Abstract
Hydrophobins are surface-active proteins produced by filamentous fungi, where they seem to be ubiquitous. They have a variety of roles in fungal physiology related to surface phenomena, such as adhesion, formation of surface layers, and lowering of surface tension. Hydrophobins can be divided into two classes based on the hydropathy profile of their primary sequence. We have studied the adhesion behavior of two Trichoderma reesei class II hydrophobins, HFBI and HFBII, as isolated proteins and as fusion proteins. Both hydrophobins were produced as C-terminal fusions to the core of the hydrolytic enzyme endoglucanase I from the same organism. It was shown that as a fusion partner, HFBI causes the fusion protein to efficiently immobilize to hydrophobic surfaces, such as silanized glass and Teflon. The properties of the surface-bound protein were analyzed by the enzymatic activity of the endoglucanase domain, by surface plasmon resonance (Biacore), and by a quartz crystal microbalance. We found that the HFBI fusion forms a tightly bound, rigid surface layer on a hydrophobic support. The HFBI domain also causes the fusion protein to polymerize in solution, possibly to a decamer. Although isolated HFBII binds efficiently to surfaces, it does not cause immobilization as a fusion partner, nor does it cause polymerization of the fusion protein in solution. The findings give new information on how hydrophobins function and how they can be used to immobilize fusion proteins.
Keywords: Protein adhesion, hydrophobin, supramolecular assembly, Trichoderma reesei
Proteins provide numerous examples of specific surface adhesion interactions, such as that of the mussel adhesive proteins, S-layer proteins, and protein domains that anchor enzymes to their solid substrates (Linder and Teeri 1997; Deming 1999; Sara and Sleytr 2000). The mechanisms by which the proteins adsorb often involve specific protein interactions based on structural features in the protein and sometimes self-assembly or major conformational changes. Depending on the type of surface, various molecular interactions—such as van der Waals interactions, hydrogen bonding, electrostatic interactions, or hydrophobic interactions—may be involved (Haynes and Norde 1994).
One quite recently discovered group of adhesive proteins are the hydrophobins, which have been found only in filamentous fungi, in which they seem to be ubiquitous (Wösten and Wessels 1997; Wösten 2001). They can be secreted into the culture medium from the fungal hyphae and migrate to interfaces where they form thin surface layers. They are also found on cell walls or spore surfaces. Different hydrophobins seem to fulfil a variety of tasks in fungal development and growth. One task that has been demonstrated is the lowering of surface energy of water, which allows the fungal hyphae to penetrate the air-water interface and grow up into the air (Wösten et al. 1999). With some pathogenic fungi, it has been shown that hydrophobins are essential for pathogenicity, apparently by making the hydrophobic cuticle hydrophilic, so that the initial attachment of the fungus can occur (Talbot et al. 1993; Ebbole 1997). In other cases, hydrophobins have structural roles, such as in the lining of air-channels of fruiting bodies of fungi (Lugones et al. 1996; van der Vegt et al. 1996; Wösten et al. 1999; Wösten and de Vocht 2000).
A very high surface activity and a propensity to self-assemble seem to be common denominators for all hydrophobins. Comparison of hydropathy plots forms the basis for dividing the hydrophobins into two classes, I and II (Wessels 1994). Although the two classes share several general properties, they seem to differ significantly in some aspects, such as the solubility of their assemblages. Whereas the class I assemblages are highly insoluble, the class II hydrophobin assemblages and adsorbed surface layers seem to be dissociated more easily, for example, by 60% ethanol, 2% sodium dodecyl sulfate (SDS), or applied pressure (Carpenter et al. 1992; Richards 1993). Although the distinction between classes can be made by comparison of primary structure, no explanation of the difference in properties can yet be made on the amino acid level. The sequences of hydrophobins in general show much variability and have no conserved segments except for eight conserved Cys residues that are arranged in a distinct pattern. In the folded conformation, the Cys residues form disulfide bridges (Yaguchi et al. 1993; Linder et al. 2001). The Cys residue pattern and the hydropathy plots together indicate that the hydrophobins consist of two domains (Wösten and Wessels 1997).
One hydrophobin that has been studied much is SC3 from the fungus Schizophyllum commune, which belongs to class I. The layer formed by the hydrophobin SC3 has been characterized, and has the property of changing the surface hydrophobicity so that it turns a hydrophilic surface hydrophobic and a hydrophobic surface hydrophilic (Wösten et al. 1993, 1994, 1995; Martin et al. 2000). The layer is easily visualized by electron microscopy and is characterized by its tightly packed rodlet pattern; therefore, it is often called a rodlet layer (Wessels 1997). The rodlet layer is very stable, and only very harsh chemicals such as pure trifluoroacetic or formic acid can dissolve it. For example, heating in a solution of 2% SDS does not affect the layer (de Vries et al. 1993). It has also been shown that large conformational changes are associated with its assembly and adsorption (de Vocht et al. 1998; Wösten and de Vocht 2000). Rodlets have been observed with other class I hydrophobins, but no rodlet type surface structures have been observed with class II hydrophobins (Wösten and de Vocht 2000).
In this work, we have studied the surface adhesion properties of two class II hydrophobins, HFBI (7.5 kD) and HFBII (7.2 kD), from the filamentous fungus Trichoderma reesei (Nakari-Setälä et al. 1996, 1997). The two hydrophobins were each linked as C-terminal fusions to the catalytic domain of a hydrolytic enzyme, endoglucanase I (EGIc) from T. reesei. The adhesion behaviors of the two fusion proteins were then compared, and the isolated EGIc (40 kD) was used as a reference (Srisodsuk et al. 1997). EGIc from T. reesei is, like many cellulolytic enzymes, a modular protein. It consists of a catalytic domain in the N terminus (368 amino acids) and a cellulose-binding domain in the C terminus (36 amino acids). The two domains are connected by a glycosylated linker (33 amino acids). The EGIc-HFBI fusion protein contained the full linker but at position 403, where the cellulose-binding domain begins, the 75 amino acid HFBI (Nakari-Setälä et al. 1996) was inserted instead of the cellulose-binding domain. Analogously, the EGIc-HFBII has the 71-amino-acid HFBII (Nakari-Setälä et al. 1997) inserted instead of the cellulose binding-domain. The EGIc reference protein used in this study is a truncated form of EGI that lacks the C-terminal cellulose-binding domain (Srisodsuk et al. 1997). We have earlier shown that the EGIc-HFBI fusion protein can be efficiently produced and secreted to the culture medium by T. reesei (T. Nakari-Setälä and M. Penttilä, in prep.). The fusion protein was shown to separate well in surfactant two-phase extraction owing to its hydrophobin fusion tag (Linder et al. 2001; Collen et al. 2002).
One advantage of using fusion proteins was that we were able to use the enzymatic activity of EGIc for assaying the functional adsorbed protein, and that the isolated EGIc could be used as a control. The binding of the two fusions, the free hydrophobins, and the control protein was measured by three techniques, surface plasmon resonance (SPR), quartz crystal microbalance (QCM), and measuring the enzymatic activity of the adsorbed protein.
Results
Purification and characterization of the fusion proteins
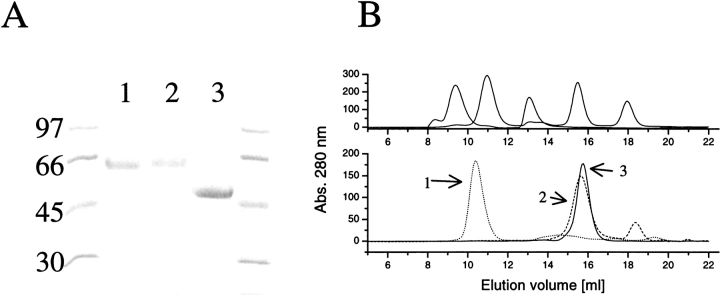
The purification of the fusion proteins gave single bands in SDS-PAGE as shown in Figure 1A ▶. In Western blots, the proteins showed binding to their respective polyclonal antibodies (anti-HFBI, anti-HFBII, and anti-EGI). Figure 1B ▶ shows the results from the gel filtration studies. The EGIc-HFBI protein had a retention time that placed it between the 669- and 440-kD standards. A decamer of EGIc-HFBI would have a calculated mass of 500 kD, which would be in agreement with the data. It could, however, be thought that a multimer with the HFBI pointing inward in a micelle-like way would not have a dense core and would therefore elute earlier than predicted from its actual mass. The EGIc-HFBI sample shown in Figure 1B ▶ was injected at a concentration of 2 mg/mL, and diluting the sample to 0.4 mg/mL did not cause any change of retention volume (data not shown).
Fig. 1.
(A) SDS-PAGE of the purified proteins EGIc-HFBI (lane 1), EGIc-HFBII (lane 2). and EGIc (lane 3). Molecular-weight markers are indicated on the side. (B) Size-exclusion chromatography of EGIc-HFBI, EGIc-HFBII, and EGIc using a Superdex 200 column. (Top) The elution volumes of the standards (669, 440, 158, 43, and 13.7 kD). (Bottom) The control EGIc (peak 3) elutes close to the EGIc-HFBII (peak 2) fusion protein, but EGIc-HFBI (peak 1) is a large multimer, putatively a decamer.
The EGIc-HFBII eluted with a retention volume corresponding to a monomer, but slightly earlier than the EGIc control corresponding to its increased mass owing to the fusion partner.
For measuring specific activities of the proteins, the soluble fluorogenic substrate 4-methylumbelliferyl β-d-cellobioside (MUG2) was used. Liberated 4-methylumbelliferrone (MU) was calculated using standard curves. At 21°C the calculated specific activities were 3.60 (±0.3) nmole/s/mg for EGIc, 3.4 (±0.3) nmole/sec/mg for EGIc-HFBI and 3.0 (±0.3) nmole/sec/mg for EGIc-HFBII.
Initial adsorption tests
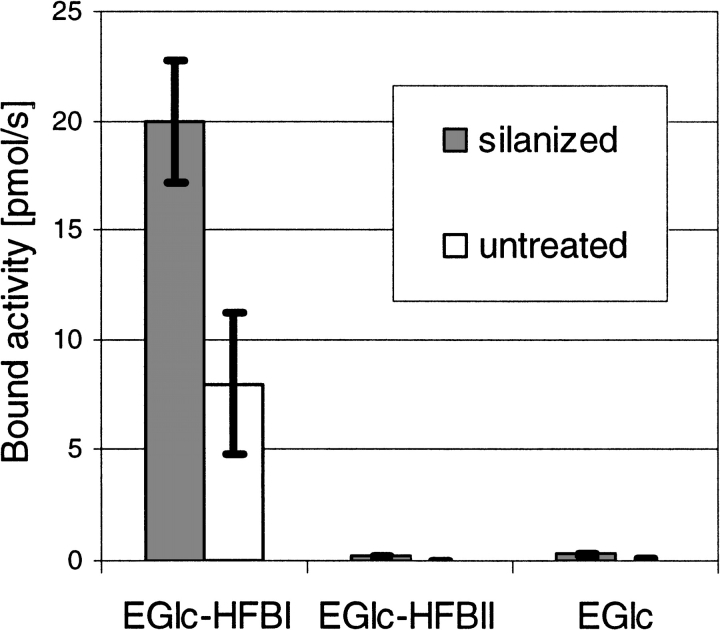
In the enzymatic activity measurements, we used the activity of EGI toward MUG2 to assay the protein. Neither hydrophobin shows any enzymatic activity on this or any other known substrate. The effect of the HFBI domain for immobilization was initially established by testing how much of EGIc-HFBI, EGIc-HFBII, and EGIc was bound to either silanized or untreated glass rods (0.2 g/L protein, 120 min, 50 mM Na-acetate buffer at pH 5, 21°C). The experiment was performed by first incubating the different types of surfaces with a protein solution, washing with buffer, and measuring bound and residual protein by the enzymatic activity of the EGIc domain. The results presented in Figure 2 ▶ show that the combination of EGIc-HFBI and the hydrophobic surface resulted in the most significant binding, whereas EGIc-HFBI bound less to nonsilanized glass. The corresponding test with the EGIc-HFBII fusion did not give results that differed significantly from the reference EGIc, indicating either that the EGIc-HFBII fusion protein does not bind or that the protein is bound very loosely and easily washed off. It was noted that especially the binding of the EGIc-HFBI to untreated glass was sensitive to how extensively the surface had been cleaned.
Fig. 2.
Bar graph showing the adhesion of EGIc-HFBI to a silanized and an untreated glass surface in comparison with the EGIc control. The adhesion is measured as bound enzymatic activity of the fusion protein and shows that the fusion with HFBI causes the protein to immobilize on a hydrophobic surface. Under the same conditions, EGIc-HFBII does not bind and is indistinguishable from the control.
Adsorption studied by surface plasmon resonance
Using SPR measured with a Biacore sensor, the kinetics of the binding was investigated. In this method, a liquid is continuously pumped over a surface, and bound mass can be followed as response units (RUs) in a sensogram, which shows the RU as a function of time or volume (for a discussion of RU and SPR response to bound mass, see Alfthan 1998; Hashimoto 2000). The rate of change in RUs thus related to the kinetics of binding, and the value of RUs is directly proportional the bound mass. In Figure 3A ▶, an example of the adsorption behavior of the two fusions and the control on an alkane thiol–coated gold surface are shown. In this example the EGIc control was first injected, then the EGIc-HFBII fusion, and finally the EGIc-HFBI fusion. EGIc has a negligible adsorption and does not show any response signal at this scale of magnification. EGIc-HFBII shows a fast binding during the sample injection, and a fast desorption during the wash step, whereas the HFBI fusion shows a somewhat slower binding and a very slow desorption. In other experiments, the order of injection and the number of injections was varied. When the HFBI-fusion was injected on a fresh surface, the level of bound protein was higher by the same amount that the HFBII fusion bound in the example in Figure 3A ▶, and correspondingly the HFBII fusion did not bind if HFBI fusion was first injected. We tested desorption with the surfactants SDS, Tween 20, C12–18EO5, and octyl-glucoside for regeneration of the surface, and found that a 5 μL injection of SDS at a concentration of 1 g/L was able to remove the bound protein. The other surfactants did not significantly reduce the level of bound protein, although surfactant injection often resulted in an unstable signal. After regeneration, the binding of the next round of protein was similar to previous ones, but sufficiently different to not allow curve fitting and calculation of binding parameters by the Biacore software. A probable cause for this is that the surface was altered by the regeneration.
Fig. 3.
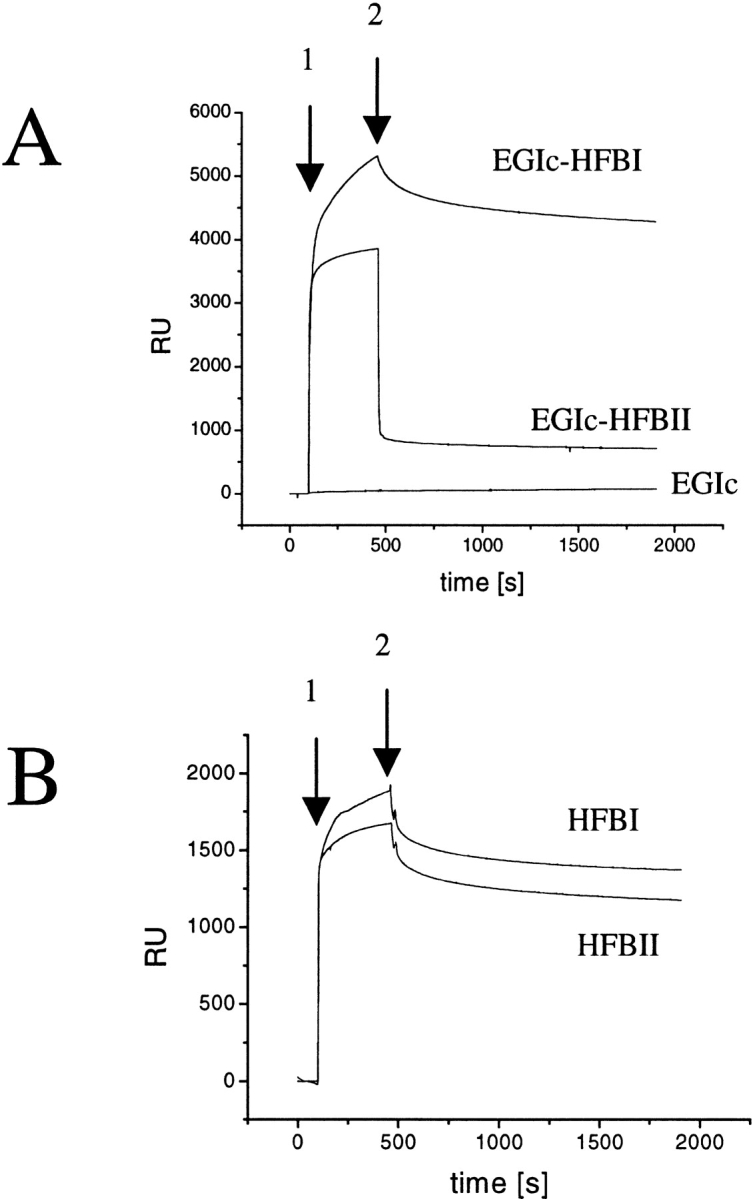
(A) Sensograms of EGIc, EGIc-HFBII, and EGIc-HFBI binding to an alkylated gold surface measured by surface plasmon resonance (SPR). The arrow 1 indicates when sample injection started; arrow 2, when sample injection stopped and buffer wash started. The EGIc control does not show any binding to the surface, EGIc-HFBII adsorbs during the injection but desorbs during the wash step, whereas EGIc-HFBI adsorbs during injection and shows a slow desorption during the wash. (B) Sensograms of native HFBI and HFBII binding to an alkylated gold surface measured by SPR. The free hydrophobins do not show the differences in binding seen when the hydrophobins are parts of fusion proteins.
In SPR, a phenomenon called the buffer or bulk effect is seen when the sample solution has a different refractive index from the running buffer. This means that a signal (RU) is seen even though there is no binding (Hashimoto 2000). In this work, the bulk effect of the EGIc control protein was negligible compared with the signal produced by the two fusion proteins. It can therefore be concluded that the RU signal of EGIc-HFBII during injection is caused by a real interaction, and not a bulk effect, but both the off- and on-rates are fast.
The EGIc-HFBI sample showed a slower on-rate binding and a much slower desorption rate than did the EGIc-HFBII sample. The bound HFBI fusion did not desorb during extended overnight buffer washes >5%. It is noteworthy that the HFBII fusion attains a very stable but low level of bound protein after the initial quick desorption step. After four subsequent injections the RU level was ∼4500 for EGIc-HFBI and ∼900 for EGIc-HFBII. The level did not rise from this during subsequent injections, even at protein concentrations >250 μg/mL. Because the response in SPR is related to the bound mass, it is possible to estimate the amount of bound protein, by the relation that ∼1000 RU corresponds to 1 mg/m2, leading to an estimate of 4.5 mg/m2 of bound protein.
Additional tests using isolated hydrophobins HFBI and HFBII are shown in Figure 3B ▶. The curves obtained were not entirely smooth and, from time to time, gave an unstable response signal. Nonetheless, it can be seen that both proteins associate with the hydrophobic surface, but HFBII had a slightly lower binding. Again, meaningful curve fitting was not possible because of the small variability in repeated runs, which was probably caused by slightly changed surface properties by the SDS used in regeneration.
Adsorption measured by quartz microbalance
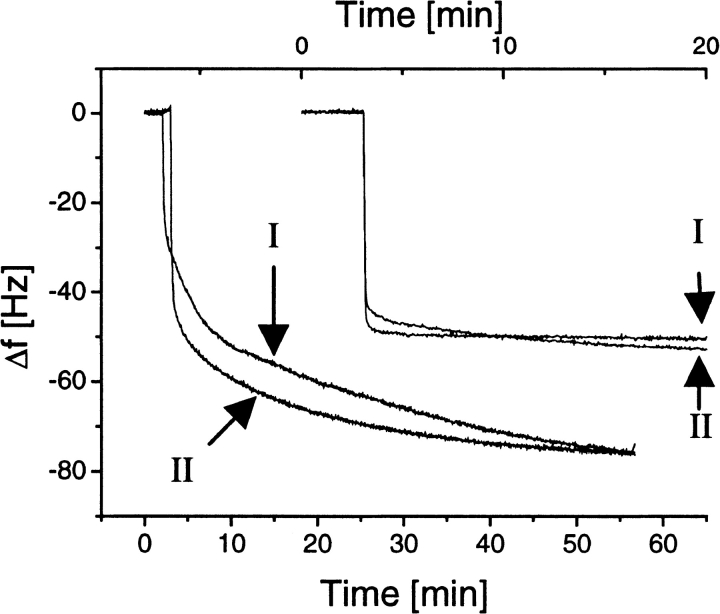
The quartz crystal microbalance (QCM) gives two types of information. The change in resonance frequency, Δf, is directly proportional to the adsorbed mass, and the dissipation change, ΔD, gives information about the rate of dampening of the signal, which is dependant on the structure of the adsorbed layer. When a voltage is applied over a quartz crystal, it will oscillate, and the frequency of the oscillation is dependent on the mass of the crystal. The adsorbed mass can be calculated from the change in resonant frequency by the Sauerbrey relation Δm = −CΔf/n, where Δm is the adsorbed mass, n is the overtone number (n = 1 for 5-MHz measurements and n = 3 for 15-MHz measurements), and C is the mass sensitivity constant (17.7 ng/cm2/Hz; Sauerbrey 1959; Hook et al. 1998c). Figure 4 ▶ shows the adsorption of HFBI and HFBII to untreated and silanized SiO2 at a protein concentration of 0.25 mg/mL. Subsequent injections at twice the concentration confirmed that saturation had been achieved. Both hydrophobins adsorb in a similar manner to each surface. There is, however, a clear distinction in the shape in the curves, indicating different modes of adsorption to the hydrophilic and hydrophobic surface. The adsorption to the hydrophobic surface is rapid, reaching the maximum level almost instantaneously, whereas the adsorption to the hydrophilic surface is slower but adsorbs with a higher capacity. A Δf of 50 Hz corresponds to 2.9 mg/m2, and 75 Hz corresponds to an absorbed mass of 4.4 mg/m2. It is possible that bi- or multilayers are formed in the case of the hydrophilic surface, whereas a thin monolayer is formed at the hydrophobic surface. If multilayers are formed, they probably are fairly rigid because the change in dissipation was similar in all cases and did not exceed 1.5 × 10−6.
Fig. 4.
Quartz crystal microbalance measurement of free hydrophobin binding to silanized and untreated quartz surface. The two curves at the upper right show binding of HFBI (I) and HFBII (II) to the silanized surface, and the two curves at the lower left show binding of the same proteins to the untreated surface. The frequency shifts are shown on the same x-axis, but the time scale has been shifted for clarity, so that the upper x-axis shows the time for binding to the silanized surface and the lower x-axis shows the binding to the untreated surface.
Figure 5A ▶ shows adsorption curves of the two fusion proteins and the control to a silanized quartz surface. EGIc-HFBI gives a maximal Δf of 240 Hz, which corresponds to an adsorbed mass of 14 mg/m2. In this measurement, the free concentration of 0.25 mg/mL was saturating, as checked by titration with higher concentrations. Because the QCM has a sample cell in which the sample is static and not flowing, we see a situation for the HFBII fusion that corresponds to the injection phase in SPR. The results showed that under an equilibrium situation the EGIc-HFBII fusion does bind to the hydrophobic surface, but not as efficiently as the EGIc-HFBI fusion.
Fig. 5.
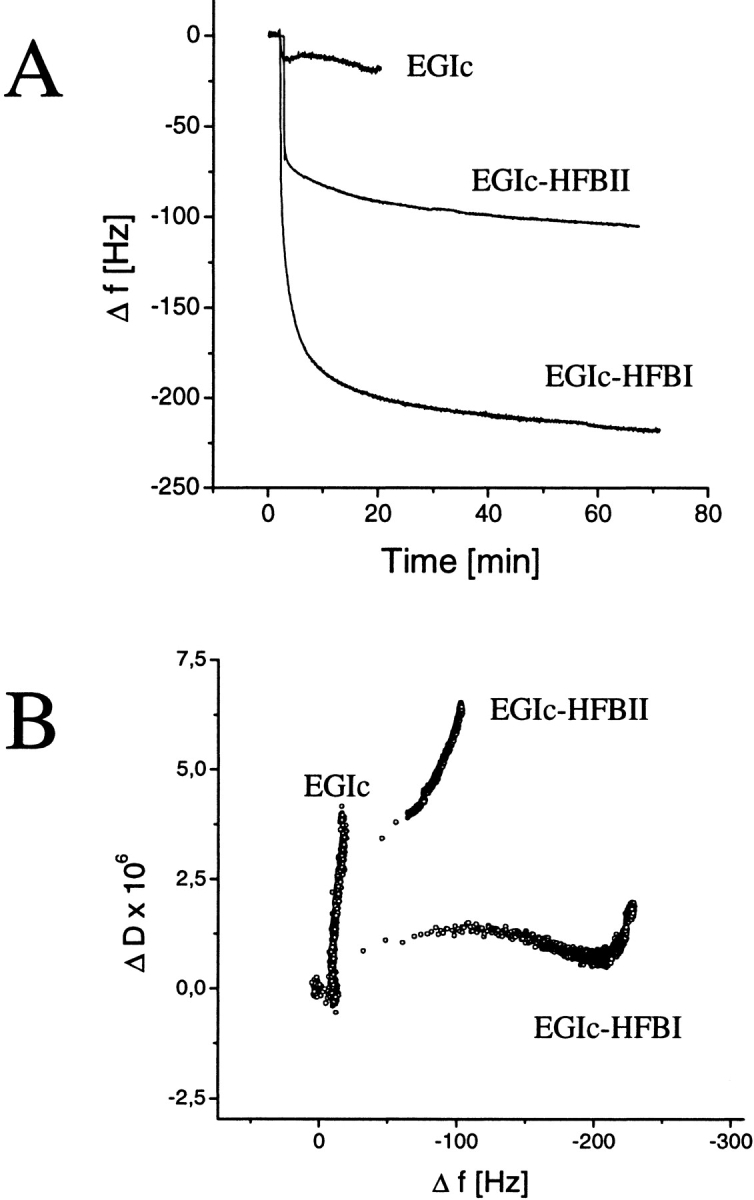
Quartz crystal microbalance measurement of EGIc and fusion protein binding. (A) Frequency shifts as a function of time during the binding of EGIc, EGIc-HFBII, and EGIc-HFBI to a silanized quartz surface. Measurements were performed in static conditions. (B) The change in energy dissipation value as a function of Δf is shown. The data show that especially the EGIc-HFBI fusion forms a dense and rigid layer.
Plotting of ΔD as a function of Δf gives information about the rigidity of the layer as a function of the adsorbed amount of protein. A small ΔD (slow dampening) indicates a rigid adsorbed layer, and a large ΔD indicates an adsorbed layer that efficiently adsorbs the vibrational energy on adsorption. From Figure 5B ▶, we see that the EGIc layer associates very loosely with the surface and gives a very steep curve, with a big change in D for a very little change in f. The EGIc-HFBI binding shows a very large change in Δf with a corresponding small change in ΔD, which indicates that a rigid layer is formed (Hook et al. 1998a–c). The HFBII fusion shows an intermediate behavior. From the plot, we can also see that the EGIc-HFBI adsorption is biphasic, with a small initial change in D that then becomes steeper closer to saturation. These data indicate that the rigidity of the bound layer increases as the layer becomes complete, whereas the last molecules to be bound as saturation is approached are more loosely attached. Usual dissipation values for globular proteins are ∼1 × 10−6 for a 20 to 40 change in f when they adsorb as a rigid layer (Hook et al. 1998a–c). In the literature, higher values have been reported, for example, in the case of immunoglobulins bound to a layer of antigen, because this type of binding leaves the long Fc part hanging out into the solution.
Adsorption analyzed by enzymatic activity
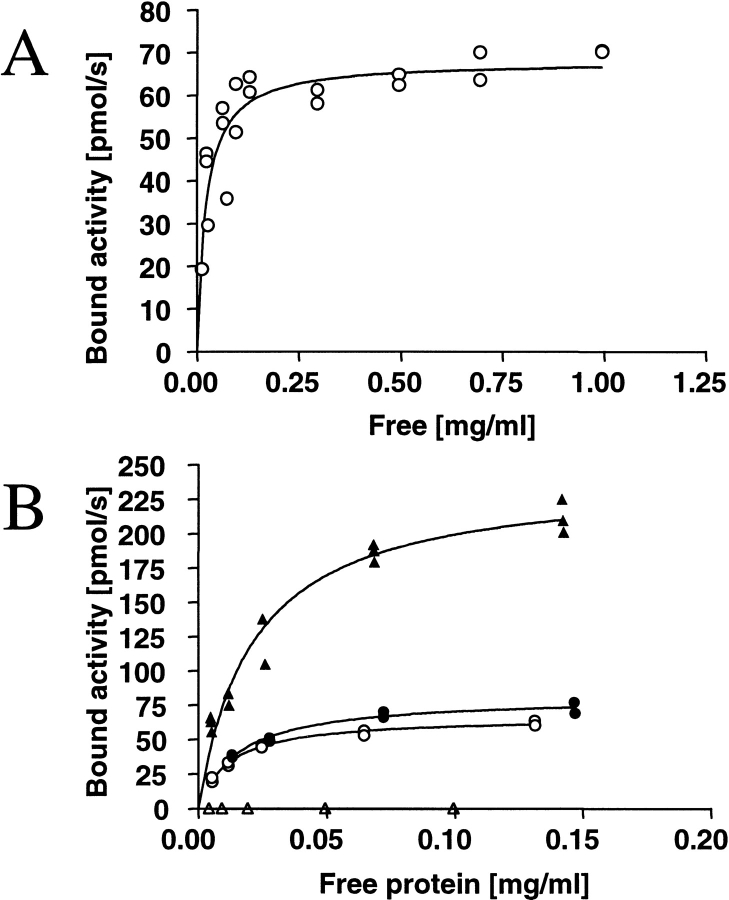
To establish the adsorption capacity, a binding isotherm was determined (Fig. 6A ▶). The isotherm was modeled with a first-order Langmuir equation, which gave a fit with R2 = 0.77. A maximum adsorption was reached with catalytic activity of 70 pmole/sec bound to the surface. For quantification, we used the specific activity for the protein, 3.4 nmole/sec/mg (21°C). From this, it can be estimated that ∼14 mg/m2 protein was immobilized on the silanized glass rods. From the isotherm data, it could be estimated that the dissociation constant (Kd) of EGIc-HFBI on silanized surface was 0.44 μM (21.8 μg/mL; Bmax = 2.8 μmole/m2).
Fig. 6.
Binding isotherms measured using the enzymatic activity of EGIc for protein quantification. (A) Binding isotherm of EGIc-HFBI fusion protein to silanized glass, measured as bound enzymatic activity of the fusion protein. A first-order Langmuir isotherm is fitted on the data giving a maximum bound activity of 2.8 μmole/m2 (67 pmole/sec) and a Kd of 0.44 μM (21.8 μg/mL). (B) Corresponding isotherms showing the binding of EGIc-HFBI to Teflon and polystyrene compared with silanized glass (polystyrene, filled triangles; Teflon, filled circles; silanized glass, open circles). The control EGIc binding to polystyrene (open triangles) is plotted, and it shows that EGIc does not bind to polystyrene in its hydrolytically active form.
Partial isotherms for the binding of the EGIc-HFBI fusion protein to Teflon and polystyrene in comparison to silanized glass are shown in Figure 7B ▶. In binding experiments with EGIc and polystyrene, no measurable enzyme activity was adsorbed.
Fig. 7.
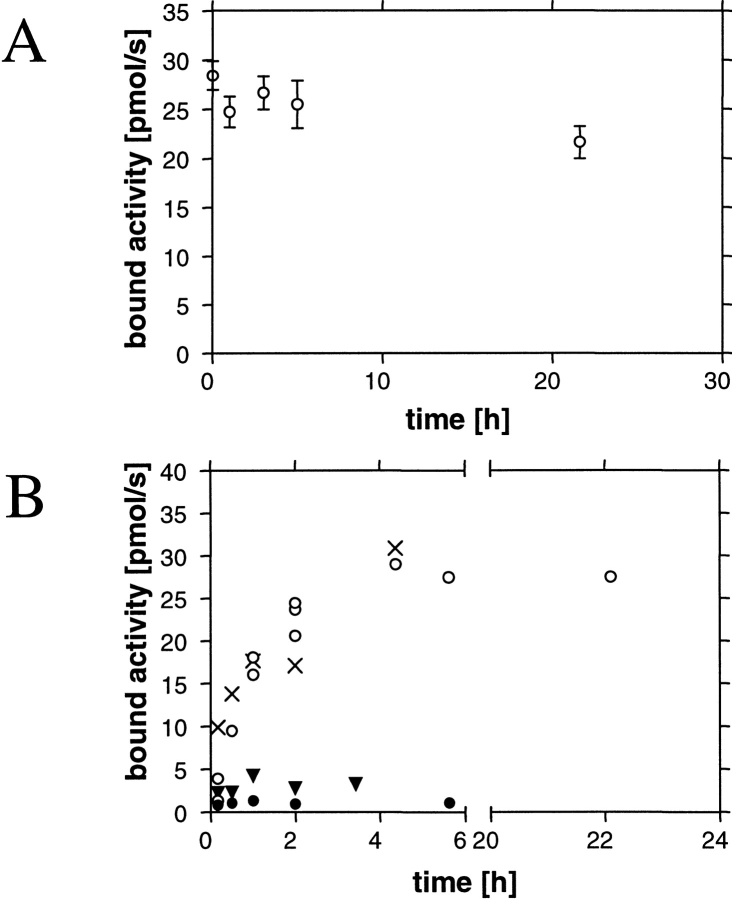
Kinetics of the binding of EGIc-HFBI to and its desorption from silanized glass. (A) Adsorption rate without (open circles) and in the presence of a 10-fold molar excess of free HFBI (filled circles) or HFBII (filled triangles). Bovine serum albumin at a sixfold excess does not effect the binding of EGIc-HFBI (X). (B) Desorption of EGIc-HFBI from a silanized glass surface by washing with an excess of buffer.
The temperature dependency of the affinity was tested by determining partial binding isotherms at 5°C, 21°C, and 35°C and comparing the initial parts of the isotherms. No significant effect of temperature was noted. Effects of changing ionic strength were determined by adding NaCl or (NH4)2SO4. A high salt concentration (1 M) resulted in a slight increase (10% to 15%) in affinity. Experiments using buffers at pH 3, 5, and 7 showed a greater effect, 25% better affinity at pH 5 than at pH 3 and 40% better at pH 5 than at pH 7 (data not shown).
The binding kinetics of EGIc-HFBI to and desorption from silanized glass are shown in Figure 7, A and B ▶. The binding is very slow and requires >5 h to reach a steady value. Simultaneously adding HFBI or HFBII in competition experiments resulted in the fusion protein being competed out as shown in Figure 7A ▶, in which both free hydrophobins were added at 10-fold molar excess. HFBI competed out the EGIc-HFBI at a corresponding ratio, whereas HFBII was slightly less efficient. The addition of a sixfold excess (w/w) of bovine serum albumin did not block the binding of the EGIc-HFBI fusion to the silanized glass.
In the desorption tests (Fig. 7B ▶), silanized glass rods that had been incubated with EGIc-HFBI were washed and then incubated for different times in a large excess of buffer (50 mM Na-acetate at pH 5). A slight desorption or inactivation can be noted over 24 h. Desorption was also studied by adding 0.5% SDS or Tween 20 to the wash buffers. Using SDS, the activity was washed away instantaneously, and using Tween 20, the bound activity was reduced to 50% in 1 to 2 h. In control experiments using soluble EGI to test the stability in the surfactants, the activity was not affected, showing that the enzyme was stable under the conditions.
Discussion
In this work, we have studied the adhesive properties of two fungal hydrophobin proteins, HFBI and HFBII from T. reesei, as partners in fusion proteins and as isolated proteins. Based on our initial observations and published observations of similar proteins, there were indications that the isolated proteins would have interesting adhesive properties. Another aim was to study if the hydrophobins could be used for protein immobilization. To test this hypothesis, two recombinant fusion protein constructs were made, where the 40-kD catalytic domain of endoglucanase EGI (EGIc) from T. reesei was fused to either HFBI (7.5 kD) or HFBII (7.2 kD) through a linker region. Characterization was made by comparing the properties of the two fusion proteins and, as a control, the isolated enzyme domain (EGIc). The types of hydrophobic surfaces were Teflon, polystyrene and alkylated glass (or quartz), or gold. The adsorption of isolated hydrophobins on hydrophilic and hydrophobic surfaces by QCM, on hydrophobic surfaces by SPR, and indirectly in competition experiments with EGIc-HFBI were made as a comparison. The main result was that HFBI can successfully be used to stably immobilize fusion proteins to hydrophobic surfaces, retaining the activity of the fusion partner. The HFBII fusion protein, on the other hand, binds but has a fast off-rate that does not allow an efficient immobilization of the fusion protein. Comparison of the free HFBI and HFBII binding by QCM and SPR showed that both hydrophobins have similar adsorption properties and kinetics on hydrophobic and hydrophilic surfaces. HFBII was also able to compete for binding with the EGIc-HFBI fusion protein. Therefore, it seems that although HFBII binds efficiently to surfaces, it does not retain this property as a fusion protein. Further studies involving the testing of different types of fusion proteins—such as N-terminal positioning, other fusion partners, or linker optimization—could of course yield fusion proteins with functional HFBII domains.
Characterization by SDS-PAGE, Western blotting, and activity measurements all showed that both fusion proteins were active, were of correct size, and reacted with their corresponding antibodies. Characterization by size-exclusion chromatography, however, revealed an other functional difference between the HFBI and HFBII fusions. We have previously shown that both HFBI and HFBII exist as tetramers in solution, and that the equilibrium can be shifted toward smaller aggregates by dilution of the sample (Torkkeli et al. 2002). HFBI seemed to dimerize at low concentrations, but HFBII seemed to exist as monomers at low concentrations. In this work, we showed that EGIc-HFBI exists as a multimer, putatively a decamer, in solution. The EGIc-HFBII, however, elutes very close to the EGIc control. It therefore seems that in the case of EGIc-HFBII, the HFBII is unable to multimerize and possibly folds back toward the EGIc domain. This result would indicate a link between adhesion properties and solution multimerization.
The reason for the poor immobilization could then be that the multimerization in some other form is involved in surface immobilization by, for example, a self-assembly process, or that the surface interacting face of HFBII is shielded by an interaction with the EGIc domain. However, the QCM and SPR results indicated that the HFBII domain in the EGIc-HFBII fusion is at least partly functional, because the fusion protein clearly interacted with the surface differently than the control protein. The reason for the different effects of the linkage could be owing to differences in the N-terminal parts of the hydrophobins. Comparison of the sequences (Linder et al., 2001) shows that the N terminus of HFBI is five amino acids longer than that of HFBII, which could support this idea. The absence of tailing of the EGIc-HFBI peak in the chromatogram also shows that the time scale of multimer-equilibrium is slow.
The rate of adsorption of EGIc-HFBI from a solution in the kinetic measurements by enzymatic activity is very slow, as shown in Figure 7A ▶. This implies the possibility that there is some other factor involved that slows the binding down. One possible explanation is that the multimers that EGIc-HFBI forms in solution need to dissociate, for example, if the protein only binds to the surface as a monomer. This would mean that the surface adhesion is slow, although it is thermodynamically favorable. The observed rate of adsorption would then be determined by the rate at which the complex breaks down and reforms. It is interesting to note that dilution of the EGIc-HFBI sample did shift the aggregation toward smaller complexes, as it did with the isolated HFBI. Apart from the size-exclusion chromatography, this explanation is supported by the observation that visible aggregates form in a solution containing EGIc-HFBI (but not EGIc-HFBII) under vigorous shaking. The aggregates dissolve readily by, for example, a quick sonication or even by applying pressure in a syringe. The experimental setup in both SPR and QCM seems to give a somewhat faster initial adsorption rate, although it was not possible to find a good enough mathematical fit to any model for the binding using the Biacore modeling software so that rates could be calculated. The faster adsorption in SPR might be caused by the shear force or the pressure caused by the flow in the measuring cell. This might decrease the degree of association of the protein and thus increase the rate of binding. From the QCM data, we observe that binding is seen during the first hour after sample application. However, prolonged incubations of several hours did not result in significant additional binding.
Desorption of bound fusion protein was studied in two types of experiments, in the enzymatic activity and the SPR experiments. Desorption as studied by the bound enzymatic activity is slow for the HFBI fusion, and in the case of the HFBII fusion, no binding was noted at all. In the SPR experiments, the HFBII fusion off-rate is fast, but a residual amount stays very stably bound, indicating that there could be two types of bound protein. Because the corresponding small residual level was not detected after washes in experiments using enzymatic activity, it is difficult to make conclusions about the nature of this residual level. In the corresponding SPR experiment with EGIc-HFBI, there is a small initial quick desorption, followed by a very slow stage.
The adsorbed HFBI-fusion protein layer is most probably a monolayer that completely covers the surface. This is indicated by the fact that a well-defined saturation level was achieved in the isotherm shown in Figure 6A ▶, and in the SPR and QCM experiments, saturating adsorption levels form as well. The maximum bound level of EGI-HFBI corresponded to 14 mg/m2; for the isolated hydrophobins, 2.9 to 4.4 mg/m2, which is in the range expected for monolayers, considering the molecular weights. In the QCM experiments, the ΔD is low, which indicates that the layer is thin and relatively rigid. Because formation of assemblages is one of the observed general properties of hydrophobins, there was a possibility that thick multilayers would have been observed. The estimate of bound protein by SPR is lower than that by the other methods, which may be caused by differences in the surface chemistry. The SPR method is also the least direct of the used methods to quantify the bound mass.
One potential advantage that the fusion protein approach provides in protein immobilization is seen in Figure 6B ▶. It is unlikely that the control EGIc actually does not bind at all to polystyrene, as the bound activity indicates. Rather, the situation is probably that any control EGIc that binds becomes denatured and thus nonfunctional. In contrast, when the fusion protein binds, the molecule is probably tethered in such a fashion that the EGIc part points outward and does not denature on the surface.
It is noteworthy that both the fusion proteins and isolated hydrophobins are water soluble up to at least 10 mg/mL, although their hydrophobic interactions seem to be important for the binding. In membrane proteins, it has been found that the Trp and Tyr residues are often found in the membrane water interface, because they have an amphiphilic character (Killan and von Heijne 2000). It has also been shown that adding Trp to a protein made it bind more efficiently to hydrophobic surfaces than the corresponding Ile additions did (Malmsten and Veide 1996). It is therefore noteworthy that HFBI has only one Tyr and no Trp residues, and HFBII has none of either. Both, on the other hand, have Phe residues, but Phe is not amphiphilic; instead, it has a clearer hydrophobic character. The Cys pattern and hydropathy plots indicate that the protein consists of two domains, possibly with a hinge region in between (Wösten and Wessels 1997; Linder et al. 2001). This indicates that a possible mechanism for multimerization is domain swapping (Bennett et al. 1995), and surface adhesion could also involve exposure of hydrophobic patches by domain movement.
It is interesting in the future to try to experimentally link adhesion to self-assembly in structure-function studies. We note that these hydrophobins easily self-associate, forming, for example, multimers in solution or microscopic fibrils (Torkkeli et al. 2002). This leads to the suggestion that that the surface adhesion also involves self-assembly, and not only a simple amphiphilic interaction as seen with, for example, surfactants, but so far we have not been able to find structural features on the adsorbed surfaces with Atomic Force Microscopy (data not shown). We are thus still unable, in lack of detailed structural data on hydrophobins, to suggest a molecular mechanism by which the adhesion occurs.
Materials and methods
Recombinant DNA constructions
The cloning of EGIc is described in Srisodsuk et al. (1997), and the cloning of EGIc-HFBI is described in Penttilä et al. (1987; PCT/FI00/00249). To make the strain producing EGIc-HFBII, we used the vector for EGIc-HFBI (pMQ113), which carries the gene cassette under the strong cbh1 promoter, as a template. The EGIc-HFBII was constructed by replacing the hfb1 sequence in pMQ113 with the hfb2 sequence. First, the hfb2-coding region from Ala-16 to the STOP codon was amplified with PCR using the primer pair 5`-CGG AGG AGC TCG ACG ACT TCG AGC AGC CCG AGC TGC ACG CAG GCT GTC TGC CCT ACC GG (sense) and 5`-TCA TTG GAT CCT TAG AAG GTG CCG ATG GC (antisense) and the vector phfb2 as a template (Nakari-Setälä et al. 1997). The underlined sequence in the sense primer encodes for amino acids 413–425 in EGI. The PCR fragment was digested with SacI and BamHI and ligated to pMQ113, which had been similarly digested. The resulting fungal expression vector, pTNS32, carried the EGIc-HFBII cassette under the regulatory control of the cbh1 promoter and terminator sequences. Before fungal transformation, pTNS32 was digested with EcoRI and SphI to release the expression cassette.
T. reesei strain QM9414 was transformed according to (Penttilä et al. 1987) using 10 μg of digested pTNS32 together with 4 μg of pARO21 (Aro et al. 2001), which confers resistance to hygromycin. Transformants were streaked three times on selective medium, then transferred to potato dextrose agar for sporulation. Spore suspensions were plated out on selective medium to obtain single spore colonies for further analysis. Hygromycin-positive transformants were screened for production of EGIc-HFBII fusion protein by analyzing culture medium samples from microtiter plate cultivations on cellulose-containing medium with HFBII specific antibodies.
Protein production and purification
EGIc was produced and purified by ion exchange chromatography as in Srisodsuk et al. (1997). For EGIc-HFBII, strain VTT-D-02790 was grown in shake flasks in minimal medium (Penttilä et al. 1987) supplemented with 3% Solka floc cellulose (James River Corporation) and 1.5% complex grain-based nitrogen source for 7 d. EGIc-HFBII was purified from the culture medium by first desalting on a Bio-Rad P6 (Bio-Rad) column with 10 mM acetate buffer (pH 5.0). The desalted fraction was then loaded on a Resource Q column (Amersham Pharmacia) and eluted with a linear gradient of zero to 0.2 M NaCl in 10 mM acetate (pH 5.0). As a final step, the EGIc-HFBII peak fraction was then loaded on a phenyl Sepharose column(Amersham Pharmacia) after first adding (NH4)2SO4 to a final concentration of 0.5 M. Elution was with 10 mM acetate (pH 5.0). The purification was followed by running SDS-PAGE and Western blotting using polyclonal antibodies raised against EGI, HFBI, and HFBII and using endoglucanase activity toward MUG2.
Bioreactor production of EGIc-HFBI was performed with strain VTT D-99702 (Collen et al. 2002; T. Nakari-Setälä, in prep.) with 40 g/L lactose in minimal medium for 4 d in a LF7 fermenter (Chemap). EGIc-HFBI was purified similarly as EGIc-HFBII, except that initially a surfactant two-phase extraction was made and the Phenyl Sepharose step was omitted. The two-phase extraction was made by adding 2% of the surfactant Agrimul NRE 1205 (Cognis) to the culture supernatant, collecting the surfactant phase, and extracting the EGIc-HFBI protein from it by addition of an equal volume of isobutanol. Desalting and ion exchange were preformed as described above.
For adsorption experiments, the buffers were exchanged with 10-DG columns (Bio-Rad). Protein concentrations were determined by adsorption at 280 nm using a molar extinction coefficient of 61,180 M−1/cm for EGIc-HFBI and 60,020 M−1/cm for EGIc and EGIc-HFBII.
Size-exclusion chromatography
A Superdex 200 column (Amersham Pharmacia Biotech) was used for size-exclusion chromatography. All proteins were injected in a volume of 100 μL and a concentration of 2 to 3 mg/mL. In addition, EGIc-HFBI was also analyzed at 0.4 g/L. The running buffer was 50 mM Na-acetate buffer (pH 5.0) containing 0.2 M NaCl. The flow was 0.5 mL/min, and detection was by UV at 280 nm. The molecular weight standards used were thyroglobulin (669 kD), ferritin (440 kD), aldolase (158 kD), ovalbumin (43 kD), and ribonuclease (13.7 kD). Molecular weight standards were obtained from Amersham Pharmacia Biotech.
Binding analyzed by enzymatic activity
Binding of EGIc-HFBI and the control EGIc to different surfaces was initially tested by immersing glass rods into 0.5 mL of 0.2 mg/mL protein solution in Eppendorf tubes. As standard conditions for adsorption, 50 mM Na-acetate (pH 5.0) and 21°C were used. The rods were made of either glass or Teflon and had a diameter of 2 mm, with rounded tips and a length of ∼3 cm. Using the same volume in the test tubes, the exposed surface of the rods was kept constant. The glass rods were washed with detergent and ethanol, rinsed with water, and either used as such or silanized with dimethyldichlorosilane (Sigma). Teflon rods were obtained from Cowie Technologies. At appropriate times, the rods were lifted out of the solution and placed in 5 mL of 50 mM Na-acetate buffer (pH 5.0) for 2 min with gentle shaking. The wash was then repeated. The amount of bound enzyme activity was assayed by immersing the rod into 0.5 mL of 0.5 mM MUG2 (M6018; Sigma) for 5 min. The reaction was terminated by lifting out the rod and adding 0.5 mL 1 M NaCO3. Measurement of activity in liquid samples were performed similarly. Liberated MU was assayed by fluorescence on a 96-well reader using a 355-nm excitation filter and 430-nm emission filter. (Multiscan, Labsystems). To make standard curves for the activity measurements, the pure reaction product MU (M 1381; Sigma) was used. Addition of NaCl or (NH4)2SO4 was used for testing the effect of ionic strength. For testing the effect of pH, 50 mM glycine (pH 3.0) or 50 mM Hepes (pH 7.0) was used. For binding isotherm determination, the protein concentration was varied accordingly. The rate of association of enzyme activity to the rod was calculated by fitting a one phase exponential association curve to the data, using Prism software (Graph Pad).
Binding analysis by surface plasmon resonance
For the study of binding by surface plasmon resonance, a Biacore biosensor (Biacore) was used. Prefabricated sensor surfaces were used, which have a layer of octadecanethiol bonded to an underlying gold surface (named HPA by the supplier). The concentration of protein was 0.2 mg/mL for all runs; 50 mM Na-acetate buffer (pH 5.0) was used as running buffer at 5 μL/min. The flow cells were washed with 1% octyl glucoside and washed extensively before sample application. The sample volume was typically 30 μL, and the desorption of protein was tested with octyl glucoside, SDS, and the nonionic surfactants Tween 20 and C12–18EO5, (Agrimul NRE 1205, Cognis GmbH) using dilutions of 10, 5, and 1 g/L and injection volumes of 5 to 25 μL.
Binding analyzed by quartz microbalance
The QCM used was a model QAFC 301 from Q-Sense (Lund), which allows measurement of both frequency and dissipation factor (Hook et al. 1998a). The quartz crystals had a resonance frequency of 5 MHz and were precoated with SiO2 (QSX 303). The crystals were washed with Hellmanex II (Hellma GmbH) detergent and used as such, or they were silanized with dimethyldichlorosilane, as described for the glass rods, and washed extensively in detergent, ethanol, and water. Protein (HFBI, HFBII, EGIc, EGIc-HFBI, and EGIc-HFBII) was diluted with 50 mM Na-acetate buffer (pH 5.0) to a concentration of 0.25 to 0.5mg/mL. After a stable baseline had been established, the sample was injected into the measurement chamber as described in Hook et al. (1998a) at a temperature of 30°C. Δf and ΔD were recorded at 5 and 15 MHz at a sampling speed of 2 Hz.
Acknowledgments
We thank Professor Per Stenius, Timo Kallio, and Dr. Juha Kekkonen for help with the QCM, and Dr. Kaija Alfthan for help with the Biacore. Riitta Suihkonen and Seija Nordberg are thanked for technical assistance, and Nina Hörhammer is thanked for help with constructing the strain producing EGIc-HFBII. Dr. John Londesborough is thanked for help and comments on the manuscript. The work was financially supported by the Academy of Finland and the National Technology Agency (Tekes).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0207902.
References
- Alfthan, K. 1998. Surface plasmon resonance biosensors as a tool in antibody engineering. Biosens. Bioelectron. 13 653–663. [DOI] [PubMed] [Google Scholar]
- Aro, N., Saloheimo, A., Ilmen, M., and Penttila, M. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276 24309–24314. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Schlunegger, M.P., and Eisenberg, D. 1995. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 4 2455–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, C.E., Mueller, R J., Kazmierczak, P., Zhang, L., Villalon, D.K., and Van Alfen, N.K. 1992. Effect of a virus on accumulation of a tissue-specific cell-surface protein of the fungus Cryphonectria (Endothia) parasitica. Mol. Plant Microbe Interact. 5 55–61. [DOI] [PubMed] [Google Scholar]
- Collen, A., Persson, J., Linder, M., Nakari-Setälä, T., Penttilä, M., Tjerneld, F., and Sivars, U. 2002. A novel two-step extraction method with detergent/polymer systems for primary recovery of the fusion protein endoglucanase I-hydrophobin I. Biochim. Biophys. Acta. 1569 139–150. [DOI] [PubMed] [Google Scholar]
- Deming, T.J. 1999. Mussel byssus and biomolecular materials. Curr. Opin. Chem. Biol. 3 100–105. [DOI] [PubMed] [Google Scholar]
- de Vocht, M.L., Scholtmeijer, K., van der Vegte, E.W., de Vries, O.M., Sonveaux, N., Wosten, H.A., Ruysschaert, J.M., Hadziloannou, G., Wessels, J.G., and Robillard, G.T. 1998. Structural characterization of the hydrophobin SC3, as a monomer and after self-assembly at hydrophobic/hydrophilic interfaces. Biophys. J. 74 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, O., Fekkes, M.P., Wösten, H.A.B., and Wessels, J.G.H. 1993. Insoluble hydrophobin complexes in the walls of Schizophyllum communae and other filamentous fungi. Arch. Microbiol. 159 330–335. [Google Scholar]
- Ebbole, D.J. 1997. Hydrophobins and fungal infection of plants and animals. Trends Microbiol. 5 405–408. [DOI] [PubMed] [Google Scholar]
- Hashimoto, S. 2000. Principles of BIACORE. In Real time analysis of biomolecular interactions (eds. K. Nagata and H. Handa), pp. 23–32. Springer Verlag, Tokyo, Japan.
- Haynes, C.A. and Norde, W. 1994. Globular proteins at solid/liquid interfaces. Colloids Surfaces 2 517–566. [Google Scholar]
- Hook, F., Rodahl, M., Brzezinski, P., and Kasemo, B. 1998a. Energy dissipation kinetics for protein and antibody-antigen adsorption under shear oscillation on a quartz crystal microbalance. Langmuir 14 729–734. [Google Scholar]
- ———. 1998b. QCM-measurements of ferritin monolayers on methyl-thiolated gold: Dependence of energy dissipation and saturation coverage on salt concentration. J. Colloid. Interface. Sci. 208 263–267. [DOI] [PubMed] [Google Scholar]
- Hook, F., Rodahl, M., Kasemo, B., and Brzezinski, P. 1998c. Structural changes in hemoglobin during adsorption to solid surfaces: effects of pH, ionic strength, and ligand binding. Proc. Natl. Acad. Sci. 95 12271–12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killan, J.A. and von Heijne, G. 2000. How proteins adapt to a membrane-water interface. Trends. Biochem. Sci. 25 429–434. [DOI] [PubMed] [Google Scholar]
- Linder, M. and Teeri, T.T. 1997. The roles and function of cellulose-binding domains. J. Biotechnol. 57 15–28. [DOI] [PubMed] [Google Scholar]
- Linder, M., Selber, K., Nakari-Setälä, T., Qiao, M., Kula, M.-R., and Penttilä, M. 2001. The hydrophobins HFBI and HFBII from Trichoderma reesei showing efficient interactions with nonionic surfactants in aqueous two-phase systems. Biomacrimolecules 2 511–517. [DOI] [PubMed] [Google Scholar]
- Lugones, L.G., Bosscher, J.S., Scholtmeyer, K., de Vries, O.M., and Wessels, J.G. 1996. An abundant hydrophobin (ABH1) forms hydrophobic rodlet layers in Agaricus bisporus fruiting bodies. Microbiology. 142 1321–1329. [DOI] [PubMed] [Google Scholar]
- Malmsten, M. and Veide, A. 1996. Effects of amino acid composition on protein adsorption. J. Colloid. Interface Sci. 178 160–167. [DOI] [PubMed] [Google Scholar]
- Martin, G.G., Cannon, G.C., and McCormick, C.L. 2000. Sc3p hydrophobin organization in aqueous media and assembly onto surfaces as mediated by the associated polysaccharide schizophyllan. Biomacromolecules 1 49–60. [DOI] [PubMed] [Google Scholar]
- Nakari-Setälä, T., Aro, N., Ilmen, M., Munoz, G., Kalkkinen, N., and Penttila, M. 1997. Differential expression of the vegetative and spore-bound hydrophobins of Trichoderma reesei: Cloning and characterization of the hfb2 gene. Eur. J. Biochem. 248 415–423. [DOI] [PubMed] [Google Scholar]
- Nakari-Setälä, T., Aro, N., Kalkkinen, N., Alatalo, E., and Penttila, M. 1996. Genetic and biochemical characterization of the Trichoderma reesei hydrophobin HFBI. Eur. J. Biochem. 235 248–255. [DOI] [PubMed] [Google Scholar]
- Penttilä, M., Nevalainen, H., Ratto, M., Salminen, E., and Knowles, J. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 61 155–164. [DOI] [PubMed] [Google Scholar]
- Richards, W.C. 1993. Cerato-ulmin: A unique wilt toxin of instrumental significance in the development of Dutch elm disease. In Dutch elm disease research: Cellular and molecular approaches (eds. M.B. Sticklen and J.L. Sherald), pp. 89–151. Springer Verlag, New York.
- Sara, M. and Sleytr, U.B. 2000. S-Layer proteins. J. Bacteriol. 182 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerbrey, G. 1959. Verwendung von Schwingquarzen zur Wargung dunner Schichten zur Mikrowagung. Z. Physik. 155 206–222. [Google Scholar]
- Srisodsuk, M., Lehtio, J., Linder, M., Margolles-Clark, E., Reinikainen, T., and Teeri, T.T. 1997. Trichoderma reesei cellobiohydrolase I with an endoglucanase cellulose-binding domain: Action on bacterial microcrystalline cellulose. J. Biotechnol. 57 49–57. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J., Ebbole, D.J., and Hamer, J.E. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 5 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkkeli, M., Serimaa, R., Ikkala, O., and Linder, M. 2002. Aggregation and self-assembly of hydrophobins from Trichoderma reesei: Low resolution structural models. Biophys. J. (in press). [DOI] [PMC free article] [PubMed]
- van der Vegt, W., van der Mei, H.C., Wosten, H.A., Wessels, J.G., and Busscher, H.J. 1996. A comparison of the surface activity of the fungal hydrophobin SC3p with those of other proteins. Biophys Chem. 57 253–260. [DOI] [PubMed] [Google Scholar]
- Wessels, J.G. 1994. Developmental regulation of fungal cell wall formation. Annu. Rev. Phytopathol. 32 413–437. [Google Scholar]
- ———. 1997. Hydrophobins: Proteins that change the nature of the fungal surface. Adv. Microb. Physiol. 38 1–45. [DOI] [PubMed] [Google Scholar]
- Wösten, H.A. 2001. Hydrophobins: Multipurpose proteins. Annu. Rev. Microbiol. 55 625–646. [DOI] [PubMed] [Google Scholar]
- Wösten, H.A. and de Vocht, M. L. 2000. Hydrophobins: The fungal coat unravelled. Biochim. Biophys. Acta. 1469 79–86. [DOI] [PubMed] [Google Scholar]
- Wösten, H.A., Schuren, F.H., and Wessels, J.G. 1994. Interfacial self-assembly of a hydrophobin into an amphipathic protein membrane mediates fungal attachment to hydrophobic surfaces. EMBO J. 13 5848–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, H.A., van Wetter, M. A., Lugones, L. G., van der Mei, H. C., Busscher, H. J., and Wessels, J. G. 1999. How a fungus escapes the water to grow into the air. Curr. Biol. 9 85–88. [DOI] [PubMed] [Google Scholar]
- Wösten, H.A.B. and Wessels, J.G.H. 1997. Hydrophobins: From molecular structure to multiple functions in fungal development. Mycoscience 38 363–374. [Google Scholar]
- Wösten, H.A.B., de Vries, O.M.H., and Wessels, J G.H. 1993. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell. 5 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, H.A.B., Ruardy, TG., van der Mei, H.C., Busscher, H.J., and Wessels, J.G.H. 1995. Interfacial self-assembly af a Schizophyllum commune hydrophobin into an insoluble amphipathic protein membrane depends on surface hydrophobicity. Colloids Surfaces 5 189–195. [Google Scholar]
- Yaguchi, M., Puszari-Carey, M., Roy, C., Surewicz, W.K., Carey, P.R., Stevenson, K.J., Richards, W.C., and Takai, S. 1993. Amino acid sequence and spectroscopic studies of Duch elm disease toxin, Cerato-ulmin. In Dutch elm disease research: Cellular and molecular approaches (eds. M.B. Sticklen and J.L. Sherald), pp. 152–170. Springer Verlag, New York.