Abstract
Studies with the homodimeric recombinant human macrophage colony-stimulating factor beta (rhM-CSFβ), show for the first time that a large number (9) of disulfide linkages can be reduced after amide hydrogen/deuterium (H/D) exchange, and the protein digested and analyzed successfully for the isotopic composition by electrospray mass spectrometry. Analysis of amide H/D after exchange-in shows that in solution the conserved four-helix bundle of (rhM-CSFβ) has fast and moderately fast exchangeable sections of amide hydrogens in the αA helix, and mostly slow exchanging sections of amide hydrogens in the αB, αC, and αD helices. Most of the amide hydrogens in the loop between the β1 and β4 sheets exhibited fast or moderately fast exchange, whereas in the amino acid 63–67 loop, located at the interface of the two subunits, the exchange was slow. Solvent accessibility as measured by H/D exchange showed a better correlation with the average depth of amide residues calculated from reported X-ray crystallographic data for rhM-CSFα than with the average B-factor. The rates of H/D exchange in rhM-CSFβ appear to correlate well with the exposed surface calculated for each amino acid residue in the crystal structure except for the αD helix. Fast hydrogen isotope exchange throughout the segment amino acids 150–221 present in rhM-CSFβ, but not rhM-CSFα, provides evidence that the carboxy-terminal region is unstructured. It is, therefore, proposed that the anomalous behavior of the αD helix is due to interaction of the carboxy-terminal tail with this helical segment.
Keywords: Macrophage colony-stimulating factor β, H/D exchange, disulfide bonds, ESI-MS, amide residue depth
Globular structures with distinct surface crevices and interior cavities provide proteins with unique three-dimensional conformations. In solution these conformations may contain flexible domains. Various analytical methods, including low temperature flash photolysis, X-ray diffraction, Mössbauer effect (Frauenfelder et al. 1988) and NMR (Tolman et al. 1997) have provided experimental evidence for the conforrmational substates attending such protein dynamics in time frames of 10−8 to 10−11 sec. An understanding of both the three-dimensional structure and dynamics of a protein is thought to be critical for establishing precise pharmacologic activity under physiological conditions (Jardetzky and Lefevre 1994). Many of these properties are not evident when proteins are crystallized, but appear in solution where proteins may access conformational substates.
Hydrogen/deuterium (H/D) exchange and kinetic analysis of protein amide hydrogen isotope exchange may be used to probe protein structure and conformational changes (Hvidt and Nielsen 1966; Englander et al. 1979; Woodward et al. 1982; Englander and Mayne 1992; Clarke and Itzhaki 1998; Raschke and Marqusee 1998). The exchange rates of peptide amide hydrogens in proteins are highly sensitive to a number of structural features (Englander et al. 1988; Wagner and Anderegg 1994; Smith 1998), but the regions where exchange takes place must be accessible to the aqueous solvent, which means that the hydrophobic core that comprises a large fraction of the volume of a folded protein may be largely inaccessible. Only through local or global unfolding or when a protein exists in molten-globule intermediate states are such sites accessible to water for H/D exchange to occur. Recent reports indicate that depth calculations of amide site residues from X-ray crystal structure data can be correlated with hydrogen exchange rates in solution (Chakravarty and Varadarajan 1999).
The solution structure of mature macrophage colony-stimulating factor (M-CSF) possesses nine disulfide bridges; three intersubunit disulfides (Cys31–Cys`31, Cys157/159–Cys`157/159) and six intramolecular disulfide bonds (Cys48–Cys139, Cys7–Cys90, Cys102–Cys146; Cys`48–Cys`139, Cys`7–Cys`90, Cys`102–Cys`146) (Pandit et al. 1992; Glocker et al. 1993). These disulfides stabilize the three-dimensional structure and properly folded rhM-CSFβ containing the correct disulfide bonds is crucial for attaining full biological activity (Das and Stanley 1982). rhM-CSFβ is of interest because the protein is a hematopoietic growth factor that regulates proliferation, differentiation, and survival of monocytes, macrophages, and their early bone marrow progenitor lineage. It also regulates cells of the female reproductive tract (Stanley et al. 1997). Human M-CSF recombinantly expressed in Escherichia coli includes residues 4–221 and is termed rhM-CSFβ. rhM-CSFβ can be recovered from insoluble inclusion bodies and renatured in vitro in high yield to a biologically active, disulfide-linked 49-kD homodimeric protein (Halenbeck et al. 1989; Pandit et al. 1992).
The fairly large size of native rhM-CSFβ makes this protein unsuitable for study by current NMR methods. The difficulty of obtaining good crystals from the purified protein is obviously an obstacle to studying this protein by high-resolution X-ray diffraction. No X-ray structural data of rhM-CSFβ have been reported. The crystal structure of the homodimer rhM-CSFα (PDB accession number: 1hmc), a truncated sequence (residues 4–158) with full biological activity, has been reported at 2.5 Å resolution (Pandit et al. 1992). Recent developments for measuring H/D exchange by mass spectrometry have provided a powerful new tool for characterizing solution structural features of proteins, without the limitation in size and solubility associated with NMR methods (Raschke and Marqusee 1998; Engen and Smith 2001).
The H/D exchange information in specific fragments of proteins is generally obtained by rapid proteolytic fragmentation followed by fast HPLC mass spectrometric analysis, as described by Smith and co-workers (Zhang and Smith 1993). If the protein is rich in disulfide bonds or resistant to digestion, as in rhM-CSFβ, the H/D information can be difficult to obtain. In the present study we describe the use of a high concentration of urea, reducing agent, and immobilized pepsin to rapidly fragment rhM-CSFβ under conditions that minimize isotopic back-exchange. The H/D exchange results of the peptic fragments from rhM-CSFβ in solution were compared with cyrstallographic factors calculated from the X-ray structure of rhM-CSFα crystal. With these methods we discovered some interesting new details about the solution structure of rhM-CSFβ.
Results
Disulfide bond reduction and peptic fragmentation
Each subunit of the rhM-CSFβ homodimer consists of 218 amino acid residues including nine cysteines. The intra- and intermolecular disulfide bonds, as well as the relatively large contact surface between the monomers, stabilize the dimer (Pandit et al. 1992). The rhM-CSFβ retains its secondary structure in acidic solution (pH 2 to ∼3) for at least a day as determined by CD (not shown). Using pepsin proteolysis alone (Zhang and Smith 1993; Zhang et al. 1996), less than 5% of rhM-CSFβ was digested. In addition to limited proteolysis, serious interferences by pepsin autocleavage fragments occurred, and unpredictable peptic cleavages along the backbone of the protein resulted with broadly distributed disulfide bonds tethering peptides together, making it difficult to identify the digested fragments. These complications resulted in unacceptably low sequence coverage for this protein.
To our knowledge there are no reported examples of successful H/D exchange studies using mass spectrometry analytical approaches on proteins with nine disulfide bonds. As a result the first objective was to find a method to reduced the disulfide bonds in the labeled protein. Under H/D exchange quench conditions (0°C, pH 2.5), before or during digestion, brief exposure to high concentrations of urea and TCEP•HCl was used to denature the protein and reduce the disulfide bonds. These reducing conditions resulted in an acidic solution (pH ∼2.5), in which rhM-CSFβ could be reduced with immobilized pepsin. The enzyme was then quickly removed. The digest concentrated and desalted in a short reverse phase column, and the peptides chromatographically introduced into the mass spectrometer as quickly as possible to minimize hydrogen back-exchange. The proteolytic fragments were assigned by liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) and liquid chromatography collision-induced dissociation tandem mass spectrometry (LC-CID-MS/MS) analysis. Peptic digestion produced ∼26 proteolytic fragments, which covered 93% of the rhM-CSFβ sequence (Fig. 1 ▶).
Fig. 1.
Amino acid sequence of rhM-CSFβ with the peptic fragments used in this study. Cysteine residues are shaded. The α-helices and β strands are illustrated based on the X-ray structure of rhM-CSFα (Pandit et al. 1992).
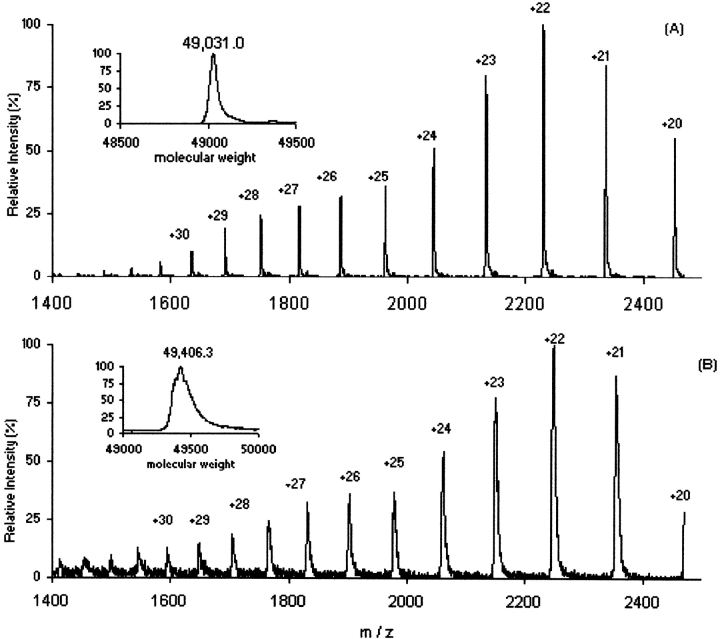
The rhM-CSFβ dimer has a total of 404 exchangeable amide hydrogens. Fully deuterated rhM-CSFβ can be obtained within 8 h in 99 atom % excess D2O (pH 6.0) after denaturation in 8 M urea-d4 at room temperature. ESI-MS gives a mass of 375.3 Da higher than the undeuterated protein (Fig. 2 ▶), indicating that 93% of the possible deuterium content is maintained in the intact protein under the experimental conditions.
Fig. 2.
ESI mass spectra with deconvoluted spectra (insets) of undeuterated rhM-CSFβ (A) and fully deuterated rhM-CSFβ (B).
The width of the peak in the deconvoluted spectrum of fully deuterated rhM-CSFβ (Fig. 2B ▶, inset) suggests there was a distribution of deuteriums in this protein, as the peak width is thought to result from the number of amide hydrogens that are only partially exchanged (Zhang et al. 1996). The broad tailing peaks (Fig. 2B ▶) also suggest that some exchangeable deuteriums in the amino acid side chains were not completely back-exchanged during LC separation. Perhaps such side chain sites were protected by the compact tertiary structure of the protein (Milne et al. 1998). The CD spectra suggested that the secondary structure of rhM-CSFβ was not greatly perturbed under the LC conditions (30%–60% acetonitrile plus TFA at pH 2.5–3.0) used. Thus, the protein did not unfold during chromatography into the mass spectrometer, but rather maintained a nativelike conformation.
The fully deuterated protein sample was used as a control (m100% control) to adjust deuterium loss during protein digestion and LC-MS analysis (Zhang et al. 1996). Back-exchange of hydrogen isotope in each peptic fragment was determined from the theoretical mass for the fully deuterated fragments and the experimental molecular mass. The extent of artifactual loss for peptic peptides was in the range of 12%–24%.
Analysis of hydrogen exchange rates for peptic fragments
Previous studies have shown that site-specific amide H/D measurements are reliable (Deng et al. 1999; Kim et al. 2001), particularly if samples are digested before freezing (Kim et al. 2001). However, the experiments performed in this study require analyses of aliquots after 10 sec, 1 min, 5 min, and so on for 5 h and a more involved procedure, that is, reduction as well as digestion was required to prepare the samples for analysis. To insure accurate H/D ratios in the early time points, quenching was done to rapidly arrest exchange by plunging the samples into liquid nitrogen and then thawing them for chemical and enzymatic processing.
The centroids of the isotopic clusters of the peptic ions derived from exchange-in and control samples were used to determine the H/D levels (Zhang and Marshall 1998). The deuterium level observed in each peptic fragment as a function of time represented the isotopic exchange behavior of a specific region in the intact native protein. For example, the deuterium content found in one representative peptic peptide covering residues amino acids 37–49 at different reaction times (Fig. 3 ▶) was fitted to equation (1) in an effort to determine the isotopic exchange rate constants and the numbers of hydrogens that exchanged with that rate. The analytical results indicate that for a total of 11 peptide amide hydrogens in this fragment, 3.5 amide hydrogens have k ≥4.15 min−1, 3.6 have k = 0.14 min−1, 2.5 have k = 0.009 min−1, and 1.4 have k ≤0.002 min−1. For convenience, the exchange rate constants at peptide amide hydrogens were grouped into fast (k >4.16 min−1), intermediate (4.16 > k > 0.1 min−1), slow (0.1 min−1 > k > 0.002 min−1) and nonexchangeable (k <0.002 min−1) categories. Peptide amide hydrogens with k >4.16 min−1 are those whose exchange half-lives are much smaller than the fastest time (10 sec) used to obtain the data, whereas the peptide amide hydogens with k <0.002 min−1 have exchange-in half-lives that are greater than the longest experimental exchange-in times used (5 h).
Fig. 3.
(A) Mass spectra for singly and doubly charged ions of peptic peptide amino acids 37–49 at various H/D exchange times. The deuterium level in this fragment was deduced from centriods of the envelope isotope peaks of both ions, averaged and corrected with two reference samples. (B) Fitted curves of representative peptic fragments according to equation 1. Data for fragment of amino acids 190–221 was not able to deconvolute by equation 1.
Thus, the rate constant categories of amide hydrogens in 26 peptic peptides covering 93% of the amide hydrogens in the backbone of rhM-CSFβ were determined. The resolution of ESI-MS for proteolytic fragmentation is generally limited by the length of the peptic peptides. Some overlapping peptic fragments were observed. By difference of overlapping sequences, improved resolution was obtained for some regions (Table 1). The b-type ions were difficult to obtain in some peptic peptides, or hard to recognize due to overlapping peaks in CID MS/MS. In addition, it is well known that the mass measurement accuracy of fragment ions is limited in the LCQ mass spectrometer. Our data indicated that the uncertainty in the measured mass was ±0.2 Da in the CID MS/MS spectra of some fragments. Site-specific amide exchange data were not reliable under these conditions, but CID MS/MS was found useful to determine exchange rate categories within narrower ranges of the rhm-CSFβ of certain peptides.
Table 1.
The number of amide hydrogens in peptic fragments of rhM-CSFβ having rate constants (in parentheses, min−1) within a specified range as determined by equation 1
| No. of amide hydrogens in different exchange rate constants | ||||||||
| Entry no. | Fragments | Total no. of NH | Average depth (A) | Fast k>4.15 min−1 | Intermediate 4.15>k>0.1 min−1 | Slow 0.1>k>0.002 min−1 | No exchange k<0.002 min−1 | Solvent accessibility %d |
| 1 | 4–16 | 12 | 4.8 | 7.0 ± 0.7 | 5.0 ± 0.8 (2.0) | 100 | ||
| 2 | 4–19 | 15 | 5.0 | 8.8 ± 0.4 | 5.2 ± 0.3 (1.6) | 1.0 ± 0.1 (0.004) | 100 | |
| 3 | 20–27 | 7 | 6.6 | 0.2 ± 0.1 | 0.8 ± 0.6 (0.03) | 6.0 ± 0.6 | 14.3 | |
| 4 | 37–49 | 11 | 5.6 | 3.4 ± 0.1 | 3.6 ± 0.4 (0.14) | 2.5 ± 0.4 (0.009) | 1.5 ± 0.4 | 86.4 |
| 5 | 37–55 | 17 | 6.2 | 2.9 ± 0.2 | 3.8 ± 0.9 (0.12) | 3.2 ± 0.8 (0.01) | 7.1 ± 0.9 | 58.2 |
| 6 | 56–62 | 6 | 5.8 | 0.2 ± 0.1 | 2.2 ± 0.1 (0.22) | 2.5 ± 0.1 (0.004) | 1.1 ± 0.1 | 81.7 |
| 7 | 63–67 | 4 | 5.3 | 0.2 ± 0.1 | 3.0 ± 0.2 (0.02) | 0.8 ± 0.2 | 80.0 | |
| 8 | 63–74 | 10 | 4.9 | 2.8 ± 0.3 | 6.3 ± 1.2 (0.03) | 0.9 ± 1.2 | 91.0 | |
| 9 | 63–76 | 12 | 5.0 | 3.0 ± 0.1 | 7.7 ± 0.8 (0.03) | 1.3 ± 0.8 | 89.2 | |
| 10 | 68–76 | 7 | 4.9 | 2.0 ± 0.1 | 4.0 ± 0.7 (0.03) | 1.0 ± 0.7 | 85.7 | |
| 11 | 77–82 | 5 | 5.7 | 0.2 ± 0.1 | 1.1 ± 0.2 (0.03) | 3.7 ± 0.2 | 26.0 | |
| 12 | 81–85 | 4 | 5.7 | 0.1 ± 0.0 | 1.5 ± 0.3 (0.027) | 2.4 ± 0.3 | 40.0 | |
| 13 | 86–105 | 19 | 5.0 | 10.1 ± 0.4 | 3.0 ± 0.5 (1.68) | 3.6 ± 0.4 (0.014) | 2.3 ± 0.5 | 87.9 |
| 14 | 95–105 | 10 | 5.2 | 4.7 ± 0.6 | 2.3 ± 0.5 (1.65) | 3.0 ± 0.5 | 70.0 | |
| 15 | 106–113 | 6 | 5.9 | 0.2 ± 0.1 | 1.7 ± 0.1 (0.27) | 1.1 ± 0.1 (0.009) | 3.0 ± 0.1 | 50.0 |
| 16 | 107–113 | 5 | 5.9 | 0.2 ± 0.1 | 0.9 ± 0.1 (0.25) | 1.5 ± 0.2 (0.01) | 2.4 ± 0.2 | 52.0 |
| 17 | 114–128 | 14 | 6.4 | 0.2 ± 0.1 | 0.8 ± 0.4 (0.05) | 13.0 ± 0.4 | 7.1 | |
| 18 | 114–135 | 21 | 5.9 | 1.8 ± 0.3 | 5.0 ± 0.4 (0.12) | 14.2 ± 0.4 | 32.4 | |
| 19 | 114–143 | 29 | 5.6 | 7.9 ± 0.4 | 7.1 ± 0.5 (0.33) | 14.0 ± 0.5 | 51.7 | |
| 20 | 150–189 | 31 | /c | 31 ± 1.5 | 100.0 | |||
| 21 | 161–189 | 22 | /c | 22 ± 0.7 | 100.0 | |||
| 22 | 190–221 | 29 | /c | 29 ± 1.2 | 100.0 | |||
| 23 | 17–19a | 3 | 6.0 | 1.9 ± 0.6 | 1.1 ± 0.4 (0.003) | 100.0 | ||
| 24 | 37–45b | 8 | 5.2 | 2.7 ± 0.6 | 4.2 ± 0.4 (0.11) | 1.0 ± 0.6 | 87.5 | |
| 25 | 46–49b | 3 | 7.2 | 0.2 ± 0.2 | 1.8 ± 0.4 (0.01) | 1.0 ± 0.4 | 66.7 | |
| 26 | 50–55a | 6 | 7.4 | 6.0 ± 0.4 | 0.0 | |||
| 27 | 63–68a | 5 | 5.2 | 0.9 ± 0.2 | 3.0 ± 0.5 (0.05) | 1.1 ± 0.5 | 78.0 | |
| 28 | 68a | 1 | 4.8 | 1.0 ± 0.3 | 100.0 | |||
| 29 | 68–74a | 6 | 4.7 | 2.9 ± 0.3 | 2.1 ± 0.6 (0.03) | 1.0 ± 0.6 | 83.3 | |
| 30 | 75–76a | 2 | 5.6 | 0.1 ± 0.1 | 1.2 ± 0.1 (0.05) | 0.7 ± 0.1 | 65.0 | |
| 31 | 86–95a | 9 | 4.8 | 5.0 ± 0.4 | 0.9 ± 0.5 (0.42) | 2.1 ± 0.3 (0.015) | 1.0 ± 0.5 | 88.9 |
| 32 | 107a | 1 | 4.1 | 1.0 ± 0.4 (0.18) | 100.0 | |||
| 33 | 129–135a | 7 | 4.8 | 1.0 ± 0.2 | 3.6 ± 0.4 (0.30) | 2.4 ± 0.4 (0.03) | 100.0 | |
| 34 | 136–143a | 8 | 5.0 | 5.2 ± 0.5 | 2.9 ± 0.8 (1.19) | 100.0 | ||
a From overlapping fragments.
b From CID MS/MS data.
c No X-ray data available.
d Solvent accessibility is a measure of the percentage of the total number of incorporated deuteriums for fast, intermediate, and slow exchange categories divided by the theoretical number of exchangeable amide hydrogens.
Thus, CID MS/MS was used mainly to create smaller fragments of the peptic peptides in an attempt to define more closely the region of exchange. Reliable results were obtained with singly charged bn ions (Deng et al. 1999). For example, 21 amide hydrogens in residues amino acids 114–135 have 1.8 fast exchanging amide hydrogens, 5 intermediate exchanging hydrogens (0.12 min−1), and 14.2 slow or nonexchangeable hydrogens as shown by MS1 data (Table 1). The doubly charged ion of this peptide yields the b15 ion when collisionally fragmented by CID, which showed an increase in mass with incubation time (Fig. 4 ▶). After adjustment for deuterium loss and gain with two reference samples, the deuterium levels versus the exchange-in time were fitted to equation (1). The results indicated that no amide hydrogens with fast or intermediate exchange rate constants were present in this segment, i.e., b1–b15. This is exactly what was found from the data obtained for the peptic peptide amino acids 114–128, i.e., the same segment, when analyzed by MS1, thereby confirming that the fast or intermediate-exchanged amide hydrogens are located in the amino acid 129–135 segment.
Fig. 4.
Tandem mass spectrum of the doubly charged ion at m/z 1357.2 representing peptic peptide amino acids 114–135 showed a series of bn ions. The b15 ion was used to analyze the b1–b15 fragment. The deuterium level in this fragment was deduced from centriods of the envelope isotope peaks and was adjusted with m0% and m100% control samples (Zhang and Smith 1993).
All 60 amide sites in the region amino acids 150–189 and amino acids 190–221 of rhM-CSFβ were fully deuterated at the first measured time point (10 sec). These amide hydrogens exhibited fast exchange rates (k >4.15 min−1), which indicates that there was substantial solvent accessibility in the unstructured region of the carboxyl terminus. The crystal structure of the carboxy-terminal region (amino acids 150–221) of rhM-CSFβ has not been reported, but the region is predicted to have a low probability of containing α-helical or β-sheet secondary structures (Zemla et al. 1999). Comparison of the native structure with the modified one in which the Cys157/159–Cys`157/159 linkage was reduced and cyanylated showed identical near UV CD spectra, tryptophan fluorescence, and biological activity, but charge state envelopes and H/D exchange studies revealed more openness in this region of the protein when the disulfides were reduced (Zhang et al. 2001). The peptide amide hydrogens in amino acids 136–143, near this flexible tail region, showed either fast or intermediate exchange rates, that is, ∼5 amide hydrogens with k >4.15 min−1 and 3 with k = 1.19 min−1, respectively (Table 1). This region must, therefore, be reasonably solvent accessible, even with the Cys139/Cys48 bridge in place.
Four long α-helices (αA, amino acids13–24; αB, amino acids 46–63; αC, amino acids 72–88; and αD, amino acids 110–130; see Fig. 1 ▶) form the structural backbone in each subunit (Fig. 5 ▶). The αB helix nearly passes through the center of each subunit. One side of the αB helix is surrounded by the αA, αC, and αD helices, whereas the other side is adjacent to the β1 and β2 sheets. The peptic fragment including residues amino acids 4–16 spans the α1 helix and covers one-third of the αA region. Many of the amide hydrogens in this fragment exchange rapidly, i.e., 7 amide hydrogens with k >4.15 min−1 and 5 with k = 2.0 min−1. Seven amide hydrogens in the region of rhM-CSFβ which includes residues amino acids 20–27 displayed nonexchanging (6 H) or slow exchanging (1 H, k = 0.03 min−1) kinetics. X-ray structural data indicate that all peptide amide hydrogens of this fragment are hydrogen-bonded to carbonyl oxygens. Four of these peptide amide hydrogens (amino acids 21–24) are near the carboxyl -terminus of the αA helix, and amide hydrogens of Gln26 and Met27 are also solvent inaccessible. The latter residues are located at the interface of the two monomers. Stable hydrogen bonding and strong interactions between the two covalently linked subunits makes it difficult for water to penetrate this region. In the fragment with residues amino acids 17–19, two amide hydrogens exchange rapidly, whereas the other exchanges slowly (k = 0.003 min−1). This small fragment must, therefore, connect flexible and nonflexible regions within the αA helix. The fragment with residues amino acids 37–45 (entry no. 24) encompasses the connecting region of the β1 strand and αB helix. All but one amide hydrogen in this region exhibited relatively large exchange rate constants (Table 1).
Fig. 5.
A ribbon plot of rhM-CSFβ based on the crystal structure of rhM-CSFα (Pandit et al. 1992). The disulfide bonds were labeled and shown by stick model. The gray color shows amino acids not identified in the digest. Dotted lines represent the carboxy-terminal strands for which there are no X-ray data. The different colors indicate the regions with fast or intermediate H/D exchange rates (red), nonexchangeable or slow exchange rates (blue), or multiple categories of exchange rates (yellow). Average depths of the chains were obtained from X-ray coordinates.
The region containing residues amino acids 46–55 (from peptic fragments amino acids 46–49 and 50–55 (Table 1) exhibited very slow H/D exchange rates. In particular, residues amino acids 50–55, which are located at the center of the αB helix, displayed essentially no deuterium exchange-in during the 300-min incubation period. X-ray data show that all amide hydrogens are involved in hydrogen bonding except for that of Val47. The peptic fragment that includes residues amino acids 56–62 is located near the edge of the αB helix. About four amide hydrogens showed slow or no exchange, whereas two hydrogens displayed intermediate exchange rates (k = 0.22 min−1). The amide hydrogen in residue Gln58 is not involved in hydrogen bonding, and more than half of the surface areas of residues Asp59 (68%) and Glu62 (59%) would be expected to be exposed to solvent as determined from X-ray data, which could account for the intermediate exchange rate.
In the regions spanning residues amino acids 75–85 (fragments with residues amino acids 75–76, 77–82, and 81–85) and amino acids 114–128 (Table 1), which encompasses most residues of αC and αD helices, respectively, the H/D exchange behavior for all amide hydrogens was either very slow (k ≈0.03 min−1) or nonexchangeable (k <0.002 min−1).
The four amide hydrogens in the fragment containing residues amino acids 63–67 are located in a turn, connecting αB and αC helices. The exchange rate constants in this fragment were small (3 H, k = 0.02 min−1; 1 H, k <0.002 min−1). This fragment is located near the interface of the two subunits. The first three amide hydrogens (Thr64, Met65, and Arg66) in this region participate in hydrogen bonding, but the fourth (Phe67) does not. The amide hydrogen of Arg68 is not involved in hydrogen bonding. The exchange rate inferred from overlapping fragments (entry nos. 7 and 27) is fast (k >4.15 min−1), and suggests that this residue is not shielded from solvent and furthermore, that it is located outside the dimer interface. Other peptic peptides that contain connecting resides of secondary structures such as amino acids 37–45, 56–62, 106–113, 95–105, and 129–135 exhibited multiple categories of H/D exchange rates (Fig. 1 ▶, Table 1) indicating that these peptides are in regions of structural variability that are both solvent penetrable and solvent impenetrable. From the H/D exchange data it is concluded that there are regions with fast, slow, and multiple exchange rates in the backbone of rhM-CSFβ (Fig. 5 ▶).
The amide hydrogens on Cys residues of the disulfide linkage in the carboxy-terminal reigion (Cys157/159–Cys`157/159) were found to exchange very rapidly (see Table 1, entry no. 20). On the other hand, in the Cys48–Cys139 (or Cys`48–Cys`139) bonds, one Cys residue (Cys139, Table 1, entry no. 34) exchanged rapidly and the other (Cys48, entry no. 25) was found to fall in the nonexchangeable category. The hydrogen on Cys7 of the disulfide linkage Cys7–Cys90 was also found to exchange rapidly (Table 1, entry nos. 1 and 2), whereas Cys90 (Table 1, entry no. 31) was difficult to characterize because there were both fast and slow exchanging hydrogens in the peptide (amino acids 86–95). It is not possible to determine amide hydrogen exchange rate constants in the Cys residues of Cys102–Cys146 because the data at one end (Table 1, entry nos. 13 and 14) do not allow distinguishing where slow and fast exchanges occurred and at the other end, the relevant peptides could not be identified.
Amide residue depth analysis
X-Ray crystallographic coordinates of atoms in the structures of proteins can be used as close approximations for the locations of these atoms in solution. The atomic motions in protein structures, when in solution, can result in the exposure of subsurface atoms to solvent, hence amide hydrogen exchange of these residues buried within the folded protein core becomes possible. Because rhm-CSFβ is biologically active under the conditions of the exchange-in experiments, the protein must have been in its native state. The different rates of exchange of amide hydrogens in regions comprising the α-helices in rhm-CSFβ may be attributed to the different depths at which these groups are located from the protein surface. Depth calculations were performed for each atom from the X-ray crystal structure of rhm-CSFα. Overall, the numbers of amide hydrogens with given exchange rate constants seem to correlate roughly with the average depth of the main chain residues in peptic peptides (Table 1). Shallower chains are more likely to contain higher numbers of fast exchanging amide hydrogens and more buried residues show slow exchange. The average depths of secondary structural regions with large, multiple, and small exchange rates are 4.8, 5.6, and 6.4 Å, respectively (Fig. 5 ▶).
The solvent accessibility (Table 1, column 9) in peptic fragments can be expressed as a percentage of the total number of hydrogen ions that had exchanged within a given time period relative to the total number of exchangeable amide hydrogen ions available (Hughes et al. 2001). This approach indicates that for hydrogen ions exchanged within 300 min there is a good correlation between solvent accessibility and depth (r = 0.88) (Fig. 6 ▶). The correlation with crystallographic B-factors (r = 0.60) is not as good. When only fast exchanging hydrogens (t1/2 <10 sec. Table 1, column 5) are considered, the correlation coefficients of solvent accessibility with depth (r = 0.71) and B-factor (r = 0.66) are closer. These results suggest that B-factors may be a less reliable measure of solvent accessibility for atoms deeper within the protein.
Fig. 6.
The solvent accessibility percentage in each fragment, which is derived from the total number of incorporated deuteriums for fast, intermediate, and slow exchange (column 9 in Table 1) over the number of theorectically exchangeable amide hydrogens, correlated with the average depth for each fragment. The errors are the standard deviation for the average depth. The line through the data was calculated using a weighted least squares fit when each data point was weighted by 1/σ2 and yielded the linear relationship of y = −0.017x + 6.604 (R = 0.88). The data used for this correlation calculation are the independent fragments with entry numbers from 1 to 19, and 24 in Table 1.
Solvent exposure analysis
In addition to the depth from the surface, the accessibility of each amino acid could also be estimated from the atomic coordinates of the single crystal structure. In this analysis the exposures of the amino acids, as calculated using the Connally method, could be grouped into three categories: (1) highly exposed (≥50Å2), (2) moderately exposed (30–50Å2), and (3) buried (<30Å2). The numbers of amino acids in each peptide that fall into these categories of exposure appear to correlate with the corresponding numbers of residues in the various categories of H/D exchange (Table 1, Fig. 7 ▶). From this analysis it is seen that peptide fragment 17 (from residues amino acids 114–128 in the αD helix) was predicted to be significantly more accessible to solvent than was observed from the H/D exchange rates. Because peptide 17 is included in both peptides 18 and 19, this deviation greatly affects the correlation between surface exposure and rates of exchange of the latter two peptides. This suggests that amino acids 114–128 in this helix behave differently in solution than predicted from the solvent exposure of the crystal structure. Otherwise, the rates of deuterium exchange at the amide hydrogens in rhm-CSFα appear to be dependent on the overall exposure of each amino acid to solvent.
Fig. 7.
Comparison of a number of amino acid residues with different solvent exposures in the crystal structure (x-axes) in rhM-CSFα with a number of amide hydrogens having different H/D exchange rates (y-axes) in rhM-CSFβ. The number of amino acids that are highly exposed are compared to the number of amide hydrogens in fast exchange (A), partially exposed compared to amide hydrogens in intermediate exchange (B), and buried compared to amide hydrogens in slow or no exchange (C) for each independent peptide fragment listed in Table 1 (entry numbers 1 to 19, and 24). The least squares fit for the data in each panel yielded linear relationships of y = 0.77x − 1.33 (R = 0.79) for A, y = 1.18x − 0.46 (R = 0.83) for B, and y = 1.25x + 0.155 (R = 0.74) for C. Fragments 17, 18, and 19 are labeled in each panel.
Discussion
The correlations between H/D exchange rates and the exposure of amino acids to the solvent, as calculated from the crystal structures (Fig. 7 ▶), are striking. The primary anomaly was seen in peptide amino acids 114–128 of the αD helix, and in the two longer peptic peptides that are extended from this helix. In this case, the surface calculations greatly overestimated the exposure of the amino acids when compared to the exchange rates in solution, suggesting that some important conformational feature could not be accounted for by the crystal structure. The most obvious explanation of this discrepancy is that the long carboxy-terminal tails, which are not present in rhm-CSFα and therefore not observed in the crystal structure, affect the exposure of the αD helix to solvent. It may reasonably be concluded that the carboxy-terminal tails, rather than extending down and away from the globular structures of the first 151 residues (Fig. 5 ▶), actually lie across the surfaces of the two globular domains. In this way the αD helix can provide an interaction surface for the apparently unstructured carboxy-terminal tails. Thus, the correlations between surface exposure and H/D exchange further show that the conformation of the protein in solution during these studies is the native form as seen in the crystal structure. Furthermore, regions that significantly deviated between the exchange rates and predictions from the crystal structure can be used to derive some general topological information concerning large regions of this protein that were not resolved in the crystal structure.
An attempt was also made to determine whether the relationship between calculated depths and amide hydrogen exchange rates could provide useful information within limited structural elements, i.e., α-helices (Fig. 8 ▶). Thus, peptide fragment 1 (amino acids 4–16) comprising a large part of the αA helix has an average depth of 4.8 Å in the crystal, and the amide hydrogens undergo rapid exchange (Table 1). At the other extreme, in fragment 26 (amino acids 50–55) located within the αB helix, all amide hydrogens are nonexchangeable (Tables 1 and 2). The average calculated depth of this peptide is 7.4 Å. The peptide fragments from helices αC and fragment 6 (amino acids 56–62) in αB are more accessible (5.6–5.8Å) to solvent (Table 2, Fig. 8 ▶). A large number of amide hydrogens in αC undergo slow exchange and more than one-third in fragment 6 (amino acids 56–62) undergo intermediate rates of exchange. In peptide fragment 17 (amino acids 114–128), 13 hydrogen ions are nonexchangeable, although the average depth of this segment is shallower (6.4 Å) than that of amino acids 50–55. This result is consistent with the anomalous behavior observed from solvent exposure analysis (Fig. 7 ▶) and supports the hypothesis that the αD helix is protected by interaction with the carboxy-terminal tail. Peptide fragment 25 (amino acids 46–49) (Table 2) has a depth of 7.2Å but two hydrogen ions exchange slowly. The result is easily explained by the fact that this segment is adjacent to the loop connecting β1 and αB, which should result in greater flexibility in the sequence. Overall, it is apparent that there is a reasonable relationship between solvent accessibility and residue depth when applied to a common secondary structural element.
Fig. 8.
Molecular model of the two subunits of rhM-CSFα. Labeled fragments in one subunit, but same color in other subunit, are shown as ribbon plots. The rest of the protein (gray area) is shown as space-filling model. Colored ribbons represent partial sequences in αB, αC, and αD helices.
Table 2.
H/D Exchange and average depth in alpha helix partial sequences of rhM-CSFβ
| Fragment | No. of NH | Non exchange | Slow exchange | Intermediate exchange | Fast exchange | Average depth (Å) | |
| αB | 46–49 | 3 | 33% | 60% | 7% | 7.2 | |
| 50–55 | 6 | 100% | 7.4 | ||||
| 56–62 | 6 | 18% | 42% | 37% | 3% | 5.8 | |
| αC | 75–76 | 2 | 35% | 60% | 5% | 5.6 | |
| 77–82 | 5 | 74% | 22% | 4% | 5.7 | ||
| 81–85 | 4 | 60% | 38% | 2% | 5.7 | ||
| αD | 114–128 | 14 | 93% | 6% | 1% | 6.4 |
Another interesting aspect is seen at the interface between the two subunits. Peptide 3 (amino acids 20–27) has an average depth of 6.6 Å and most of the seven hydrogen ions are nonexchangable (Table 1). Residues 24–27 are in a loop connecting the αA helix with the β1 sheet and might be expected to exchange at least slowly. These residues, however, are at the interface of the covalently linked subunits and form intersubunit hydrogen bonds, which limits their exchange. The residues of peptide fragment 7 (amino acids 63–67) in the loop connecting αB and αC helices are closer to the surface (5.3 Å), but most of the amide hydrogens exchange slowly because these residues are also located near the interface between the two subunits.
Conclusion
A novel method has been developed using TCEP•HCl and urea in conjunction with pepsin digestion to unfold and obtain peptic fragments from a protein, i.e., rhm-CSFβ, with nine disulfide linkages so that amide site H/D analysis can be performed by mass spectrometry. The H/D label information in the protein is preserved by simultaneous reduction and peptic digestion. This experimental procedure, when used for in proteins with intractable disulfides, can be used to provide H/D exchange rate data from the proteolytic peptides, which when deconvoluted into a sum of rate expressions (equation 1) becomes a powerful alternative to site-specific amide hydrogen exchange information (Kim et al. 2001).
In the present study, analysis of the exchange rates obtained by mass spectrometry permitted an examination of the structural characteristics of rhm-CSFβ as reflected by solvent accessibility of the amide hydrogens. Mapping of the results onto the X-ray structure of the related rhm-CSFα allowed the correlation of solvent accessibility with B-factor and residue depth. Solvent exposure analysis estimated from the atomic coordinates of the crystal structure of rhm-CSFα compared with H/D analysis of the solution structure of rhm-CSFβ revealed a structural feature in the latter that would not have been detected by NMR because of its size. These studies have demonstrated the unique power of H/D exchange and analysis by mass spectrometry for characterizing structural features of proteins in solution.
Materials and methods
Materials
Purified biologically active rhM-CSFβ was obtained as a lyophilized powder from Dr. C. Cowgill (Chiron Corp. Emeryville, CA), which was formulated in 20 mM citrate containing 0.5% sodium chloride and 1% mannitol. Before deuterium exchange, the freshly dissolved buffered protein solution was exchanged for H2O buffer (10 mM phosphate at pH 6.8) with a Biomax membrane ultrafilter (5 K MW cutoff; Millipore Co.) and equilibrated for 1 h at room temperature. Immobilized pepsin suspension and Tris(2-carboxyethylphosphine) hydrochloride (TCEP) were from Pierce (Rockford, IL). Urea-d4 (98 atom % D) and deuterium oxide (99.9 atom % D) were purchased from Aldrich Chemical Co. Immobilized pepsin was washed three times and resuspended as original ratio in 0.25 M TCEP solution (with 50% H2O/D2O). It was stored in an ice bath before use. All other chemicals used in this study were of the highest quality commercially available.
Hydrogen exchange and protein fragmentation
Deuterium exchange-in was initiated by diluting10 μL of a 1.6 mM solution of rhM-CSFβ into 250 μL D2O buffer (10 mM phosphate at pD 6.8). At various exchange times (10 sec. to 5 h), a 6-μL aliquot of labeled rhM-CSFβ was taken and quenched by adding to a 20-μL solution of ice-cold 8 M urea-d4/1 M TCEP• HCl in roughly 50% H2O/D2O and immediately frozen in liquid nitrogen until analysis was performed, usually within 3 days. The pH was measured at this point and found to be 2.5 because of the addition of TCEP• HCl. Aliquots were thawed rapidly and maintained at 0°C for 6 min to complete the reduction of the nine disulfide bonds. At this point 20 μL of the sample solution was then added to 45 μL of ice-cold (0°C) immobilized pepsin suspension (protein:pepsin = 1:3 molar ratio) in a microfilterfuge sample tube with a membrane at the bottom (Rainin Instrument Co.). Digestion was carried out for 3 min in an ice bath. The digestion products were forced through a microporous membrane by centrifugation (10–15 sec) and collected in a prechilled receiver tube. The peptic peptides were injecteded into tandem C18 column system (0.32 × 25 mm, 0.32 × 100 mm, Luna C18, 5 μ, in-house packed) for fast analysis by ESI mass spectrometry (Finigan LCQ). Initially, the first column is disconnected from the second column to allow for loading and desalting of the digest (50 μL) in as short a time as possible (1 min) using 3% acetonitrile, 0.05%TFA, 100 μL/min for 0.5 min, then 200 μL/min for another 0.5 min. Then, the second column was connected to achieve separation of peptides and introduction into the mass spectrometer. The elution continued for 9 min with 10%–70% acetonitrile, 0.05%TFA with a 7-min. gradient (8 μL/min). The injector and columns were submerged in an ice bath to minimize back-exchange. Triplicate samples were obtained at each time point. The mean value of the deuterium content for each proteolytic fragment was determined.
Calculation of rate groups
The H/D exchange reaction was carried out at pH 6.8. The amide deuterium levels for each peptide were determined from the mass difference between the deuterated and the nondeuterated fragments. Exchangeable hydrogens on amino or carboxy-terminal ends and on amino acid side chains are rapidly exchanged during HPLC separation when using an aqueous solvent even under quench conditions (0°C, pH 2.5) (Englander et al. 1985; Zhang and Smith 1993). H/D exchange rate constants in fragments, converted from time course data, were calculated by nonlinear least squares analysis with Origin 6.0 (Microcal Software, Inc.). The sum of exponentials is described by the following equation, from a series of first-order rate data (Englander and Kallenbach 1983).
 |
(1) |
where D is the deuterium content of a peptide with N peptide amide hydrogens and ki is the rate constant for exchange on each peptide hydrogen (Zhang et al. 1996). To assess and correct for back-exchange, two control samples, i.e., undeuterated rhM-CSFβ (m0%) and fully deuterated rhM-CSFβ (m100%), were digested and analyzed using the same conditions (Zhang and Smith 1993). The deuterium content in the peptic peptides was deduced from their molecular masses. The fully deuterated rhM-CSFβ was obtained by denaturing rhM-CSFβ in D2O containing 8 M urea-d4 and allowing exchange for 8 h at room temperature, and then renaturing in D2O buffer (10 mM phosphate, pD 6.8) by removing urea-d4 with a Biomax membrane ultrafilter.
Calculation of factors from X-ray structure of rhM-CSFα relating H/D exchange rates and correlation
Exchange rates of amide protons in a protein are controlled by several structural factors, including intramolecular hydrogen bonding and the extent of shielding from solvent. Therefore, the protein secondary structure is an important determinant of amide H/D exchange. The static X-ray structure of rhM-CSFα, whose sequence is identical with those of rhM-CSFβ over the range amino acids 4–148, was used for calculation. The number of amide hydrogens involved in α-helices and β-sheets was calculated from the coordinate file of rhM-CSFα (Mizuguchi et al. 1998). Hydrogen bonds were identified with HBPLUS (McDonald and Thornton 1994). The depth of the residue has been defined in the paper of Chakravarty and Varadarajan (1999). Briefly, the depth of an atom in the protein crystal is its average distance from the nearest surface water molecule computed from rotation of the protein through a random angle about its axis. A number of iterations, i.e., rotation, translation to generate a configuration of water molecules around the protein, and the removal of water molecules from cavities, are performed to convergence, therefore the coefficient of variance of depth for each atom is less than a predetermined value.
The correlation between solvent accessibility in fragments and average depth or average B-factor was calculated with program Origin 6.0 (Microcal Software, Inc.) using a weighted least squares fit when each date point was weighted by 1/σ2.
Solvent exposure calculations
The exposure of each amino acid in the single crystal structure of rhm-CSFα was calculated by the Connolly rolling ball method (Connolly 1983). In this analysis hydrogen atoms were first added to the atomic coordinates of the crystal structure using standard geometries. The solvent exposures were then calculated using a probe radius of 1.4Å and 5 dots. Both procedures were performed using the program Insight II (Biosym/MSI San Diego). Residues with solvent-exposed surface areas ≥Å2 were considered highly exposed, those with exposures between 30 and 50 Å2 were considered partially exposed, and those with <30Å2 were considered buried. The number of amino acids in each exposure category was summed for each peptic fragment.
Acknowledgments
We thank Dr. Cynthia Cowgill of Chiron Inc. for rhM-CSFβ, Dr. Sung-Hou Kim for the complete set of rhM-CSFα coordinates file, and Dr. Chakravarty for the depth calculation. We thank Dr. Shing Ho for his assistance with calculations on accessible surface areas and helpful discussions. This work was supported in part by National Institutes of Environmental Health Sciences (NIEHS) grant nos. ES 00040 to M.L.D. and M.I.S., and ES 00210.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0204402.
References
- Chakravarty, S. and Varadarajan, R. 1999. Residue depth: a novel parameter for the analysis of protein structure and stability. Structure 7 723–732. [DOI] [PubMed] [Google Scholar]
- Clarke, J. and Itzhaki, L.S. 1998. Hydrogen exchange and protein folding. Curr. Opin. Struct. Biol. 8 112–118. [DOI] [PubMed] [Google Scholar]
- Connolly, M. L. 1983. Solvent-accessible surfaces of proteins and nucleic acids. Science 221 709–13. [DOI] [PubMed] [Google Scholar]
- Das, S.K. and Stanley, E.R. 1982. Structure–function studies of a colony stimulating factor (CSF-1). J. Biol. Chem. 257 13679–13684. [PubMed] [Google Scholar]
- Deng, Y., Pan, H., and Smith, D.L. 1999. Selective isotope labeling demonstrates that hydrogen exchange at individual peptide amide linkages can be determined by collision-induced dissociation mass spectrometry. J. Am. Chem. Soc. 121 1966–1967. [Google Scholar]
- Engen, J.R. and Smith, D.L. 2001. Investigating protein structure and dynamics by hydrogen exchange MS. Anal. Chem. 73 256A–265A. [DOI] [PubMed] [Google Scholar]
- Englander, S.W. and Kallenbach, N.R. 1983. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16 521–655. [DOI] [PubMed] [Google Scholar]
- Englander, S.W. and Mayne, L. 1992. Protein folding studied using hydrogen-exchange labeling and two-dimensional NMR. Annu. Rev. Biophys. Biomol. Struct. 21 243–265. [DOI] [PubMed] [Google Scholar]
- Englander, J.J., Calhoun, D.B., and Englander, S.W. 1979. Measurement and calibration of peptide group hydrogen-deuterium exchange by ultraviolet spectrophotometry. Anal. Biochem. 92 517–524. [DOI] [PubMed] [Google Scholar]
- Englander, J.J., Englander, S.W., Louie, G., Roder, H., Tran, T., and Wand, A.J. 1988. In Protein hydrogen exchange, dynamics, and energetics. (eds. R.H. Sarma and M.H. Sarma), pp. 107–117 Adenine Press, Schenectady, NY.
- Englander, J.J., Rogero, J.R., and Englander, S.W. 1985. Protein hydrogen exchange studied by the fragment separation method. Anal. Biochem. 147 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder, H., Parak, F., and Young, R.D. 1988. Conformational substates in proteins. Annu. Rev. Biophys. Biophys. Chem. 17 451–479. [DOI] [PubMed] [Google Scholar]
- Glocker, M.O., Arbogast, B., Schreurs, J., and Deinzer, M.L. 1993. Assignment of the inter- and intramolecular disulfide linkages in recombinant human macrophage colony stimulating factor using fast atom bombardment mass spectrometry. Biochemistry 32 482–488. [DOI] [PubMed] [Google Scholar]
- Halenbeck, R., Kawasaki, E., Wrin, J., and Koths, K. 1989. Renaturation and purification of biologically active recombinant human macrophage colony-stimulating factor expressed in E. coli. Bio/Technol. 7 710–715. [Google Scholar]
- Hughes, C.A., Mandell, J.G., Anand, G.S., Stock, A.M., and Komives, E.A. 2001. Phosphorylation causes subtle changes in solvent accessibility at the interdomain interface of methylesterase CheB. J. Mol. Biol. 307 967–976. [DOI] [PubMed] [Google Scholar]
- Hvidt, A. and Nielsen, S.O. 1966. Hydrogen exchange in proteins. Adv. Protein Sci. 21 287–386. [DOI] [PubMed] [Google Scholar]
- Jardetzky, O. and Lefevre, J.-F. 1994. Protein dynamics. FEBS Lett. 338 246–250. [DOI] [PubMed] [Google Scholar]
- Kim, M.Y., Maier, C.S., Reed, D.J., and Deinzer, M.L. 2001. Site-specific amide hydrogen/deuterium exchange in E. coli thioredoxins measured by electrospray ionization mass spectrometry. J. Am. Chem. Soc. 123 9860–9866. [DOI] [PubMed] [Google Scholar]
- McDonald, I.K. and Thornton, J.M. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238 777–793. [DOI] [PubMed] [Google Scholar]
- Milne, J.S., Mayne, L., Roder, H., Wand, A.J., and Englander, S.W. 1998. Determinants of protein hydrogen exchange studied in equine cytochrome c. Protein Sci. 7 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, K., Deane, C.M., Blundell, T.L., Johnson, M.S., and Overington, J.P. 1998. Protein sequence–structure representation and analysis. Bioinformatics 14 617–623. [DOI] [PubMed] [Google Scholar]
- Pandit, J., Bohm, A., Jancarik, J., Halenbeck, R., Koths, K., and Kim, S.H. 1992. Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor. Science 258 1358–1362. [DOI] [PubMed] [Google Scholar]
- Raschke, T. and Marqusee, S. 1998. Hydrogen exchange studies of protein structure. Curr. Opin. Biotech. 9 80–86. [DOI] [PubMed] [Google Scholar]
- Smith, D.L. 1998. Local structure and dynamics in proteins characterized by hydrogen exchange and mass spectrometry. Biochemistry (Moscow) 63 285–293. [PubMed] [Google Scholar]
- Stanley, E.R., Berg, K.L., Einstein, D.B., Lee, P.S., Pixley, F.J., Wang, Y., and Yeung, Y.G. 1997. Biology and action of colony-stimulating factor-1. Mol. Reprod. Dev. 46 4–10. [DOI] [PubMed] [Google Scholar]
- Tolman, J.R., Flanagan, J.M., Kennedy, M.A., and Prestegard, J.H. 1997. NMR evidence for slow collective motions in cyanometmyoglobin. Nature Struct. Biol. 4 292–297. [DOI] [PubMed] [Google Scholar]
- Wagner, D.S. and Anderegg, R.J. 1994. Conformation of cytochrome c studied by deuterium exchange-electrospray ionization mass spectrometry. Anal. Chem. 66 706–711. [DOI] [PubMed] [Google Scholar]
- Woodward, C., Simon, I., and Tuchsen, E. 1982. Hydrogen exchange and the dynamic structure of proteins. Mol. Cell Biochem. 48 135–160. [DOI] [PubMed] [Google Scholar]
- Zemla, A., Venclovas, C., Fidelis, K., and Rost, B. 1999. A modified definition of Sov, a segment-based measure for protein secondary structure prediction assessment. Proteins 34 220–223. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. and Marshall, A.G. 1998. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 9 225–233. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. and Smith, D.L. 1993. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 2 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.H., Yan, X., Maier, C.S., Schimerlik, M.I., and Deinzer, M.L. 2001. Structural comparison of recombinant human macrophage colony stimulating factor beta and a partially reduced derivative using hydrogen deuterium exchange and electrospray ionization mass spectrometry. Protein Sci. 10 2336–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Post, C.B., and Smith, D.L. 1996. Amide hydrogen exchange determined by mass spectrometry: application to rabbit muscle aldolase. Biochemistry 35 779–791. [DOI] [PubMed] [Google Scholar]