Abstract
We have used two fluorescent probes, NBD and dansyl, attached site-specifically to the serpin plasminogen activator inhibitor-1 (PAI-1) to address the question of whether a common mechanism of proteinase translocation and full insertion of the reactive center loop is used by PAI-1 when it forms covalent SDS-stable complexes with four arginine-specific proteinases, which differ markedly in size and domain composition. Single-cysteine residues were incorporated at position 119 or 302 as sites for specific reporter labeling. These are positions ∼30 Å apart that allow discrimination between different types of complex structure. Fluorescent derivatives were prepared for each of these variants using both NBD and dansyl as reporters of local perturbations. Spectra of native and cleaved forms also allowed discrimination between direct proteinase-induced changes and effects solely due to conformational change within the serpin. Covalent complexes of these derivatized PAI-1 species were made with the proteinases trypsin, LMW u-PA, HMW u-PA, and t-PA. Whereas only minor perturbations of either NBD and dansyl were found for almost all complexes when label was at position 119, major perturbations in both wavelength maximum (blue shifts) and quantum yield (both increases and decreases) were found for all complexes for both NBD and dansyl at position 302. This is consistent with all four complexes having similar location of the proteinase catalytic domain and hence with all four using the same mechanism of full-loop insertion with consequent distortion of the proteinase wedged in at the bottom of the serpin.
Keywords: Plasminogen activator inhibitor 1, serpin, inhibition mechanism, proteinase, t-PA, u-PA, fluorescence
Serpins such as plasminogen activator inhibitor-1 (PAI-1) inhibit target proteinases by a unique conformational change-based mechanism that leads to a kinetically trapped SDS-stable covalent intermediate (Lawrence et al. 1995; Gettins et al. 1996). Some of the controversy over the precise chemical nature of the intermediate and the possible mechanism of kinetic trapping has been resolved by a recent X-ray-structure determination of the covalent complex of one serpin, α1-proteinase inhibitor (α1PI), with the serine proteinase trypsin (Huntington et al. 2000) This study showed that the covalent intermediate is, in fact, the long proposed acyl ester (Jörnvall et al. 1979; Longas and Finlay 1980) and that full insertion of the cleaved reactive center loop into β-sheet A of the serpin had occurred. In addition, the structure provided the first clear structural insight into the possible basis for kinetic trapping by showing that the active site of the proteinase had been distorted in a major way, with the P1 side chain removed from the S1 specificity pocket, the active-site serine moved away from the histidine of the catalytic triad by about 3 Å, and the oxyanion hole, which is needed for stabilization of the tetrahedral intermediate that is formed en route to hydrolysis of the acyl ester, disrupted.
Although such an X-ray structure represents a big step forward in understanding the properties and behavior of serpins, it still leaves open the question of how general such a structure is for other serpin–proteinase pairs, that is, whether partial rather than full insertion occurs with larger proteinases that might more readily be obstructed by helix F (which overlays β-sheet A) in their passage from one end of the serpin in the region of the reactive center loop to the opposite pole 70 Å away. This question is particularly important because the proteinase used for the X-ray study of a serpin:proteinase complex, trypsin, is not a normal physiological target for α1PI. In addition, a recent study on the chymotrypsin:α1–antichymotrypsin pair made the claim that, in that particular case, the covalent complex involved no large-scale movement of the proteinase or insertion of the reactive center loop (O'Malley and Cooperman 2000).
We have previously successfully used fluorescence probes on the serpin as reporters of the location of the proteinase in a covalent serpin:proteinase complex. The model of the complex that we obtained using this approach gave a placement of the proteinase relative to α1PI (Stratikos and Gettins 1999) that was subsequently shown to be correct by the X-ray structure of the same complex.
In this study we have used the same kind of proximity perturbation fluorescence approach that we successfully used in our earlier study with α1PI to examine the nature of the covalent complexes of PAI-1 with a series of structurally related, physiologically relevant proteinases and to permit evaluation of the effect of differences in the proteinase on the nature of the covalent complex formed. The four proteinases were chosen to provide a range of sizes and domain organization, though with the same arginine specificity and the same trypsin fold for the catalytic domain in each case. Of the four proteinases, trypsin is the smallest, LMW u-PA is a larger but still single domain proteinase, and HMW u-PA and t-PA are multidomain larger proteinases.
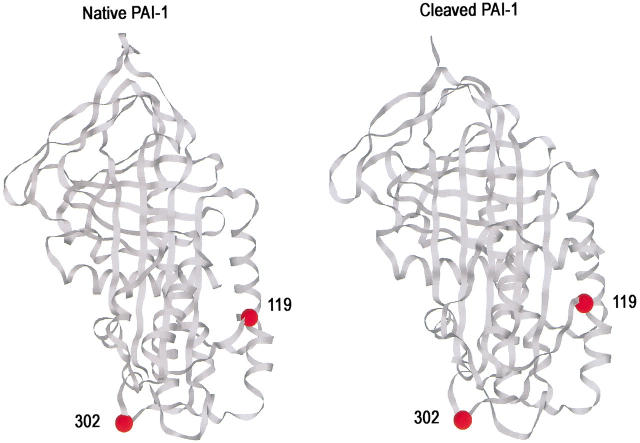
Single cysteines were separately engineered at each of two sites in PAI-1 to serve as attachment sites for fluorescent reporters of proteinase proximity in complexes of different structure. These were positions 119 and 302 (Fig 1 ▶). Position 302 is equivalent to position 314 in α1PI, which, when labeled with dansyl, showed high sensitivity of fluorescence to proximity of proteinase in complex. This sensitivity was explained by the X-ray structure of the α1PI:trypsin complex, which showed residue 314 to be located within the contact interface between the serpin and the proteinase. The 302 position in PAI-1, therefore, was thought likely to be an excellent location for a reporter of nearby proteinase if complexes resembled that of α1PI with trypsin. Position 119 is 27 Å away from residue 302, close to helix F in the native structure and, therefore, might be sensitive to complex structures in which only partial-loop insertion and proteinase movement had taken place. NBD and dansyl were each used as reporters at both labeling positions for all four complexes. In this way we had four separate reporter fluorophores for a given complex. In addition, spectra of native and reactive center-loop cleaved forms allowed identification of those spectral changes that resulted solely from internal conformational changes within PAI-1. We found that only fluorophores at position 302 showed consistent and dramatic sensitivity to complex formation, whereas reporters at position 119 showed almost no effects of complex formation. This strongly suggests that all complexes resulted in closely similar location of the proteinase catalytic domain relative to the body of the serpin, at a position similar to that found both by X-ray crystallography (Huntington et al. 2000) and fluorescence (Stratikos and Gettins 1999) for the α1PI:trypsin pair. More generally it therefore seems likely that serpin–proteinase pairs that form covalent, SDS-stable complexes all use the same method of full-loop insertion and proteinase translocation as the means of distorting the proteinase active site and hence kinetically trapping the intermediate.
Fig. 1.
Structures of native and cleaved PAI-1 showing the two positions of fluorescent label incorporation (residues 119 and 302). Ribbon diagrams were drawn using Rasmol with coordinates for native PAI-1 (left) from structure 1b3k (Sharp et al. 1999) and for cleaved PAI-1 (right) from 9pai (Aertgeerts et al. 1995). The reactive center loop is at the top. The location of the proteinase in the α1PI:trypsin complex is at the bottom of the structure, impinging upon residue 314, which is the equivalent of residue 302 in PAI-1.
Results
Fluorescence emission spectra of native and cleaved PAI-1 species
To be able to distinguish between changes in reporter fluorescence due to direct proximity perturbation and those due to internal conformational changes resulting from insertion of the reactive center loop into β-sheet A and also to gauge the sensitivity of each position and label to such conformational changes, we recorded spectra of both native and reactive center-loop cleaved forms of each of the four PAI-1–fluorphore conjugates.
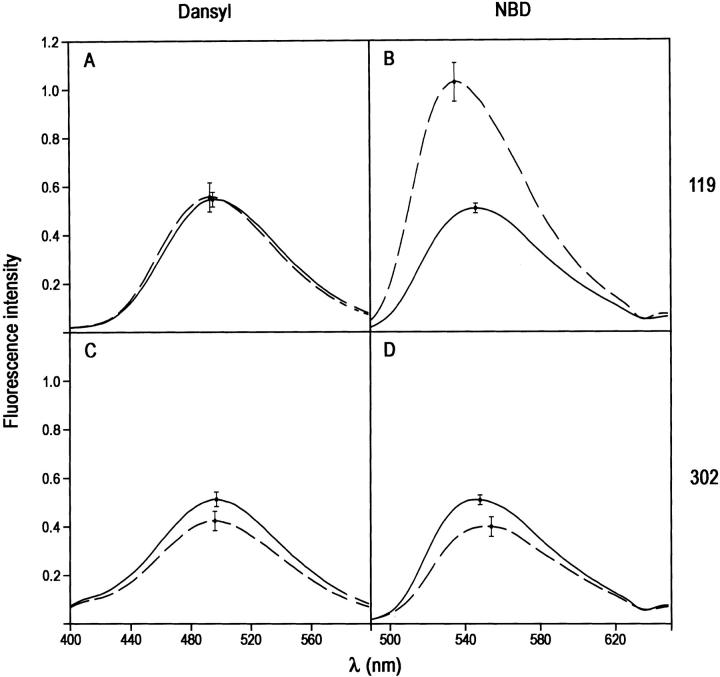
Native PAI-1 species with dansyl attached at position 119 or 302 (Fig. 1 ▶) gave similar emission spectra in terms of intensity and wavelength maxima (Fig. 2A,C ▶; Table 1). The maxima, at 495–6nm, are consistent with some degree of solvent exposure (Larsson et al. 1987). Upon cleavage of the reactive center loop and insertion of the cleaved loop into β-sheet A there was little change in wavelength maximum for either labeling position and no change in intensity for label at position 119 (Fig. 2 ▶). For position 302 there was an ∼20% reduction in dansyl emission intensity for the reactive center-loop inserted form (Fig. 2C ▶). NBD showed a different responsiveness to loop insertion. As with dansyl label, the NBD spectra in native PAI-1 were similar in intensity and wavelength maximum for the 119 and 302 positions (Fig. 2B,D). Upon reactive center-loop cleavage, label at position 119 gave a very large change in intensity (∼100% enhancement) and in the wavelength maximum (11-nm blue shift; Fig. 2B ▶), whereas label at position 302 showed a 20% reduction and a 6-nm red shift (Fig. 2D ▶).
Fig. 2.
Effect of reactive center-loop cleavage and insertion on the fluorescence emission spectra of PAI-1 species labeled at either position 119 or 302 with the fluorophore IAEDANS or NBD. Each panel shows spectra from native (solid line) and cleaved (dashed line) species. (A) 119C-dansyl-PAI-1; (B) 119C-NBD-PAI-1; (C) 302C-dansyl-PAI-1; (D) 302-NBD-PAI-1. Spectra have been normalized to the same label concentration. Error bars at the wavelength maximum show the range of variation in intensity seen in separate experiments.
Table 1.
Spectral properties and SI values of NBD and dansyl-labeled PAI-1 species
| Raw data | Data corrected for SI | ||||||
| Position and fluorophore | λmax | Fluorescence intensity at λmax | SI | λmax | Fluorescence intensity at λmax | % Change | |
| 119 dansyl | Native | 495 ± 1 | 0.55 ± 0.03 | ||||
| Cleaved | 493 ± 2 | 0.56 ± 0.06 | |||||
| Trypsin | 492 ± 0 | 0.58 ± 0.02 | 2.0 | 488 ± 2 | 0.61 ± 0.07 | 9 | |
| LMW u-PA | 493 ± 1 | 0.56 ± 0.03 | 1.2 | 491 ± 3 | 0.58 ± 0.05 | 3 | |
| HMW u-PA | 494 ± 2 | 0.57 ± 0.03 | 1.2 | 491 ± 5 | 0.63 ± 0.05 | 13 | |
| t-PA | 489 ± 1 | 0.64 ± 0.04 | 1.3 | 484 ± 1 | 0.71 ± 0.06 | 27 | |
| 302 dansyl | Native | 497 ± 2 | 0.51 ± 0.03 | ||||
| Cleaved | 496 ± 3 | 0.42 ± 0.04 | |||||
| Trypsin | 487 ± 1 | 0.69 ± 0.04 | 2.2 | 486 ± 1 | 0.97 ± 0.09 | 131 | |
| LMW u-PA | 488 ± 3 | 0.71 ± 0.05 | 2.0 | 482 ± 1 | 1.07 ± 0.1 | 155 | |
| HMW u-PA | 485 ± 1 | 0.70 ± 0.07 | 1.9 | 481 ± 1 | 1.02 ± 0.2 | 143 | |
| t-PA | 480 ± 1 | 0.82 ± 0.03 | 1.8 | 475 ± 2 | 1.22 ± 0.09 | 190 | |
| 119 NBD | Native | 546 ± 2 | 0.51 ± 0.02 | ||||
| Cleaved | 535 ± 1 | 1.03 ± 0.08 | |||||
| Trypsin | 536 ± 1 | 0.89 ± 0.03 | 2.4 | 534 ± 2 | 0.87 ± 0.2 | −15 | |
| LMW u-PA | 538 ± 0 | 0.75 ± 0.04 | 1.1 | 538 ± 1 | 0.79 ± 0.06 | −23 | |
| HMW u-PA | 537 ± 2 | 0.76 ± 0.06 | 1.1 | 534 ± 2 | 0.95 ± 0.1 | −8 | |
| t-PA | 538 ± 0 | 0.75 ± 0.04 | 1.1 | 535 ± 1 | 1.03 ± 0.1 | 0 | |
| 302 NBD | Native | 548 ± 1 | 0.51 ± 0.02 | ||||
| Cleaved | 554 ± 3 | 0.40 ± 0.04 | |||||
| Trypsin | 548 ± 2 | 0.34 ± 0.04 | 2.0 | 545 ± 2 | 0.24 ± 0.08 | −40 | |
| LMW u-PA | 542 ± 1 | 1.14 ± 0.07 | 1.3 | 542 ± 1 | 1.62 ± 0.1 | 305 | |
| HMW u-PA | 544 ± 2 | 1.1 ± 0.1 | 1.3 | 543 ± 1 | 1.95 ± 0.2 | 388 | |
| t-PA | 545 ± 2 | 0.83 ± 0.04 | 1.3 | 546 ± 1 | 1.58 ± 0.1 | 295 | |
These results indicate that loop insertion per se causes local structural changes at both 302 and 119 positions that are sensed by one or both of the reporter fluorophores but particularly strongly sensed by NDB at position 119. This is not unexpected, for reactive center-loop cleavage of PAI-1 results in insertion of the cleaved loop into β-sheet A as a central strand, with consequent movement of the β-sheet relative to under- and overlying structures. It also indicates that the two fluorophores are responsive to changes in their environment at the labeling positions chosen.
SI values of fluorescent PAI-1 derivatives
When a serpin reacts with a proteinase it does so using a branched pathway, with one branch leading to covalent complex and the other to cleaved serpin (Patston et al. 1991). The stoichiometry of inhibition (SI) of the reaction measures the relative fluxes along the two pathways and is defined as (k3 + k4)/k4, where k3 and k4 are the rate constants for the substrate and inhibitory branches of the pathway, respectively (Patston et al. 1991; Cooperman et al. 1993). When k4 ⋍ k3, the predominant product is covalent complex and the SI is close to 1. The SI increases significantly from 1 only as k3 approaches the value of k4. Under such conditions, the products of reaction include significant amounts of cleaved serpin in addition to the covalent complex. To be able to extract the fluorescence emission spectra of covalent complexes generated by in situ reaction of derivatized PAI-1 with different proteinases, it, therefore, was necessary to determine the SI for each PAI-1:proteinase pair, in addition to the emission spectra of native and cleaved PAI-1 described above, so that contributions from these species could be subtracted from the spectrum of the reaction mixture. This was important for reactions where SI values were significantly greater than 1.
SI values were determined under conditions of reaction identical to those used for each fluorescence experiment by incubating each dansyl- or NBD-labeled PAI-1 with each of the proteinases, trypsin, low and high molecular weight u-Pas, and t-PA, and running an SDS polyacrylamide gel immediately after the end of the incubation. The distribution of components was determined from the fluorescence intensity of each of the complex bands, as described above. In this way, the distribution of products should exactly match that used for the fluorescence measurements. We also independently analyzed products from each fluorescence experiment and obtained reasonable agreement between the two sets of SI measurements. However, we considered the first measure of SI to be a more accurate measurement of the composition in the cuvette at the time the fluorescence spectra were recorded. Furthermore, the good reproducibility of the spectra for a given PAI-1:proteinase reaction suggested that the SI was not subject to large experimental variation.
SI values for each PAI-1 derivative in combination with each proteinase are given in Table 1. For all four PAI-1 derivatives, the SI with trypsin was similar but elevated over the value for unlabeled wild-type PAI-1, emphasizing the need to determine SI values for each derivative rather than using values for unlabeled wild-type PAI-1. The values of 2–2.4 reflect the presence of 50%–60% of cleaved PAI-1 in the reaction mixtures. With the further exception of dansyl-labeled 302C PAI-1 in complex with each of the other three proteinases, all of the other SIs were close to 1. The 302 dansyl-labeled species showed much higher SIs with each proteinase (1.8–2.2) compared with either the equivalent NBD-labeled samples or the 119 dansyl-labeled samples (1.1–1.3). This further highlights the sensitivity of the outcome of the serpin-inhibition mechanism to any perturbation of the structure, including surface modifications that might slow down movement of the proteinase as the reactive center loop inserts into β-sheet A and hence affect the competition between trapping and cleavage reactions. In addition it provides independent support for the dansyl at position 302 being close to the proteinase in the covalent complex. Not reported in the table are the SI values for reaction of the four PAI-1 derivatives with thrombin. Thrombin was originally included in the series of proteinases as being a physiologically relevant proteinase intermediate in size between trypsin and u-PA. However, the SIs were found to be high (values of 4–6), as has previously been found for wild-type PAI-1 (Lawrence et al. 2000). Such high SI values made it problematic to deconvolute spectra of reaction mixtures to obtain spectra of the covalent complex with acceptable reliability because the complex represents only a relatively small fraction of the total species present. Accordingly, the fluorescence spectra reported below do not include those of thrombin–PAI-1 complexes.
Fluorescence spectra of trypsin: PAI-1 covalent complexes
The effect of formation of covalent complex between PAI-1 and the small single-domain proteinase trypsin (Mr 23 kD) on the fluorescence spectra of both NBD and dansyl at each of the positions 119 and 302 was next examined. Because all evidence on the nature of covalent PAI-1:proteinase complexes supports some degree of loop insertion into β-sheet A (Lawrence et al. 2000) comparison of the spectra of complex, both here for trypsin and later for other proteinases, are to the spectra of the equivalent reactive center-loop cleaved PAI-1 species.
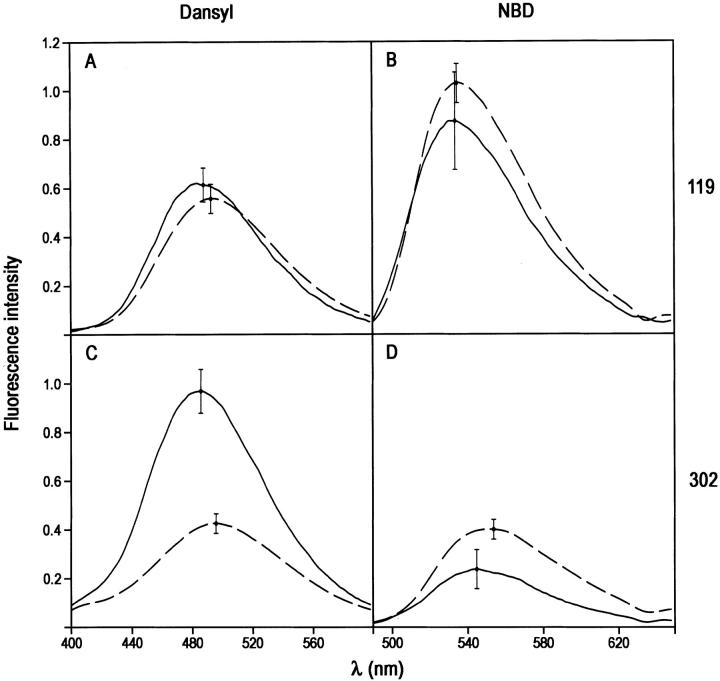
The fluorescence emission spectra of the trypsin–PAI-1 complex and cleaved PAI-1 were the same within experimental error for dansyl at position 119 (Fig. 3A ▶; Table 1). Similarly for NBD label at position 119 in covalent complex, there was no difference within experimental error from cleaved PAI-1 (Fig. 3B ▶; Table 1). In contrast, both dansyl and NBD at position 302 showed large perturbations compared with spectra of the equivalent cleaved PAI-1. 302C-dansyl PAI-1 gave a 131% enhancement and a 10-nm blue shift relative to cleaved PAI-1 (Fig. 3C ▶; Table 1), whereas 302C-NBD gave a 40% reduction in fluorescence intensity, accompanied by a 9-nm blue shift (Fig. 3D ▶; Table 1). Taken together, these results show a consistent pattern of perturbation of fluorophore at position 302, both in intensity and in wavelength maximum, but no perturbation at position 119, strongly suggesting a location for the relatively small trypsin moiety very close to position 302 but at the same time distant from position 119, as was found previously for the α1PI:trypsin complex.
Fig. 3.
Effect of formation of covalent trypsin:PAI-1 complex on the fluorescence emission spectra of NBD and dansyl fluorophores attached to positions 119 or 302 of cysteine-substituted PAI-1 species. (A) Complex with 119C-dansyl-PAI-1; (B) complex with 119C-NBD-PAI-1; (C) complex with 302C-dansyl-PAI-1; (D) complex with 302C-NBD-PAI-1. The spectra of trypsin:PAI-1 complexes were obtained from spectra of reaction mixtures of PAI-1 with trypsin and have been corrected for the contributions of any native or cleaved PAI-1 present and then normalized to the intensity for 100% complex at this concentration.
Fluorescence spectra of LMW u-PA: PAI-1 covalent complexes
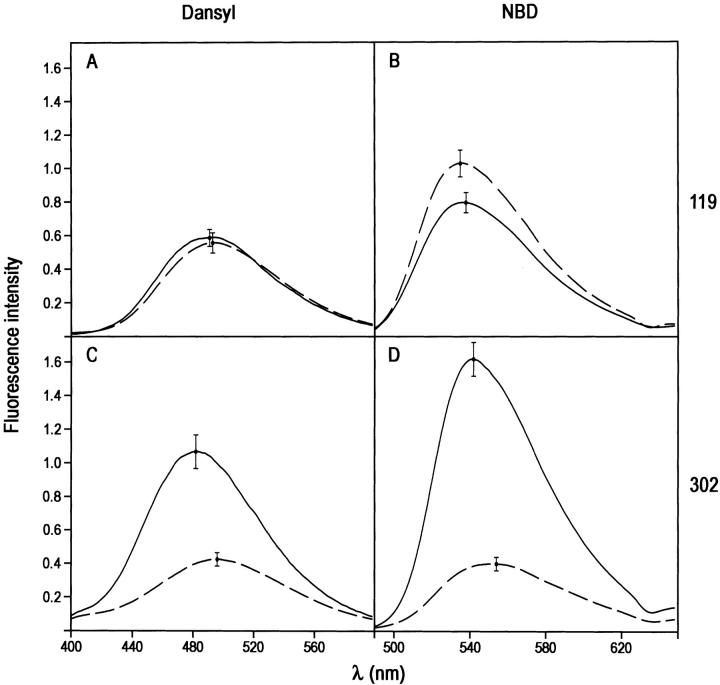
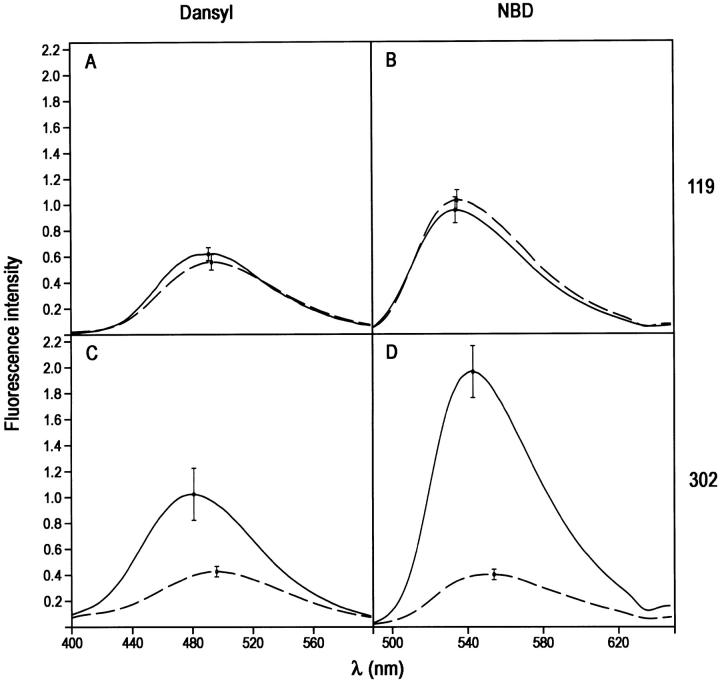
For the PAI-1 complex with the slightly larger but still single domain proteinase LMW u-PA (Mr 32 kD), a similar pattern of fluorescence perturbation was seen as for the trypsin complex, though there were differences in the magnitudes of the perturbations at position 302. Thus, neither dansyl nor NBD at position 119 showed significant difference in emission spectra in covalent complex compared with cleaved PAI-1 (Fig. 4A,B ▶; Table 1). In contrast, both dansyl and NBD at position 302 gave very large perturbations, with an enhancement in intensity of 155% and a blue shift of 14 nm for dansyl (Fig. 4C ▶) and a remarkable 305% enhancement and a blue shift of 12 nm for NBD (Fig. 4D ▶).
Fig. 4.
Effect of formation of covalent LMW u-PA:PAI-1 complex on the fluorescence emission spectra of NBD and dansyl fluorophores attached to positions 119 or 302 of cysteine-substituted PAI-1 species. (A) Complex with 119C-dansyl-PAI-1; (B) complex with 119C-NBD-PAI-1; (C) complex with 302C-dansyl-PAI-1; (D) complex with 302-NBD-PAI-1. The spectra of LMW u-PA:PAI-1 complexes were obtained from spectra of reaction mixtures of PAI-1 with trypsin and have been corrected for the contributions of any native or cleaved PAI-1 present and then normalized to the intensity for 100% complex at this concentration.
Fluorescence spectra of HMW u-PA: PAI-1 covalent complexes
HMW u-PA and LMW u-PA are closely related proteinases for they represent different extents of proteolytic processing of the same precursor zymogen. They differ only by the presence of two N-terminal domains, in addition to the large catalytic domain in HMW u-PA. These are a kringle domain and an EGF-like domain (Lawrence et al. 2000). Despite these extra domains, the effect of complex formation between HMW u-PA (Mr 54 kD) and fluorescently labeled PAI-1 species was almost identical to that seen with the LMW form of the proteinase that lacks these extra domains. Thus, complex formation caused insignificant alteration in emission spectra of either dansyl or NDB at position 119 (Fig. 5A,B ▶), whereas both fluorophores at position 302 gave very large enhancements and blue shifts (Fig. 5C,D ▶; Table 1).
Fig. 5.
Effect of formation of covalent HMW u-PA:PAI-1 complex on the fluorescence emission spectra of NBD and dansyl fluorophores attached to positions 119 or 302 of cysteine-substituted PAI-1 species. (A) Complex with 119C-dansyl PAI-1–; (B) complex with 119C-NBD-PAI-1; (C) complex with 302C-dansyl-PAI-1; (D) complex with 302-NBD-PAI-1. The spectra of HMW u-PA:PAI-1 complexes were obtained from spectra of reaction mixtures of PAI-1 with trypsin and have been corrected for the contributions of any native or cleaved PAI-1 present and then normalized to the intensity for 100% complex at this concentration.
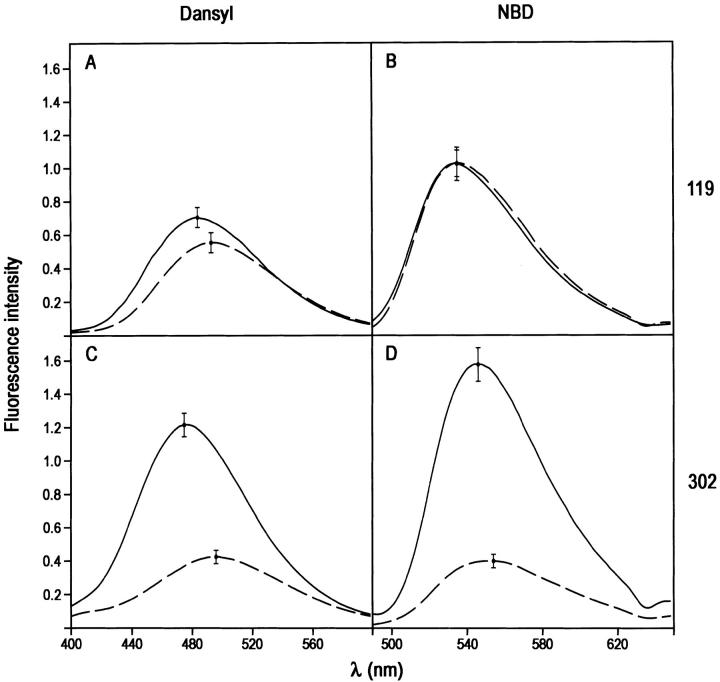
Fluorescence spectra of t-PA: PAI-1 covalent complexes
Tissue-type urokinase, or t-PA, is the largest and most complex of the four proteinases used in this study. In addition to the catalytic domain the protein contains two kringle domains, an EGF-like domain, and a finger region, all N-terminal to the catalytic domain (Pennica et al. 1983). This gives a total molecular weight of 68,000 daltons and a protein that is longer than PAI-1 itself. Despite this added complexity, higher molecular weight, and greater length, the complexes that t-PA formed with labeled PAI-1 species caused almost the same kind of fluorescence changes as both HMW u-PA and LMW u-PA. Thus, label at position 119 was either insensitive (NBD label) or only slightly sensitive (dansyl label) to complex formation, whereas label at position 302 showed large enhancements and blue shifts for both the dansyl and NBD labels (Fig. 6 ▶). In the case of the 302C-dansyl label the blue shift was 21 nm, compared to 15 nm for the two urokinase species. The enhancement (27%) and blue shift (9 nm) with dansyl at position 119, although much smaller, are thought to be real and may reflect the much longer t-PA molecule being able to also reach and perturb the fluorophore at this position through interaction with the N-terminal finger or EGF domains.
Fig. 6.
Effect of formation of covalent t-PA:PAI-1 complex on the fluorescence emission spectra of NBD and dansyl fluorophores attached to positions 119 or 302 of cysteine-substituted PAI-1 species. (A) Complex with 119C-dansyl-PAI-1; (B) complex with 119C-NBD-PAI-1; (C) complex with 302C-dansyl-PAI-1; (D) complex with 302-NBD-PAI-1. The spectra of t-PA:PAI-1 complexes were obtained from spectra of reaction mixtures of PAI-1 with trypsin and have been corrected for the contributions of any native or cleaved PAI-1 present and then normalized to the intensity for 100% complex at this concentration.
Discussion
In this study we have used perturbation of fluorescence of exogenous surface fluorophores at two locations to identify the location of the catalytic domain of four structurally related proteinases in kinetically trapped covalent complex with the same serpin inhibitor PAI-1 and hence to determine whether similar or different structures obtain in each of the four complexes. Position 302 was chosen as one attachment site based on its equivalence in the structure to residue 314 in α1-proteinase inhibitor, which we had earlier shown to be a location at which attached fluorophore was extremely sensitive to the presence of trypsin in covalent complex (Stratikos and Gettins 1999). This location was subsequently shown from a crystal structure to be at the interface between the serpin and proteinase very close to the active site (Huntington et al. 2000). PAI-1 with cysteine at position 119 was expected to serve as a sensitive reporter for proteinase in complexes where the proteinase had moved only part way from the original location of the reactive center loop to the distal end, as a result of only partial insertion of the reactive center loop in the covalent complex. By using two different fluorophores at each of the two labeling sites we, thus, had four independent reporters for a given complex.
For each of the four proteinases examined, a consistent pattern of perturbation was found in which reporter groups at position 119 showed minimal proteinase-specific effects, whereas both reporter groups at position 302 showed very large proteinase-specific perturbations in fluorescence intensity and wavelength maximum. Furthermore the wavelength changes were in each case blue shifts, consistent with decreased solvent exposure of the fluorophore. Because these perturbations are measured relative to the spectra of reactive center-loop cleaved and inserted PAI-1, they represent changes beyond those due to internal structural rearrangements within the body of the serpin caused by loop insertion, and are, therefore, almost certainly due to direct proximity effects of the proteinase. This consistent pattern of proteinase-dependent perturbation is best and most simply explained by the catalytic domain of each proteinase being close enough to residue 302 to give the observed very large perturbations seen, while at the same time being far enough away from fluorophores at position 119 that, except for the much longer t-PA, fluorophores at that site undergo no significant perturbation. This very strongly suggests that the type of structure found for the α1PI:trypsin complex is also found in the complexes of PAI-1 with each of the four arginine-specific proteinases examined here, since such a structure would place residue 302 in contact with the surface of the catalytic domain of each proteinase, close to the active site. Such a structure would account for major perturbation of any fluorophore covalently attached to this site, including large blue shifts of the spectra resulting from partial burial of the fluorophore, while at the same time being too far away from position 119 to give significant perturbations. In turn, this would require that the reactive center loop had fully inserted in each complex to permit the proteinase to move to the distal end of β-sheet A from its initial docking site with the native PAI-1.
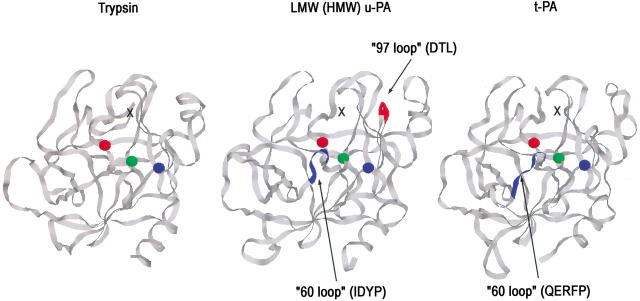
Whereas this explanation is the only one that is straightforwardly consistent with the pattern of perturbations seen for all four fluorescent species of a given PAI-1:proteinase pair, it raises two points that still need to be addressed. One point is why the magnitudes of the spectral changes in both NBD and dansyl fluorescence at position 302 are proteinase-specific. In fact, such proteinase specificity of the spectral perturbations is very easily understood in terms of differences in the surface residues of the proteinase that are likely to be close to position 302 of PAI-1 if the complex structure resembles that of α1PI:trypsin. Thus, inspection of the crystal structure of the α1PI:trypsin complex (Huntington et al. 2000) shows that residue 314 of α1PI (here equivalent to residue 302) is very close to Asn 97 and to Cys 58 of the trypsin. In both u-PA and t-PA there are insertion loops at residue 60, with different sequence in each case (IDYP in the case of u-PA and QERFP in the case of t-PA). u-PA also has a second insertion loop at residue 97 of DTL (Fig. 7 ▶). Finally, the triad of residues 95–97 differ markedly in composition between the three proteinases (NSN in trypsin, SAD in u-PA, and the highly negatively charged DDD in t-PA). Based on a similar positioning of residue 302 on PAI-1 relative to the active site of each of these proteinases (Fig. 7 ▶), the local environment of the dansyl or NBD fluorophores in each of the three types of complex (trypsin, u-PA, and t-PA) would most probably be sufficiently different than the nature of the spectral perturbation would also be different. In this regard it is very significant that the spectra of complexes of labeled PAI-1 with LMW u-PA and HMW u-PA are identical within experimental error. Whereas each of these has an identical catalytic domain, they differ markedly in size, 32 kD versus 54 kD due to the additional domains in HMW u-PA. The identity of the spectra of complexes thus suggests that the spectral changes arise exclusively from contact effects of the catalytic domains and that the catalytic domains of each urokinase are not only in the same location relative to PAI-1, but in identical orientation, despite the presence of an N-terminal extension in HMW u-PA that consists of a kringle domain and an EGF domain.
Fig. 7.
View of the putative contact surfaces of trypsin, LMW- and HMW u-PA, and t-PA in covalent complexes with PAI-1, looking from the serpin toward the proteinase catalytic domain. The black crosses mark the possible location of residue 302 of PAI-1 in each complex based on the location of the structurally equivalent residue 314 of α1PI in the covalent α1PI:trypsin complex. Residues marked as red, green, and blue spheres are the catalytic triad serine 195, histidine 57, and aspartate 102, respectively. The 60 and 97 insertion loops are shown as blue or red ribbons. The structures were drawn with Rasmol 2.7 using the pdb coordinate files 1SMF for trypsin (Li et al. 1994), 1LMW for u-PA (Spraggon et al. 1995), and 1RTF for t-PA (Lamba et al. 1996).
The second question that needs to be answered is why the t-PA:PAI-1 complex also gives rise to a small, but probably real, perturbation of dansyl at position 119. It should first be emphasized, however, that the perturbation is small compared to that of dansyl at position 302 in the same complex (27% vs. 190% enhancement) and involves a blue shift of only 9 nm versus 21 nm. In addition NBD independently shows that the proteinase strongly perturbs the 302 position (295% enhancement and 8-nm blue shift) but not the 119 position. It seems likely, though unprovable without additional information, that the much greater extended length of t-PA compared with HMW u-PA allows one or another of the N-terminal domains to curl back from the bottom of the serpin and reach the dansyl group at position 119. The insensitivity of NBD at position 119 to the presence of t-PA, if there is indeed proximity of one of the N-terminal extension domains to dansyl at this position, may result either from the shorter linker between cysteine sulfhydryl and fluorophore in the NBD derivative (ethylene diamine) than in the dansyl derivative (acetyl aminoethylamino), keeping it farther away from the t-PA N-terminal domains, or else from differences in chemical characteristics of the two fluorophores (negatively charged vs. neutral for dansyl compared to NBD) influencing their interaction with these domains. Calorimetric data suggest that there is a sufficiently loose arrangement of kringle 2 relative to the catalytic domain to permit this, whereas the estimated length of an extended t-PA is quite sufficient to allow this (Novokhatny et al. 1991).
The significance of these findings with respect to the serpin-inhibition mechanism is that we have shown that four additional serpin–proteinase complexes involving a different serpin than α1PI all appear to have the same structural organization as the α1PI:trypsin pair. With our new structural data, there are thus now examples of the same structure for two different serpins with the same proteinase (α1PI and PAI-1 with trypsin) and of one serpin (PAI-1) with four different proteinases (those examined here). Although this is still not an exhaustive demonstration of a single common structure for all possible covalent complexes, it lends strong support to the idea that, where other common behavior exists, such as kinetic evidence for operation of the branched pathway and SDS gels of a stable covalent intermediate, the structure of the covalent complex is the same as here. In that regard it is surprising that a recent study on the α1-antichymotrypsin:chymotrypsin pair should conclude that the covalent complex has the proteinase close to the initial docking site at the top of the serpin (O'Malley and Cooperman 2000). This requires that there can be little or no loop insertion and hence that the steric compression mechanism of inactivating the proteinase cannot operate. This goes against most other biochemical evidence for this serpin–proteinase pair (Rubin et al. 1990; Cooperman et al. 1993; Schechter et al. 1993), which instead indicate that it subscribes to the same suicide-substrate pathway as those serpin:proteinase pairs examined here and the α1PI:trypsin pair examined structurally elsewhere (Stratikos and Gettins 1999; Huntington et al. 2000).
An important second conclusion of the study is that it further shows the usefulness of our fluorescence perturbation method for readily checking the structural organization of other serpin:proteinase complexes using only two carefully chosen positions for attachment of a reporter group. Although either X-ray crystallography or our recent use of NMR spectroscopy (Peterson et al. 2000; Peterson and Gettins 2001) would in all cases be the most definitive proof of structure, the fluorescence proximity perturbation approach has been well validated for the α1PI:trypsin pair and is the most readily applicable to the widest range of complexes that might also be available only in small amounts and not necessarily be crystallizable.
Materials and methods
Proteins and reagents
PAI-1 mutants with a single cysteine substitution at either position 119 (Ser 119 Cys) or 302 (Phe 302 Cys) were constructed using the Transformer Site-Directed Mutagenesis Kit (Clontech) according to the manufacturer's instructions and isolated in a fully active form as described by Kvassman and Shore (1995). The purified mutants were labeled with either NBD or IAEDANS (Molecular Probes) as described by Shore et al. (1995). HMW u-PA was from Molecular Innovations, LMW u-PA was isolated from Abbokinase (Abbott Laboratories) by chromatography on benzamidine-Sepharose (Amersham Pharmacia), and t-PA (Activase™) was from Genentech.
Preparation of fluorescent derivatives
Reactions of PAI-1 with proteinase and all subsequent measurements were carried out in 20 mM sodium phosphate buffer (pH 7.4), containing 100 mM NaCl. Complexes were generated by reacting 2 μM NBD- or dansyl-labeled PAI-1 variants with a slight excess of trypsin, LMW u-PA, HMW u-PA, or t-PA. Reactions were carried out at room temperature. Different incubation times were used for the reactions based on the different second-order rate constants for the inhibition of each of the proteinases by PAI-1 (Lawrence et al. 1990). Excess trypsin was inhibited by addition of FFRCK, whereas excess LMW u-PA, HMW u-PA, and t-PA were inhibited by addition of PMSF. Reaction mixtures were diluted to a final concentration of 0.41 μM PAI-1 and the emission fluorescence spectra recorded as described below.
Cleaved PAI-1 was made by incubating 10 μM PAI-1 with 1 μM PPE for 20 min at room temperature, followed by addition of 1 mM PMSF to quench the reaction. The fluorescence emission spectra of 0.41-μM cleaved PAI-1 variants were recorded as described below.
Characterization of SIs
SI values, used for deconvolution of measured emission spectra into component spectra from cleaved and complexed PAI-1, as well as residual unreacted PAI-1, were calculated from the relative amounts of complexed, cleaved, and native serpin present after reaction with proteinase, estimated from SDS-PAGE of reaction mixtures. To ensure that the SI values calculated in this way were appropriate for this deconvolution, PAI-1 was reacted with proteinases under conditions identical to those used for the acquisition of fluorescence emission spectra. Complexes were stable during the acquisition of the spectra, as shown by the invariance of spectra of a given sample, recorded consecutively two or more times.
For SI determination, samples were run on 10% SDS-PAGE. Fluorescent bands corresponding to complex, cleaved, or native PAI-1 were visualized using a NucleoTech Gel Documentation System and their intensities quantified. SI values were calculated using the values obtained for the percentage of the cleaved and complex PAI-1 present in the reaction mixtures. With the exception of thrombin, which gave large values, the SIs for all proteinases were in the range of 1.1–2.4.
Fluorescence measurements
All fluorescence measurements were made on an SLM8000 scanning fluorometer. Spectra were recorded at 25°C. Dansyl spectra were recorded by exciting at 340 nm and scanning emission from 490 to 650 nm, with excitation slits set to 4 nm and emission slits set to 8 nm. NBD spectra were acquired by exciting at 480 nm and scanning from 400 to 600nm, with all slits set to 4 nm. All spectra were scanned in 2-nm steps, with an integration time of 4 sec for dansyl and 5 sec for NBD spectra. All reported spectra are the average of three or more independent measurements. Spectra of covalent complexes were obtained by subtraction from the experimental spectra of contributions from any native or cleaved PAI-1 present at the end of the reaction.
Acknowledgments
This study was upported by grants HL49234 and HL64013 (P.G.W.G.) from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
PAI-1, plasminogen activator inhibitor-1
NBD, N,N`-dimethyl-N-(acetyl)-N`-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) ethylene diamine
IAEDANS, 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1sulfonic acid
P1, P2, etc., designation of residues in the reactive center loop using the nomenclature of Schechter and Berger (1967) in which the scissile bond is between residues P1 and P1`, residues N-terminal to this are designated P2, P3, etc., and those C-terminal P2`, P3`, etc.
u-PA, urokinase-type plasminogen activator
t-PA, tissue-type plasminogen activator
LMW, low molecular weight
HMW, high molecular weight
SI, stoichiometry of inhibition
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.4320102.
References
- Aertgeerts, K., De Bondt, H.L., De Ranter, C.J., and Declerck, P.J. 1995. Mechanisms contributing to the conformational and functional flexibility of plasminogen activator inhibitor-1. Nature Struct. Biol. 2 891–897. [DOI] [PubMed] [Google Scholar]
- Cooperman, B.S., Stavridi, E., Nickbarg, E., Rescorla, E., Schechter, N.M., and Rubin, H. 1993. Antichymotrypsin interaction with chymotrypsin-partitioning of the complex. J. Biol. Chem. 268 23616–23625. [PubMed] [Google Scholar]
- Gettins, P.G.W., Patston, P.A., and Olson, S.T. 1996. In Serpins: Structure, function and biology. R.G. Landes Co., Austin, TX.
- Huntington, J.A., Read, R.J., and Carrell, R.W. 2000. Structure of a serpin–protease complex shows inhibition by deformation. Nature 407 923–926. [DOI] [PubMed] [Google Scholar]
- Jörnvall, H., Fish, W.W., and Björk, I. 1979. The thrombin cleavage site in bovine antithrombin. FEBS Lett. 106 358–362. [DOI] [PubMed] [Google Scholar]
- Kvassman, J.-O. and Shore, J.D. 1995. Purification of human plasminogen activator inhibitor (PAI-1) from Escherichia coli and separation of its active and latent forms by hydrophobic chromatography. Fibrinolysis 9 215–221. [Google Scholar]
- Lamba, D., Bauer, M., Huber, R., Fischer, S., Rudolph, R., Kohnert, U., and Bode, W. 1996. The 2.3Å crystal structure of the catalytic domain of recombinant tissue-type plasminogen activator. J. Mol. Biol. 258 117–135. [DOI] [PubMed] [Google Scholar]
- Larsson, L.-J., Lindahl, P., Hallén-Sandgren, C., and Björk, I. 1987. The conformational changes of α-2-macroglobulin induced by methylamine and trypsin. Biochem. J. 243 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, D.A., Strandberg, L., Ericson, J., and Ny, T. 1990. Structure–function studies of the SERPIN plasminogen activator inhibitor type 1. Analysis of chimeric strained loop mutants. J. Biol. Chem. 265 20293–20301. [PubMed] [Google Scholar]
- Lawrence, D.A., Ginsburg, D., Day, D.E., Berkenpas, M.B., Verhamme, I.M., Kvassman, J.-O., and Shore, J.D. 1995. Serpin–protease complexes are trapped as stable acyl-enzyme intermediates. J. Biol. Chem. 270 25309–25312. [DOI] [PubMed] [Google Scholar]
- Lawrence, D.A., Olson, S.T., Muhammad, S., Day, D.E., Kvassman, J.-O., Ginsburg, D., and Shore, J.D. 2000. Partitioning of serpin–proteinase reactions between stable inhibition and substrate cleavage is regulated by the rate of serpin reactive center loop insertion into β-sheet A. J. Biol. Chem. 275 5839–5844. [DOI] [PubMed] [Google Scholar]
- Li, Y., Huang, Q., Zhang, S., Liu, S., Chi, C., and Tang, Y. 1994. Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: Chemical synthesis, refolding and crystallogrpahic analysis of its complex with trypsin. J. Biochem. 116 18–25. [DOI] [PubMed] [Google Scholar]
- Longas, M.O. and Finlay, T.H. 1980. The covalent nature of the human antithrombin III-thrombin bond. Biochem. J. 189 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novokhatny, V.V., Ingham, K.C., and Medved, L.V. 1991. Domain structure and domain–domain interactions of recombinant tissue plasminogen activator. J. Biol. Chem. 266 12994–13002. [PubMed] [Google Scholar]
- O'Malley, K.M. and Cooperman, B.S. 2000. Formation of the covalent chymotrypsin:antichymotrypsin complex involves no large-scale movement of the enzyme. J. Biol. Chem. 276 6631–6639. [DOI] [PubMed] [Google Scholar]
- Patston, P.A., Gettins, P., Beechem, J., and Schapira, M. 1991. Mechanism of serpin action: Evidence that C1 inhibitor functions as a suicide substrate. Biochemistry 30 8876–8882. [DOI] [PubMed] [Google Scholar]
- Pennica, D., Holmes, W.E., and Kohr, W.J. 1983. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature 301 214. [DOI] [PubMed] [Google Scholar]
- Peterson, F.C. and Gettins, P.G.W. 2001. Insight into the mechanism of serpin–proteinase inhibition from 2D [1H-15N] NMR studies of the 69kDa α1-proteinase inhibitor Pittsburgh–trypsin covalent complex. Biochemistry 40 6284–6292. [DOI] [PubMed] [Google Scholar]
- Peterson, F.C., Gordon, N.C., and Gettins, P.G.W. 2000. Formation of non-covalent serpin–proteinase complex involves no conformational change in the serpin. Use of 1H-15N HSQC NMR as a sensitive non-perturbing monitor of conformation. Biochemistry 39 11884–11892. [DOI] [PubMed] [Google Scholar]
- Rubin, H., Wang, Z.M., Nickbarg, E.B., McLarney, S., Naidoo, N., Schoenberger, O.L., Johnson, J.L., and Cooperman, B.S. 1990. Cloning, expression, purification, and biological activity of recombinant native and variant human α1-antichymotrypsin. J. Biol. Chem. 265 1199–1207. [PubMed] [Google Scholar]
- Schechter, I. and Berger, A. 1967. On the size of the active site of proteases. Biochem. Biophys. Res. Commun. 27 157–162. [DOI] [PubMed] [Google Scholar]
- Schechter, N.M., Jordan, L.M., James, A.M., Cooperman, B.S., Wang, Z.M., and Rubin, H. 1993. Reaction of human chymase with reactive site variants of α-1-antichymotrypsin-modulation of inhibitor versus substrate properties. J. Biol. Chem. 268 23626–23633. [PubMed] [Google Scholar]
- Shore, J.D., Day, D.E., Francis-Chmura, A.M., Verhamme, I., Kvassmann, J., Lawrence, D.A., and Ginsburg, D. 1995. A fluorescent probe study of plasminogen activator inhibitor-1. J. Biol. Chem. 270 5395–5398. [DOI] [PubMed] [Google Scholar]
- Spraggon, G.S., Phillips, C., Nowak, U.K., Ponting, C.P., Saunders, D., Dobson, C.M., Stuart, D.I., and Jones, E.Y. 1995. The crystal structure of the catalytic domain of human urokinase-type plasminogen activator. Structure 3 681. [DOI] [PubMed] [Google Scholar]
- Stratikos, E. and Gettins, P.G.W. 1999. Formation of the covalent serpin–proteinase complex involves translocation of the proteinase by more than 70Å and full insertion of the reactive center loop into β-sheet A. Proc. Natl. Acad. Sci. 96 4808–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]