and diversity of genes encoding C. elegans nAChR subunits with similarities to the homomer-forming vertebrate α7 subunits and the identification of related non-α subunits, only found in C. elegans to date, suggest that at least some of these subunits may contribute to heteromers in vivo.
Keywords: Nicotinic acetylcholine receptors, Caenorhabditis elegans, ion channel, molecular modeling, α7 subunit
Nicotinic acetylcholine receptors (nAChRs) are pentameric membrane proteins which mediate fast synaptic transmission at neuromuscular junctions and in the nervous system of many organisms (Changeux and Edelstein 1998). The vertebrate nAChR gene family is diverse, with separate sets of genes encoding the currently known 5 muscle and 12 neuronal subunit isoforms. Subunits of nAChRs fall into two main categories: α subunits are defined by adjacent cysteines which contribute to the ACh binding site; non-α subunits lack this motif (Changeux and Edelstein 1998). Most vertebrate nAChR isoforms are expressed in the nervous system, and among these the homomer-forming α7–9 subunits are the most divergent.
Caenorhabditis elegans provided the first viable mutants resistant to the cholinergic anthelmintic compound, levamisole (Lewis et al. 1980). Subsequent molecular and genetic analyses revealed that several of the levamisole resistance mutations were in genes encoding structural components of heteromeric nAChRs (Fleming et al. 1997). Other C. elegans nAChR subunits have been identified by cross-species hybridization [acr-2 (Squire et al. 1995), acr-16 (previously referred to as Ce21) (Ballivet et al. 1996), and acr-3 (Baylis et al. 1997)] and genetic screens [deg-3 (Treinin and Chalfie 1995) and des-2 (Treinin et al. 1998)]. Also, 8 novel nAChR α subunits were identified by Mongan et al. (1998) from the genome sequence of C. elegans (The C. elegans Sequencing Consortium, 1998), making a total of 13 α subunits, by far the largest number of nAChR α subunits reported for any organism.
ACR-16, a nematode homolog of α7 (Ballivet et al. 1996), like vertebrate α7 subunits (Couturier et al. 1990), can form functional homomeric receptors when expressed in Xenopus laevis oocytes. The amino acid sequence of the C. elegans homomer-forming subunit ACR-16 is 47% identical to chicken α7, and aspects of the pharmacology of these recombinant nematode and vertebrate receptors have been compared (Ballivet et al. 1996; Raymond et al. 2000). Four additional α subunits and two non-α subunits most similar to ACR-16 have been identified in C. elegans (Mongan et al. 1998). Thus, although ACR-16 is capable of forming a homomeric nAChR when expressed in Xenopus oocytes (Ballivet et al. 1996), heteromeric receptors containing other ACR-16-like subunits cannot be ruled out in vivo.
A second group of C. elegans nAChR subunits, distinct from the ACR-16 group, with most similarity to the α7–9 vertebrate neuronal nAChR subunits (Mongan et al. 1998), has also been found in C. elegans. The first member identified was DEG-3 (Treinin and Chalfie 1995). Two additional DEG-3-like subunits have been identified, ACR-5 (Mongan et al. 1998) and DES-2 (Treinin et al. 1998). The DEG-3-like subunits (Treinin and Chalfie 1995; Mongan et al. 1998) are unique so far to C. elegans. It was shown that DEG-3 and DES-2 can form functional heteromeric nAChRs when coexpressed in Xenopus oocytes (Treinin et al. 1998). However, physiological and genetic studies of these receptor subunit-encoding genes suggest that other subunits participate in DEG-3-containing channels in vivo (Treinin et al. 1998).
Here we report the transcription of five novel DEG-3-like nAChR subunits. This includes the first report of a non-α nAChR subunit relative of DEG-3. The transcription of two additional ACR-16-like nAChR subunits is also demonstrated, making ACR-16-like subunits the largest nAChR subunit group identified to date in any organism. Initial clues to aspects of functional diversity in this large nAChR gene family have been sought using molecular modeling of the second transmembrane domain (M2), a region known to influence channel selectivity and gating (Wilson et al. 2000). Of particular interest is the finding that the residues which are oriented away from the channel lumen and therefore interact directly with other transmembrane domains show much greater variability than those residues exposed to the pore.
Results
RT-PCR demonstration of nAChR subunit-gene transcription
The C. elegans genome sequence databases were screened with a range of nAChR protein sequences using the BLAST algorithm. Genes encoding nAChR α subunits were identified among these sequences by the presence of the vicinal cysteine motif corresponding to residues 192 and 193 of the Torpedo α subunit (Noda et al. 1982). RT-PCR was used to confirm the transcription of six novel nAChR α subunits and one putative non-α subunit (Fig. 1 ▶). The partial cDNA products were gel-purified and sequenced to confirm their identity. All fall into either the ACR-16 or DEG-3-like nAChR subunit groups (Fig. 2 ▶, Table 1). ACR-22 is most similar to the DEG-3-like α subunits but lacks the vicinal cysteine motif found in α subunits and must therefore be designated a non-α subunit. Together with the 19 subunits previously reported (Treinin and Chalfie 1995; Ballivet et al. 1996; Fleming et al. 1997; Mongan et al. 1998), this is by some margin the largest known family of nAChR subunits (20 α and 7 non-α subunits).
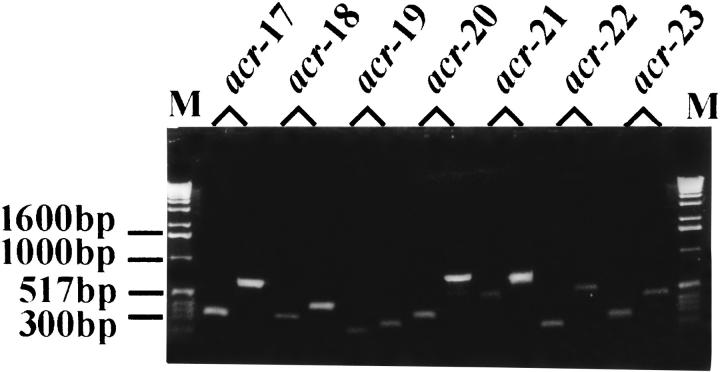
Fig. 1.
RT-PCR confirms transcription of novel nAChR subunits. PCR products are shown after electrophoresis on a 1.3% agarose-TBE gel. The gel image shows pairs of adjacent cDNA and genomic PCR products corresponding to the following nAChR subunits: acr17 (F53E10.2); acr-18 (F28F8.1); acr<-19 (C31H5.3); acr-20 (R06A4); acr-21 (F27B3); acr-22 (F48E3.7), and acr-23 (F59B1.9).
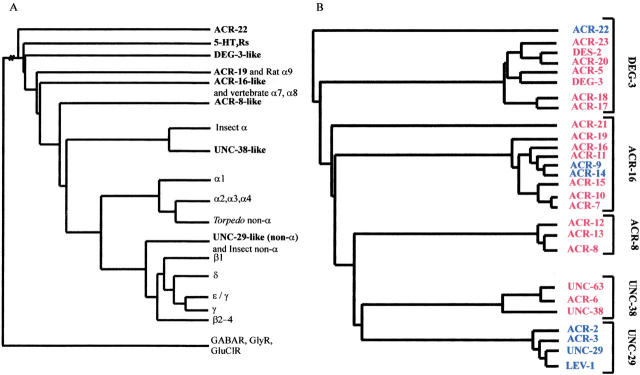
Fig. 2.
(A) An alignment of all nAChR subunit sequences available in public databases and a selection of anionic selective GABAR, GlyR, and GluClR was generated using default parameters of ClustalW (Thompson et al. 1994). The anionic channels were selected as the out-group to root the tree. The global tree was constructed using the Protdist (Dayhoff PAM algorithm) and Neighbor programs of the PHYLIP package. For reasons of clarity, related groups of vertebrate, insect, and C. elegans subunits groups are illustrated by single branches in A. The insect branch includes the two available Myzus persicae sequences (X81887, X81888) in addition to the Manduca sexta (Y09795), Locusta migratoria (AJ000390, AJ000391, AJ000393), Heliothis virescens (AJ000399) and Drosophila melanogaster ALS (X07194), SBD (X55676) and Dα3 (Y15594) subunits. UNC-38-like homologs have been isolated from the parasitic nematodes Ascaris suum (AJ011382), Trichostrongylus columbriformis (U56903), Haemonchus contortus (U72490), and a non-α subunit, which is more similar to UNC-38 than other non-α subunits, from Onchocerca volvulus (L20465). (B) This tree shows the C. elegans groups expanded to reveal all C. elegans nAChR subunits. Five distinct groups of C. elegans subunits can be recognized: four groups of mainly α subunits; DEG-3-like, ACR-16-like, UNC-38-like, and ACR-8-like nAChR α subunits, in addition to the UNC-29-like group containing only non-α subunits. The alignment and global tree used to generate this Fig. are available on request from the authors.
Table 1.
Percent similarity and identity between the predicted protein sequences of novel nAChR subunits and key C. elegans subunits
| % Identity of derived protein sequences | |||||||||||||
| DEG-3 | ACR-16 | ACR-8 | UNC-38 | ACR-17 | ACR-18 | ACR-19 | ACR-20 | ACR-21 | ACR-22 | ACR-23 | UNC-63 | UNC-29 | |
| DEG-3 | ∼ | 28 | 22 | 24 | 30 | 25 | 25 | 43 | 33 | 27 | 27 | 25 | 23 |
| ACR-16 | 37 | ∼ | 31 | 37 | 31 | 28 | 40 | 34 | 45 | 23 | 22 | 37 | 44 |
| ACR-8 | 32 | 41 | ∼ | 38 | 24 | 24 | 30 | 28 | 30 | 23 | 40 | 41 | 33 |
| UNC-38 | 34 | 48 | 51 | ∼ | 26 | 21 | 32 | 30 | 39 | 23 | 22 | 52 | 39 |
| ACR-17 | 41 | 41 | 36 | 38 | ∼ | 47 | 27 | 32 | 36 | 26 | 23 | 27 | 24 |
| ACR-18 | 35 | 37 | 34 | 29 | 59 | ∼ | 25 | 30 | 35 | 25 | 24 | 26 | 28 |
| ACR-19 | 35 | 49 | 40 | 41 | 39 | 32 | ∼ | 27 | 41 | 23 | 20 | 34 | 28 |
| ACR-20 | 56 | 43 | 40 | 42 | 42 | 38 | 41 | ∼ | 34 | 28 | 49 | 29 | 29 |
| ACR-21 | 46 | 56 | 43 | 51 | 47 | 47 | 54 | 46 | ∼ | 28 | 26 | 40 | 33 |
| ACR-22 | 40 | 35 | 38 | 35 | 40 | 34 | 33 | 44 | 40 | ∼ | 19 | 25 | 25 |
| ACR-23 | 37 | 35 | 40 | 34 | 30 | 31 | 29 | 57 | 37 | 31 | ∼ | 25 | 20 |
| UNC-63 | 36 | 47 | 53 | 63 | 39 | 36 | 42 | 43 | 53 | 35 | 34 | ∼ | 41 |
| UNC-29 | 34 | 34 | 47 | 50 | 34 | 31 | 40 | 41 | 46 | 35 | 29 | 52 | ∼ |
| % Similarity of derived protein sequences | |||||||||||||
The GAP analysis program (Wisconsin Package, version 9.1, Genetics Computer Group (GCG), Madison, WI) was used to determine the percentage similarity and identity between the predicted protein sequences of novel nAChR subunits and key C. elegans subunits, DEG-3, ACR-16, ACR-8, UNC-38, and UNC-29. Default parameters were used (gap creation penalty of 12 and gap extension penalty of 4).
Extensive DEG-3-like and ACR-16-like groups of C. elegans nAChR subunits
We identified genes encoding four novel α subunits (ACR-17, ACR-18, ACR-20, ACR-23) and one putative non-α subunit (ACR-22) most similar to DEG-3 (Fig. 2 ▶). Unlike the des-2 and deg-3 α subunit genes, which are adjacent on chromosome V and under the control of the same promoter (Treinin et al. 1998), acr-17, acr-18, and acr-23 are widely distributed on the same chromosome, while the genes encoding ACR-20 and ACR-22 are located on chromosomes II and X, respectively.
Two additional α subunit members of the ACR-16-like subunits of C. elegans (ACR-19 and ACR-21) were identified. This is currently the largest nAChR subunit group in C. elegans (nine members total) and includes two non-α subunits, ACR-9 and ACR-14. The genes encoding ACR-19 and ACR-21 are located on chromosomes I and III, respectively. ACR-21 is the most diverged member of this group, and its predicted amino acid sequence derived from analysis of the genome sequence is most similar (55%) to the homomer-forming vertebrate α9 subunit, which is itself the most distant relative of vertebrate α7 nAChRs (Elgoyhen et al. 1994).
Molecular modeling of the channel lining M2 regions
Molecular modeling of homo-pentameric M2 helix bundles derived from the six novel nAChR α subunits revealed that the physicochemical properties of the pore-lining residues were highly conserved between C. elegans and all other nAChR subunits, with the exception of the previously described ACR-8-like subunits (Mongan et al. 1998). The protein interface residues of the DEG-3-like subunits were also investigated (Fig. 3 ▶). There is greater variability in residues predicted to interact with the transmembrane protein environment of these subunits than in residues exposed to the channel lumen. Although understanding of the role of transmembrane residues is increasing (Bouzat and Barrantes 1997), little is known of the physical interactions between the M2 helices and the surrounding transmembrane domains.
Fig. 3.
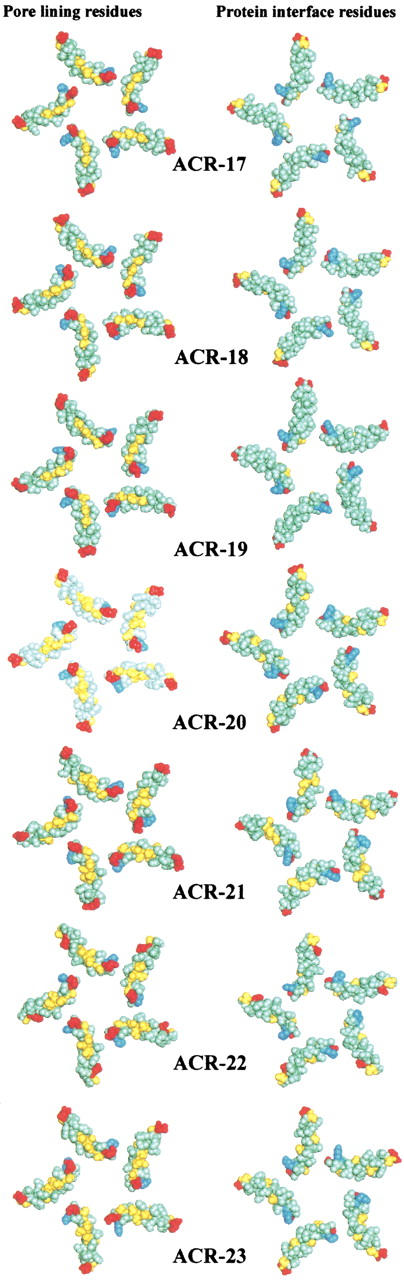
Pore-lining and protein interface residues of the M2 α helix were generated using PORELINING (G.R. Smith and M.S.P. Sansom, unpubl.). The M2 helices are rotated into a plane to form a disk, one surface of which corresponds to the pore-lining surface of the M2 helices (left-hand diagrams) and the other surface of which (right-hand diagrams) corresponds to the protein interface residues. The color code for residues is: gray, apolar; yellow, polar and neutral; red, acidic; blue, basic.
Furthermore, variability in M4 residues, which are oriented away from the pore but which interact with the lipid environment, has recently been shown to strongly influence channel function (Bouzat et al. 2000; Tamamizu et al. 2000). This shows that transmembrane helices not exposed to the channel lumen can also play a role in channel gating. The unexpected variability of residues at the protein interface of the M2 domains of the C. elegans nAChR subunits may give rise to subtle differences in the allosteric transitions of each helix relative to neighboring transmembrane domains, resulting in distinct channel characteristics.
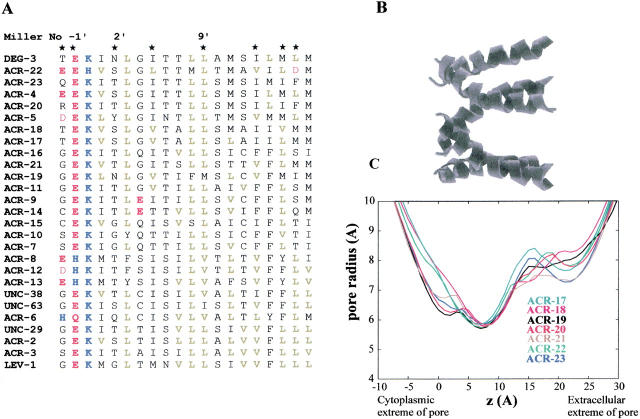
When pore models were compared in terms of their pore radius profiles (Fig. 4 ▶), in each case the narrowest region was close to the 2` residue and had a van der Waals radius of approximately 6Å, corresponding to an effective radius of about 3Å when pore solvation is taken into account (Sankararamakrishnan et al. 1996). The pore diameters of DEG-3 and ACR-19 are smaller relative to the other novel C. elegans nAChR subunits due to the presence of the asparagine residue at the 2` position in place of the more common small and/or polar residues (T, S, or G; Fig. 4 ▶). The 2` residue is believed to form the narrow region of the pore, and substitution of this residue increases or decreases the channel conductance depending on the volume of the side chain of the substituted residue (Villarroel et al. 1991; Villarroel and Sakmann 1992).
Fig. 4.
(A) An alignment of the predicted amino acid sequences of all C. elegans nAChR subunits was calculated using ClustalW. Only the sequence of M2, the second transmembrane region, is illustrated. Amino acid residues are numbered according to Miller (1989) and are colored according to side chain properties: acidic (red), basic (blue), and aliphatic (green). (B) Molecular models of M2 homo-pentameric α-helix bundles were generated by simulated annealing/molecular dynamics using restraints derived from structural and mutagenesis data (Sankararamakrishnan et al. 1996; Adcock et al. 1998). The upper diagram shows an M2 helix bundle, with the peptide backbone shown as helical ribbons. The N-termini of the helices (corresponding to the intracellular mouth of the pore) are shown on the left-hand side of the diagram. (C) Pore radius profiles determined for the M2 α-helix bundle models. The HOLE algorithm (Smart et al. 1996) was used to determine pore radius profiles.
Discussion
This study has identified seven novel candidate nAChR subunit-encoding genes (ACRs-17 to -23) in the nematode C. elegans. They possess features common to nAChR subunits: the di-cysteine loop (Fig. 5 ▶); aromatic residues in loops A, B, C, and D which are important for ligand binding (Fig. 5 ▶; for review, see Corringer et al. 2000); a glutamate residue at the −1` position of M2 (Fig. 4 ▶) which has been shown to be involved in cation selectivity and calcium permeability; and four putative transmembrane regions (Figs. 4, 6 ▶ ▶). Overall, conservation is stronger in the M2 region with the seven novel subunits described in this study, showing 25%–60% identity with the M2 region of the ACR-16 subunit. For the other three transmembrane regions, identities of 21%–53% (M1), 6%–50% (M3), and 11%–32% (M4) were observed. Six of the novel subunits (ACRs-17 to 21 and ACR-23) possess two adjacent cysteine residues in loop C, marking them as α subunits, whereas ACR-22 lacks the vicinal cysteine motif and is therefore designated a non-α subunit. The combined results from this and other laboratories (Treinin and Chalfie 1995; Ballivet et al. 1996; Fleming et al.1997; Mongan et al. 1998) demonstrate an extensive and diverse gene family of nAChR subunits (20 α and 7 non-α subunits), the largest currently known for any organism.
Fig. 5.
An alignment of the predicted amino acid sequences of the N-terminal (extracellular) domains of all C. elegans nAChR subunits was calculated using Clustal W and presented by the GENEDOC program (available from its authors at http://www.cris.com/~ketchup/genedoc.shtml). The conserved di-cysteine loop and the loops contributing to the ACh binding site are shown. Residues are numbered according to the chicken α7 nAChR subunit.
Fig. 6.

An alignment of the predicted amino acid sequences of the M1, M3, and M4 regions of the novel seven C. elegans nAChR subunits was calculated using Clustal W. The corresponding sequences of ACR-16 and DEG-3 are included for comparison. Amino acid residues are colored according to side chain properties: acidic (red), basic (blue), and aliphatic (green). At present there is no M4 sequence for the ACR-17 subunit. Since we have shown that this subunit is transcribed, the predicted protein sequence for ACR-17 in the C. elegans databases is likely to be incomplete.
Compared to vertebrate nAChR sequences, the seven subunits showed greatest homology with homomer-forming subunits, most notably α7. Several nAChR subunits can form functional homomeric receptors when expressed in Xenopus oocytes. These include vertebrate α7 (Couturier et al. 1990), α8 (Schoepfer et al. 1990; Gerzanich et al. 1994), α9 (Elgoyhen et al. 1994), Schistocerca migratoria αL1 (Marshall et al. 1990), and C. elegans ACR-16 (= Ce21) (Ballivet et al. 1996). The chicken α7 (Couturier et al. 1990) and α8 subunits form pharmacologically distinct homomeric nAChRs when expressed in Xenopus oocytes (Gerzanich et al. 1994). However, these recombinant nAChRs differ pharmacologically from native chicken nAChRs containing the α7 subunit (Anand et al. 1993).
There is some evidence that α7 homomers can form functional receptors in vivo (Drisdel and Green 2000). However, other studies have indicated that the α7 subunit may be part of more complex receptors. For instance, the functional properties of α7-containing nAChRs were shown to be dependent on cell type (Cuevas and Berg 1998). In contrast to rat hippocampal α7-nAChRs (Séguéla et al. 1993), the α7-containing nAChRs present on rat intracardiac ganglia desensitize slowly and recover rapidly from 125I-α-bungarotoxin (α-bgt) blockade (Cuevas and Berg 1998). It was speculated that such modification of receptor functional properties could arise from the combination of α7 with as yet unknown nAChR subunits to form heteromeric receptors or from cell-specific or location-specific regulatory interactions. The ability of α7 to coassemble with the β3 subunit when expressed in Xenopus oocytes demonstrates the possibility of the α7 subunit combining with other nAChR subunits to form functional heteromeric receptors (Palma et al. 1999). Indeed, α-bgt binding studies showed that nAChRs composed of both α7 and α8 subunits constitute 17% of 125I-α-bgt-binding protein (Keyser et al. 1993), indicating that at least some native chicken nAChRs containing α7 are heteromers in vivo. Moreover, physiological studies of embryonic chick sympathetic neurons indicated that the α7 subunit contributed to at least three subtypes of native nAChRs, leading to the proposal that the α7 subunit was forming heteromeric channels with other nAChR subunits (Yu and Role 1998). Thus, the subunit makeup of native α7-containing receptors as well as their functional diversity remains to be fully elucidated.
The C. elegans ACR-16-like group is now the largest collection of nAChR subunits resembling the neuronal α7-like homomer-forming subunits of vertebrates found in any organism to date. Our present finding of non-α subunit members of this subunit group (see also Mongan et al. 1998) is therefore of interest. This suggests that although ACR-16 is capable of forming a homomeric nAChR when expressed in Xenopus oocytes (Ballivet et al. 1996), there is the possibility that heteromeric receptors containing other ACR-16-like subunits exist in vivo, as is the case for DEG-3 and DES-2 (Yassin et al. 2001).
To date, the ACR-16 homomer and the DEG-3/DES-2 heteromer are the only subunits of the ACR-16 and DEG-3 groups to have been functionally expressed (Tables 1, 2). Other C. elegans nAChR subunits have been expressed as heteromeric channels in Xenopus oocytes (Table 2). These subunits are members of the UNC-38 and UNC-29 groups (Fig. 2B ▶), which share close homology with vertebrate muscle nAChR subunits (Mongan et al. 1998). It will be of interest to express more ACR-16-like α subunits and see whether they, like ACR-16 and the vertebrate α7 subunits, are able to form functional homomeric channels. In addition, coexpression of ACR-16-like non-α subunits with other members of this group may help determine the influence of α7-like non-α subunits on generating functional diversity. It is worth noting that in ACR-22, the highly conserved 0` lysine of M2 is replaced by histidine (Fig. 4 ▶). Functional studies with this subunit may contribute to our understanding of the effect of variations within the channel lining region on receptor activity. Sequence comparisons revealed that the newly discovered ACR-21, the most divergent ACR-16-like α subunit, is most similar to the α9 subunit. If functional nAChRs containing ACR-21 can be generated, it will be of interest to see if they share the atypical "mixed" pharmacology exhibited by vertebrate α9 homomeric nAChRs (Elgoyhen et al. 1994; Rothlin et al. 1999).
Table 2.
Expression of C. elegans nAChRs
| Receptor | Heterologous expression | Expression in C. elegans | Reference |
| DEG-3/DES-2 | Cation channel preferentially activated by choline | Sensory neurons including nonsynaptic regions | Yassin et al. 2001 |
| ACR-5 | — | DB/VB motor neurons | Esmaeili et al. 2002 |
| ACR-16 | Homomeric cation channel, levamisole is antagonist | — | Ballivet et al. 1996 Raymond et al. 2000 |
| UNC-38/UNC-29/LEV-1 | Cation channel, levamisole is agonist | UNC-29: body and head muscles, central neurophil | Fleming et al. 1997 |
| UNC-63/UNC-29/LEV-1 | Cation channel, levamisole is agonist | UNC-63: body muscles, motor and head ganglia neurons LEV-1: body muscles, motor neurons | E. Culetto and D.B. Sattelle, pers. comm. |
| UNC-38/ACR-2 | Cation channel, levamisole is agonist | — | Squire et al. 1995 |
| UNC-38/ACR-3 | Cation channel, levamisole is agonist | — | Baylis et al. 1997 |
The α subunits are underlined.
To identify pore-lining and protein interface residues in the novel C. elegans α subunits, molecular models of homo-pentameric M2 helix bundles were generated. It is interesting to note that residues oriented away from the channel lumen show much greater variability than those directly exposed to the pore. Most studies have focused on the contribution of the channel lining residues to the functional characteristics of the receptor (for review, see Hucho et al. 1996). However, the finding of such variability in residues which can interact directly with the transmembrane "superstructure" composed of M1, M3, and M4 may reflect a role for these residues in allosteric transitions required for channel gating.
This study adds to our current understanding of nAChR subunit diversity (Le Novère and Changeux 1995; Tsunoyama and Gojobori 1998) and has contributed to the characterization of the largest known family of nAChR subunit genes. Analysis of the predicted amino acid sequence of the subunits has highlighted diversity in residues known to contribute to the channel lining region. The observed molecular diversity among C. elegans nAChR α subunits suggests greater functional diversity in the actions of ACh than previously envisaged.
It has been shown that a mutation in the M2 region of the DEG-3 subunit leads to neuronal degeneration in C. elegans, probably due to increased receptor activity leading to Ca2+-mediated toxicity (Treinin and Chalfie 1995). Interestingly, mice homozygous for a similar mutation in the α7 subunit die within 24 hours of birth and show cell death in neuronal cells, perhaps also due to increased intracellular Ca2+ levels (Orr-Urtreger et al. 2000). In addition, receptors with the DEG-3 and DES-2 subunits have been localized to nonsynaptic regions where they are thought to perform a chemosensory role (Yassin et al. 2001). The α7 subunit is also expressed in non-neuronal cells such as cells of the thymus (Navaneetham et al. 1997) and also in bronchial epithelial cells (Wang et al. 2001). It will be of interest to investigate further the functions of these non-neuronal α7-containing nAChRs. Thus, studies of nAChRs from C. elegans may contribute to our understanding of the function of similar receptors in higher organisms.
A preliminary search of the larger Drosophila genome sequence revealed a smaller nAChR subunit gene family with seven α subunits identified to date and three candidate non-αs (Littleton and Ganetzky 2000; M. Grauso, pers. comm.). Nevertheless, it may be that functional diversity is maintained in Drosophila through alternative processing of nAChR gene transcripts, as is well established in the case of Drosophila GABA receptors (Hosie et al. 1997). In the future it will be of interest to determine whether genomes of higher eukaryotes reveal a similarly complex nAChR gene family.
Materials and methods
Identification of novel nAChR α subunits in databases
The complete genome sequence of C. elegans is available at URL: http://www.sanger.ac.uk and http://genome.wustl.edu/gsc. The C. elegans databases were searched with the following representative nAChR α subunits: Drosophila melanogaster ALS (X07194); Torpedo californica muscle α subunit (J00963); chicken α7 (X68586); DEG-3 (U19746); ACR-16 (X83887); UNC-38 (X98600) and ACR-8 (Z50029) using BLAST (Altschul et al. 1990). Putative nAChR α subunits were identified by the presence of the adjacent cysteine residues in addition to the cysteine-loop which defines members of this superfamily of receptors.
Reverse transcription and polymerase chain reaction (RT-PCR)
Genomic DNA was prepared as described in detail elsewhere (Mongan et al. 1998). Total RNA was isolated from mixed-stage cultures of C. elegans using TRIzol (Life Technologies). Messenger RNA was purified using an Oligotex Direct mRNA isolation kit (QIAGEN) and first-strand cDNA synthesized using Superscript II (Life Technologies). The cDNA was diluted to 100 μL with sterile water, phenol-extracted and ethanol-precipitated. cDNA pellets were resuspended in 100 μL of sterile ultrapure water and stored at −20°C. Subunit-specific oligonucleotide primers were designed to flank the sequence predicted to encode the vicinal Cys motif and M2, and also span an intron/exon boundary. In this way cDNA and genomic PCR products differ in size. The sequences of primers used are as follows:
Subunit-specific primers were employed in PCR reactions using either cDNA or genomic DNA in a 50 μL total reaction volume composed of 1 × PCR buffer supplemented to 2.5 mM MgCl2 (Promega), 0.2 μM each primer, and 0.2 mM dNTP mix (Pharmacia). Either 1 μL first-strand cDNA or 0.1 μg genomic DNA was added as template. Negative control PCRs were carried out without the addition of template DNA for all amplifications. The following conditions were used for 40 cycles in a Perkin Elmer (480 model) thermocycler: 94°C for 5 min, prior to addition of 2.5 units of Taq polymerase (Promega); 94°C for 30 sec, 55°C for 30 sec, 72° for 90 sec. All PCR products were analyzed by electrophoresis in a 1.3% agarose-TBE gel. RT-PCR products were gel purified using a QIAquick gel extraction kit (QIAGEN) and sequenced by the dye termination method using an ABI automated sequencer. The most abundant amplification products were gel purified, as low levels of nonspecific amplification products may interfere with direct sequencing of PCR products. The RT-PCR products were sequenced and intron-exon boundaries confirmed by comparison with the genomic sequence.
Molecular modeling and pore radius calculations
The restrained molecular dynamics (MD) methods employed have been described in detail (Sansom et al. 1995; Sankararamakrishnan et al. 1996; Adcock et al. 1998). Briefly, MD simulations with experimentally derived restraints taken from 9Å resolution cryoelectron microscopy images (Unwin 1995) and from pore-lining side chains identified by site-directed mutagenesis (Hucho et al. 1996) were used to generate models of homopentameric M2 helix bundles for the C. elegans nAChR sequences. MD simulations were performed using Xplor V3.1 (Brünger 1992) with the CHARMM PARAM19 (Brooks et al. 1983) parameter set. Display and examination of models were carried out using Quanta98 (Molecular Simulations), and diagrams of structures drawn using Molscript (Kraulis 1991). Simulations were carried out on Silicon Graphics workstations. Pore-lining residues in the generated models were identified using diagrams produced by rotating the constituent helices onto a plane, forming a disc of helices with one surface corresponding to their pore-lining faces and the other surface revealing the M2 residues which interact directly with the other transmembrane domains. In these diagrams, the innermost ring of side chains corresponds to the intracellular end and the outermost ring corresponds to the extracellular end of the pore.
The "HOLE" algorithm (Smart et al. 1996) was employed to estimate the pore dimensions of the nAChR models generated by SA/MD. HOLE determines progressively the radius of the largest sphere that can be accommodated without overlap with the van der Waals radii of residues exposed to the channel lumen. The net result is a series of dimensions determined by measuring the radius of a flexible sphere "squeezing" through the ion channel.
Computational sequence analysis
The Genefinder program was developed (P. Green and L. Hillier, unpublished software) to identify potential protein coding genes within genome data. The program uses statistical criteria based on genomic features characteristic of protein encoding sequences including splice consensus sites, translation start and stop sequences, G-C characteristics, and codon biases (Wilson 1999). The computer-generated gene predictions are then inspected manually, and EST and protein homologies are used to improve the final annotation, presented in ACeDB. Predicted proteins are then compared with existing protein databases and matches recorded in the public C.elegans databases. This method of gene prediction has proven to have greater than 70% accuracy, although errors do occur in the identification of translational start sites and intron-exon boundaries. Genefinder has identified 19,099 predicted genes on the C. elegans genome (The C. elegans Genome Sequencing Consortium, 1998).
Novel nAChR subunits were identified using BLAST analysis of the C. elegans genomic and protein databases, and alignments were constructed using ClustalW (Thompson et al. 1994). The relationship between protein sequences may be inferred by detailed analysis of multiple alignments. Although our analyses were based on predicted protein sequences, these novel nAChR subunits were identified using BLAST homology searches, which found the features highly conserved in known nAChR subunits. Hence only small regions of highly divergent sequence may not have been identified by our searches, which should not alter the overall result of our analyses.
Phylogenetic studies have investigated the molecular evolution of ligand-gated ion channels (LGICs) (Ortells and Lunt 1995), the nAChR subunit gene family (Le Novère and Changeux 1995; Tsunoyama and Gojobori 1998), avermectin-sensitive GluClR (Vassilatis et al. 1997), and inhibitory ligand-gated ion channels (Xue 1998). The Phylip package [PHYLIP (Phylogeny Inference Package) version 3.5c, distributed by the author, J. Felsenstein, Department of Genetics, University of Washington, Seattle, USA] can be used to construct trees representing the phylogenetic relationship between sequences. However, in the present study the programs were employed to determine the sequence relationship of novel nAChR subunits relative to a functionally distinct out-group containing ionotropic receptors that form anion channels (GluClRs, GABAR, and GlyRs). As in our previous study (Mongan et al. 1998), the Protdist program of the Phylip package was used to compute a distance measurement for the protein sequences, the Dayhoff PAM matrix. The neighbor-joining (NJ) method was then used to produce an unrooted tree by examining the distance matrix calculated by Protdist. Although the tree generated by the NJ method was unrooted, the inclusion of a functionally distinct but structurally related group of sequences in the dataset (termed the out-group) allowed the tree to be rooted and the final relationship determined.
Acknowledgments
The authors gratefully acknowledge the financial support of the Medical Research Council (MRC) (D.B.S., A.K.J.) and the Biotechnology and Biological Sciences Research Council (BBSRC) (D.B.S.), the Marie Curie Fellowship program of the E.U. (N.P.M.), and the Wellcome Trust (M.S.P.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.3040102.
References
- Adcock, C., Smith, G.R., and Sansom, M.S.P. 1998. Electrostatics and the ion channel selectivity of ligand gated ion channels. Biophys. J. 75 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Anand, R., Peng, X., and Lindstrom, J. 1993. Homomeric and native α7 acetylcholine receptors exhibit remarkably similar but non-identical pharmacological properties, suggesting that the native receptor is a heteromeric protein complex. FEBS Letts. 327 241–246. [DOI] [PubMed] [Google Scholar]
- Ballivet, M., Alliod, C., Bertrand, S., and Bertrand D. 1996. Nicotinic acetylcholine receptors in the nematode Caenorhabditis elegans.J. Mol. Biol. 258 261–269. [DOI] [PubMed] [Google Scholar]
- Baylis, H.A., Matsuda, K., Squire, M.D., Fleming, J.T., Harvey, R., Darlison, M.G., Barnard, E.A., and Sattelle, D.B. 1997. ACR-3, a Caenorhabditis elegans nicotinic acetylcholine receptor subunit: Molecular cloning and functional expression. Receptors Channels 5 149–158. [PubMed] [Google Scholar]
- Bouzat, C. and Barrantes, F.J. 1997. Assigning functions to residues in the acetylcholine receptor channel region. Mol. Membr. Res. 14 167–177. [DOI] [PubMed] [Google Scholar]
- Bouzat, C., Barrantes, F., and Sine, S. 2000. Nicotinic receptor fourth transmembrane domain—Hydrogen bonding by conserved threonine contributes to channel gating kinetics. J. Gen. Physiol. 115 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. 1983. CHARMM: A program for macromolecular energy, minimization and dynamics calculations. J. Comp. Chem. 4 187–217. [Google Scholar]
- Brünger, A.T. 1992. X-PLOR Version 3.1. A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Changeux, J.P. and Edelstein, S.J. 1998. Allosteric receptors after 30 years. Neuron 21 959–980. [DOI] [PubMed] [Google Scholar]
- Corringer, P.J., Le Novère, N., and Changeux, J.P. 2000. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 40 431–458. [DOI] [PubMed] [Google Scholar]
- Couturier, S., Bertrand, D., Matter, J.M., Hernandez, M.C., Bertrand, S., Millar, N., Valera, S., Barkas, T., and Ballivet, M. 1990. A neuronal nicotinic acetylcholine receptor subunit α7 is developmentally regulated and forms a homo-oligomeric channel blocked by α-BTX. Neuron 5 847–856. [DOI] [PubMed] [Google Scholar]
- Cuevas, J. and Berg, D.K. 1998. Mammalian nicotinic receptors with α7 subunits that slowly desensitize and rapidly recover from α-bungarotoxin blockade. J. Neurosci. 24 10335–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel, R.C. and Green, W.N. 2000. Neuronal α-bungarotoxin receptors are α7 subunit homomers. J. Neurosci. 20 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgoyhen, A.B., Johnson, D.S., Boulter, J., Vetter, D.E., and Heinemann, S. 1994. α9: An acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell 79 705–715. [DOI] [PubMed] [Google Scholar]
- Esmaeili, B., Ross, J.M., Neades, C., Miller, D.M., III, and Ahringer, J. 2002. Dev. 129 853–862. [DOI] [PubMed] [Google Scholar]
- Fleming, J.T., Squire, M.D., Barnes, T.M., Tornøe, C.T., Matsuda, K., Sulston, J.E., Barnard, E.A., Sattelle, D.B., and Lewis, J.T. 1997. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29 and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. 17 5843–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerzanich, V., Anand, R., and Lindstrom, J. 1994. Homomers of α8 and α7 subunits of nicotinic receptors exhibit similar channel but contrasting binding site properties. Mol. Pharmacol. 45 212–220. [PubMed] [Google Scholar]
- Hosie, A.M., Aronstein, K., Sattelle, D.B., and ffrench-Constant, R.H. 1997. Molecular biology of insect neuronal GABA receptors. TINS 20 578–583. [DOI] [PubMed] [Google Scholar]
- Hucho, F., Tsetlin, V.I., and Machold, J. 1996. The emerging three dimensional structure of α receptor: The nicotinic acetylcholine receptor. Eur. J. Biochem. 239 539–557. [DOI] [PubMed] [Google Scholar]
- Keyser, K.T., Britto, L.R.G., Schoepfer, R., Whiting, J., Cooper, J., Conroy, W., Brozozowska-Prechtl, A., Karten, H.J., and Lindstrom, J. 1993. Three subtypes of α-bungarotoxin-sensitive nicotinic acetylcholine receptors are expressed in chick retina. J. Neurosci. 13 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Le Novère, N. and Changeux, J.P. 1995. Molecular evolution of the nicotinic acetylcholine receptor: An example of multigene family in excitable cells. J. Mol. Evol. 40 155–172. [DOI] [PubMed] [Google Scholar]
- Lewis, J.A., Wu, C.H., Berg, H., and Levine, J.H. 1980. The genetics of levamisole resistance in the nematode Caenorhabditis elegans.Genetics 95 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton, J.T. and Ganetzky, B. 2000. Ion channels and synaptic organization: Analysis of the Drosophila genome. Neuron 26 35–43. [DOI] [PubMed] [Google Scholar]
- Marshall, J., Buckingham, S.D., Shingai, R., Lunt, G.G., Goosey, M.W., Darlison, M.G., Sattelle, D.B., and Barnard, E.A. 1990. Sequence and functional expression of a single α subunit of an insect nicotinic acetylcholine receptor. EMBO J. 9 4391–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C. 1989. Genetic manipulation of ion channels: A new approach to structure and mechanism. Neuron 2 1195–1205. [DOI] [PubMed] [Google Scholar]
- Mongan, N.P., Baylis, H.A., Adcock, C., Smith, G.R., Sansom, M.S.P., and Sattelle, D.B. 1998. An extensive and diverse nicotinic acetylcholine receptor α subunit gene family in Caenorhabditis elegans.Receptors Channels 6 213–228. [PubMed] [Google Scholar]
- Noda, M., Takahashi, H., Tanabe, T., Toyosato, M., Furutani, Y., Hirose, T., Asai, M., Inayama, S., Miyata, T., and Numa, S. 1982. Primary structure of a subunit precursor of Torpedo californica acetylcholine receptor deduced from cDNA sequence. Nature 299 793–797. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger, A., Broide, R.S., Kasten, M.R., Dang, H., Dani, J.A., Beaudet, A.L., and Patrick, J.W. 2000. Mice homozygous for the L250T mutation in the alpha7 nicotinic acetylcholine receptor show increased neuronal apoptosis and die within 1 day of birth. J. Neurochem. 74 2154–2166. [DOI] [PubMed] [Google Scholar]
- Ortells, M.O. and Lunt, G.G. 1995. Evolutionary history of the ligand-gated ion-channel superfamily of receptors. Trends Neurosci. 18 121–127. [DOI] [PubMed] [Google Scholar]
- Palma, E., Maggi, L., Barabino, B., Eusebi, F., and Ballivet, M. 1999. Nicotine acetylcholine receptors assembled from the alpha 7 and beta 3 subunits. J. Biol. Chem. 274 18335–18340. [DOI] [PubMed] [Google Scholar]
- Raymond, V., Mongan, N.P., and Sattelle, D.B. 2000. Actions of cholinergic anthelmintics and ivermectin on recombinant homomeric nicotinic acetylcholine receptors, chicken α7 and Caenorhabditis elegans ACR-16. Neuroscience 101 785–791 [DOI] [PubMed] [Google Scholar]
- Rothlin, C.V., Katz, E., Verbitsky, M., and Elgoyhen, A.B. 1999. The α9 nicotinic acetylcholine receptor shares pharmacological properties with type A γ-aminobutyric acid, glycine and type 3 serotonin receptors. Mol. Pharmacol. 55 248–254. [DOI] [PubMed] [Google Scholar]
- Sankararamakrishnan, R., Adcock, C., and Sansom, M.S.P. 1996. The pore domain of the nicotinic acetylcholine receptor: Molecular modeling, pore dimensions and electrostatics. Biophys. J. 71 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom, M.S.P., Sankararamakrishnan, R., and Kerr, I.D. 1995. Modeling membrane proteins using structural restraints. Nat. Struct. Biol. 2 624–631. [DOI] [PubMed] [Google Scholar]
- Schoepfer, R., Conroy, W.G., Whiting, P., Gore, M., and Lindstrom, J. 1990. Brain α-bungarotoxin binding protein cDNAs and MAbs reveal subtypes of this branch of the ligand-gated ion channel gene superfamily. Neuron 5 35–48. [DOI] [PubMed] [Google Scholar]
- Séguéla, P., Wadiche, J., Dineley-Miller, K., Dani, J.A., and Patrick, J.W. 1993. Molecular cloning, functional properties and distribution of rat brain α7: A nicotinic cation channel highly permeable to calcium. J. Neurosci. 13 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, O.S., Neduvelil, J.G., Wang, X., Wallace, B.A., and Sansom, M.S.P. 1996. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14 354–360. [DOI] [PubMed] [Google Scholar]
- Squire, M.D., Baylis, H.A., Fleming, J.T., Barnard, E.A., and Sattelle, D.B. 1995. Molecular cloning and functional co-expression of a Caenorhabditis elegans nicotinic acetylcholine receptor subunit acr-2.Receptors Channels 3 107–115. [PubMed] [Google Scholar]
- Tamamizu, S., Guzman, G.R., Santiago, J., Rojas, L.V., McNamee, M.G., and Lasalde-Dominicci, J.A. 2000. Functional effects of periodic tryptophan substitutions in the alpha M4 transmembrane domain of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 39 4666–4673. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treinin, M. and Chalfie, M. 1995. A mutated acetylcholine receptor subunit causes neuronal degeneration in C. elegans.Neuron 14 871–877. [DOI] [PubMed] [Google Scholar]
- Treinin, M., Gillo, B., Liebman, L., and Chalfie, M. 1998. Two functionally dependent acetylcholine subunits are encoded in a single Caenorhabditis elegans operon. Proc. Natl. Acad. Sci. 95 15492–15495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Genome Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282 2012–2018. [DOI] [PubMed] [Google Scholar]
- Tsunoyama, K. and Gojobori, T. 1998. Evolution of nicotinic acetylcholine receptor subunits. Mol. Biol. Evol. 15 518–527. [DOI] [PubMed] [Google Scholar]
- Unwin, N. 1995. Acetylcholine receptor channel imaged in the open state. Nature 373 37–43. [DOI] [PubMed] [Google Scholar]
- Vassilatis, D.K., Elliston, K.O., Paress, P.S., Hamelin, M., Arena, J.P., Schaeffer, J.M., van der Ploeg, L.H.T, and Cully, D.F. 1997. Evolutionary relationship of the ligand gated ion channels and the avermectin sensitive glutamate gated chloride channels. J. Mol. Evol. 44 501–508. [DOI] [PubMed] [Google Scholar]
- Villarroel, A.S., Herlitze, S., Koenen, M., and Sakmann, B. 1991. Location of a threonine residue in the α-subunit M2 transmembrane segment that determines the ion flow through the acetylcholine receptor channel. Proc. R. Soc. Lond. B. Biol. Sci. 243 69–74. [DOI] [PubMed] [Google Scholar]
- Villarroel, A. and Sakmann, B. 1992. Threonine in the selectivity filter of the acetylcholine receptor channel. Biophys J. 62 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Pereira, E.F., Maus, A.D., Ostlie, N.S., Navaneetham, D., Lei, S., Albuquerque, E.X., Conti-Fine, B.M. 2001. Human bronchial epithelial and endothelial cells express alpha7 nicotinic acetylcholine receptors. Mol. Pharmacol. 60 1201–1209. [DOI] [PubMed] [Google Scholar]
- Wilson, G.G., Pascual, J.M., Brooijmans, N., Murray, D., and Karlin, A. 2000. The intrinsic electrostatic potential and the intermediate ring of charge in the acetylcholine receptor channel. J. Gen. Physiol. 115 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.K. 1999. How the worm was won: The genome sequencing project. TIGS 15 51–58. [DOI] [PubMed] [Google Scholar]
- Xue, H. 1998. Identification of major phylogenetic branches of inhibitory ligand gated channel receptors. J . Mol. Evol. 47 323–333. [DOI] [PubMed] [Google Scholar]
- Yassin, L., Gillo, B., Kahan, T., Halevi, S., Eshel, M., and Treinin, M. 2001. Characterization of the deg-3/des-2 receptor: A nicotinic acetylcholine receptor that mutates to cause neuronal degeneration. Mol. Cell. Neurosci. 17 589–599. [DOI] [PubMed] [Google Scholar]
- Yu, C.R., and Role, L.W. 1998. Functional contribution of the alpha7 subunit to multiple subtypes of nicotinic receptors in embryonic chick sympathetic neurons. J. Physiol. 590 651–665. [DOI] [PMC free article] [PubMed] [Google Scholar]