Abstract
Rat μ class glutathione transferases M1-1 and M2-2 are homodimers that share a 78% sequence identity but display differences in stability. M1-1 is more stable at the secondary and tertiary structural levels, whereas its quaternary structure is less stable. Each subunit in these proteins consists of two structurally distinct domains with intersubunit contacts occurring between domain 1 of one subunit and domain 2 of the other subunit. The chimeric subunit variants M(12), which has domain 1 of M1 and domain 2 of M2, and its complement M(21), were used to investigate the conformational stability of the chimeric homodimers M(12)-(12) and M(21)-(21) to determine the contribution of each domain toward stability. Exchanging entire domains between class μ GSTs is accommodated by the GST fold. Urea-induced equilibrium unfolding data indicate that whereas the class μ equilibrium unfolding mechanism (i.e., N2 ↔ 2I ↔ 2U) is not altered, domain exchanges impact significantly on the conformational stability of the native dimers and monomeric folding intermediates. Data for the wild-type and chimeric proteins indicate that the order of stability for the native dimer (N2) is M2-2 > M(12)-(12) M1-1 ∼ M(21)-(21), and that the order of stability of the monomeric intermediate (I) is M1 > M2 ∼ M(12) > M(21). Interactions involving Arg 77, which is topologically conserved in GSTs, appear to play an important role in the stability of both the native dimeric and folding monomeric structures.
Keywords: Glutathione transferase, domain exchange, stability, quaternary structure, intermediate
In nature, multimeric proteins appear to be the norm rather than the exception, providing a wide range of protein structures and functions. Benefits for the existence of multimeric proteins include reduced solvent-exposed surface areas, increased stability of the individual subunit structures, and the formation of novel functions at the interfaces (Larsen et al. 1998). Interactions at the subunit interface of multimeric proteins can also assist in their folding to functional conformations. Furthermore, subunit structures of large multimeric proteins are usually divided into compact domains. Individual domains can possess specific functional properties (Rossmann and Argos 1981) and play an important role in protein folding (Wetlaufer 1973), assembly, and stability (Jaenicke and Lilie 2000).
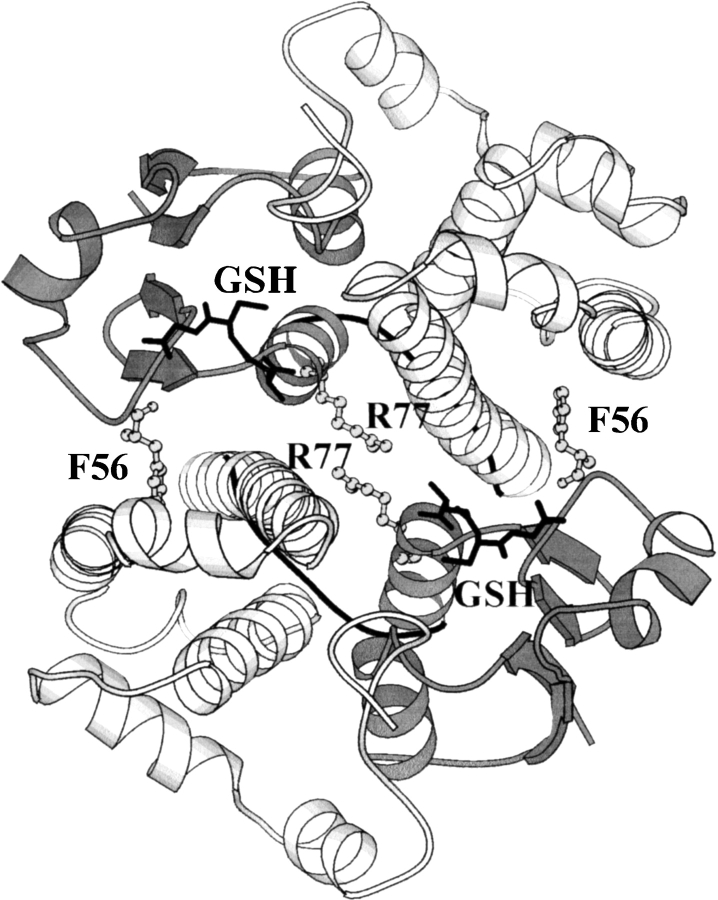
The cytosolic glutathione transferases (GSTs; EC 2.5.1.18) belong to a superfamily of multifunctional proteins that are grouped into various species-independent gene classes (Dirr et al. 1994; Armstrong 1997). The enzymes exist as stable homo- and heterodimers with a conserved archetypical fold (for review, see Dirr et al. 1994; Wilce and Parker 1994; Armstrong 1997). Each GST subunit consists of two domains. Domain 1 has a thioredoxin-like fold with a βαβαββα topology, whereas domain 2 consists solely of α-helices, as shown in Figure 1 ▶ for class μ rGSTM1-1 (Ji et al. 1992). The two domains are connected by a short linker sequence. The active site on each subunit is positioned between the domains in which glutathione and the electrophilic substrates are bound by domain 1 and domain 2, respectively. In spite of their structural homology, only subunits within a given GST gene class can associate to form homo- or heterodimers. Dimerization involves specific contacts between domain 1 of one subunit and domain 2 of the neighboring subunit. These interactions contribute significantly toward stabilizing the tertiary structures of individual subunits (Dirr 2001). The dimeric structure is required to maintain functional conformations at the active site on each subunit and the nonsubstrate ligand-binding site at the dimer interface (Sayed et al. 2000; Dirr 2001).
Fig. 1.
Ribbon representation of the structure of rat glutathione transferase M1-1 (Ji et al. 1992) looking down the noncrystallographic twofold axis of symmetry. Domain 1 and domain 2 are in dark and light gray, respectively, with the linker region connecting the two domains in black. The positions of Phe 56 and Arg 77 at the dimer interface as well as glutathione bound to domain I of each subunit are indicated.
What is unknown is the role played by individual domains in maintaining the stability of GST dimers and their monomers. At equilibrium, the class μ isozymes M1-1 and M2-2 follow a three-state folding mechanism 2U ↔ 2I ↔ N2, but display significant differences in their stabilities (Hornby et al. 2000). M1-1 is more stable at the secondary and tertiary structural levels, whereas its quaternary structure is less stable than M2-2. M1-1 and M2-2 share an overall 78% sequence identity (82.7% in domain 1 and 75.6% in domain 2) with sequence variation clustered mainly in four regions of the primary structure (Zhang et al. 1992). Sequence variable regions are involved in determining the catalytic properties of M1-1 and M2-2 (Zhang et al. 1992).
Herein, we describe the conformational stability and equilibrium folding of two domain-exchanged chimeric isozymes, M(12)-(12) and M(21)-(21). The former chimera has domain 1 from M1 and domain 2 from M2, whereas the latter chimera has domain 1 from M2 and domain 2 from M1 (The catalytic properties of both chimeric proteins and the crystal structure of the M(12)-(12) chimera will be described in a separate manuscript. (J. Chen, G. Xiao, G.L. Gilliland, and R.N. Armstrong, in prep.). The crystal structure was solved at a resolution of 1.70 Å with R = 0.179 and Rfree = 0.212. The crystallographic coordinates have been deposited in the Protein Data Bank under the file name 1B4P). Urea-induced unfolding of the chimeric proteins was monitored under equilibrium conditions by use of tryptophan fluorescence, circular dichroism, and glutaraldehyde cross-linking. The data indicate that although the class μ equilibrium unfolding mechanism (i.e., N2 ↔ 2I ↔ 2U) is not altered, domain exchanges impact significantly on the conformational stability of the native dimers and monomeric-folding intermediates. The order of stability for the native dimer (N2) is M2-2 > M(12)-(12) > M1-1 ∼ M(21)-(21), and the order of stability of the monomeric intermediate (I) is M1 > M2 ∼ M(12) > M(21). Interactions involving Arg 77, which is topologically conserved in GSTs, appear to play an important role in the stability of both the native dimeric and folding monomeric structures.
Results
Spectroscopic properties
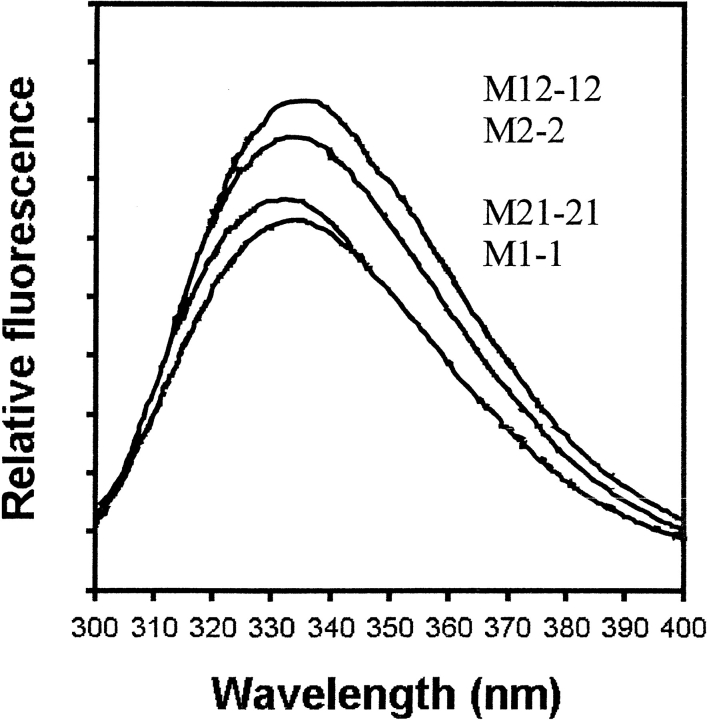
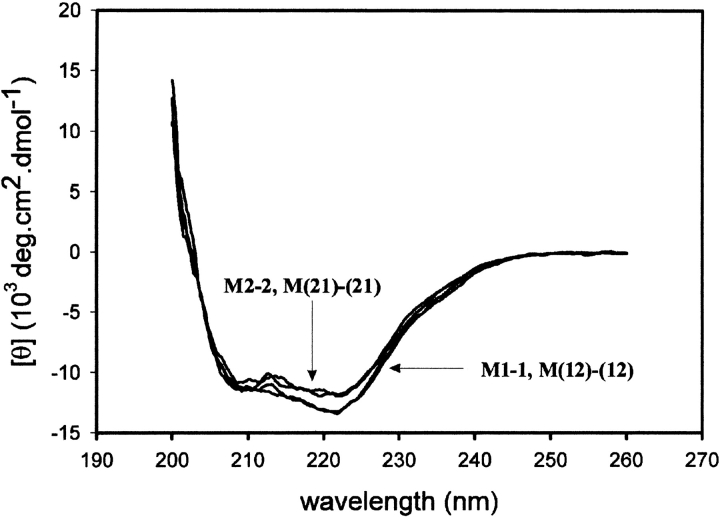
The wild-type and chimeric class μ isozymes contain four tryptophan residues per subunit at conserved positions (i.e., Trp 7, Trp 45, Trp 146, and Trp 214). The tryptophan fluorescence emission spectra of the wild-type and chimeric enzymes are shown in Figure 2 ▶. M1-1, M2-2, and M(12)-(12) display an emission maximum at 335 nm, whereas M(21)-(21) has a maximum at 332 nm. Proteins sharing a common domain 2 have similar fluorescence intensities. From the crystal structures of M1-1 (Ji et al. 1992) and M(12)-(12), there are few differences between the immediate environments around the tryptophans, although Trp 214 of M1-1 is in a more polar environment than that of M(12)-(12), which may result in a more quenching environment for domain 2 from M1. Like the wild-type proteins (Hornby et al. 2000), unfolding of the chimeras in 8 M urea resulted in an increase in fluorescence intensity and a shift in emission maximum to 355 nm (data not shown). The far-UV CD spectra of the wild-type and chimeric proteins are similar (Fig. 3 ▶) with ellipticity minima at 208 and 222 nm, typical of proteins with a high α-helical content.
Fig. 2.
Tryptophan emission spectra (excitation at 295 nm) of the wild-type enzymes M1-1 and M2-2, and the two domain exchanged chimeras. All proteins were at a dimer concentration of 5 μM.
Fig. 3.
Far-UV circular dichroism of wild-type enzymes M1-1 and M2-2, and the two domain exchanged chimeras.
Equilibrium unfolding
The unfolding of the chimeras was reversible as determined by tryptophan fluorescence spectra. Recoveries of enzyme activity was 95% and 82% for M(21)-(21) and M(12)-(12), respectively. The unfolded chimeric constructs are thus able to refold reversibly into structurally and functionally native forms. These recovery data are comparable with those for the wild-type enzymes (Hornby et al. 2000), as well as those of α (Wallace et. al. 1998) and π (Erhardt and Dirr 1995) class enzymes, which are the closest evolutionary neighbors of class μ. Furthermore, there has been no evidence of hysteresis in the equilibrium unfolding/refolding of the GSTs (L.A. Wallace and H.W. Dirr, unpubl.). The urea-induced unfolding curves for M(21)-(21) are shown in Figure 4 ▶. The fluorescence-unfolding curve is biphasic (Fig. 4A ▶), the first phase resulting in an increase in the emission intensity at 332 nm, followed by a decrease during the second phase. The emission maximum shifted from 332 to 338 nm during the first transition and from 338 to 355 nm during the second. The position of the first transition was dependent on the protein concentration (inset in Fig. 4A ▶). In contrast, the CD unfolding curve is monophasic and corresponds to the second fluorescence transition (Fig. 4B ▶). This unfolding behavior is similar to that displayed by wild-type M1-1 and M2-2 (Hornby et al. 2000), indicating that M(21)-(21) also follows a three-state unfolding mechanism at equilibrium (N2 ↔ 2I ↔ 2U). The distribution of the three states present during unfolding, shown in Figure 4C ▶, were obtained by fitting the unfolding data in Figure 4A ▶ to a three-state process (Hornby et al. 2000). The appearance of the unfolded monomer coincides with the CD transition in Figure 4B ▶. The unfolding behavior of M(12)-(12) was similar to that of M(21)-(21), as shown in Figure 5 ▶. However, unlike M(21)-(21), the first fluorescence transition displayed a very weak protein-concentration dependence (Fig. 5A ▶, inset). M(12)-(12) is dimeric in the absence of denaturant as determined by SEC-HPLC (data not shown) and cross-linking with glutaraldehyde (Fig. 6 ▶). It appears that tryptophan fluorescence in M(12)-(12) is not sensitive to the dissociation event as shown for M(21)-(21). The absence of a protein-concentration dependence for dimer dissociation/association has also been reported for creatine kinase (Fan et al. 1998) and for Ure2, which has a carboxy-terminal homology to GSTs (Perrett et al. 1999). The first and second fluorescence transitions for M(12)-(12) were accompanied by a shift in the emission maximum from 335 to 338 nm and from 338 to 355 nm, respectively. The distribution of the conformational states present during M(12)-(12) unfolding is shown in Figure 5C ▶, based on a three-state mechanism. The values of the thermodynamic parameters for the dissociation and unfolding of the chimeras and wild-type class μ GSTs are presented in Table 1.
Fig. 4.
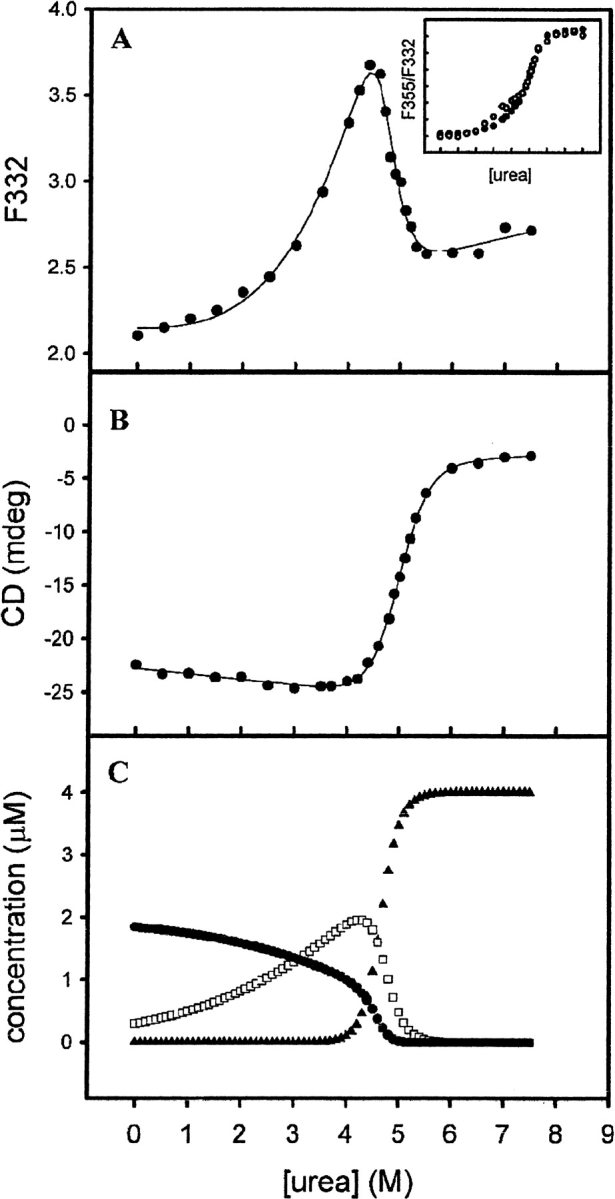
(A) Equilibrium unfolding of 2 μM M(21)-(21) monitored by tryptophan fluorescence (λexcitation = 295 nm, λemission = 332 nm). (Inset) Fuorescence-unfolding transitions of 2 μM (•) and 0.2 μM (○) protein, plotted as a ratio of the fluorescence at 355 nm and 332 nm (λexcitation = 295nm). (B) Urea-induced unfolding transition of 2 μM M(21)-(21) monitored by ellipticity at 222 nm. Solid lines in A and B are curve fits from which thermodynamic parameters were derived. (C) Concentrations of N2 (•), I (□), and U(▴) as calculated from the curve fit in A. All unfolding was done at 20°C in Buffer 1.
Fig. 5.
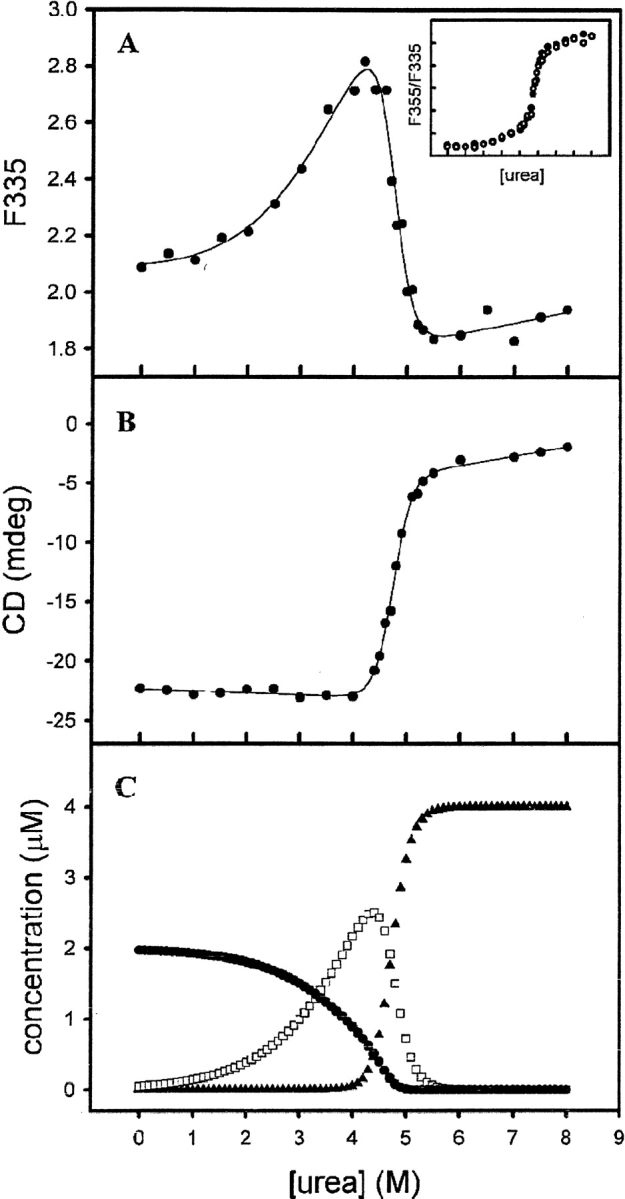
(A) Equilibrium unfolding of 2 μM M(12)-(12) monitored by tryptophan fluorescence (λexcitation = 295 nm, λemission = 335 nm). (Inset) Fluorescence unfolding transitions of 2 μM (•) and 0.2 μM (○) protein, plotted as a ratio of the fluorescence at 355 and 335 nm (λexcitation = 295 nm). (B) Urea-induced unfolding transition of 2 μM M(12)-(12) monitored by ellipticity at 222 nm. Solid lines in A and B are curve fits from which thermodynamic parameters were derived. (C) Concentrations of N2 (•), I (□), and U(▴) as calculated from the curve fit in A. Conditions are identical to those described in Fig. 4 ▶.
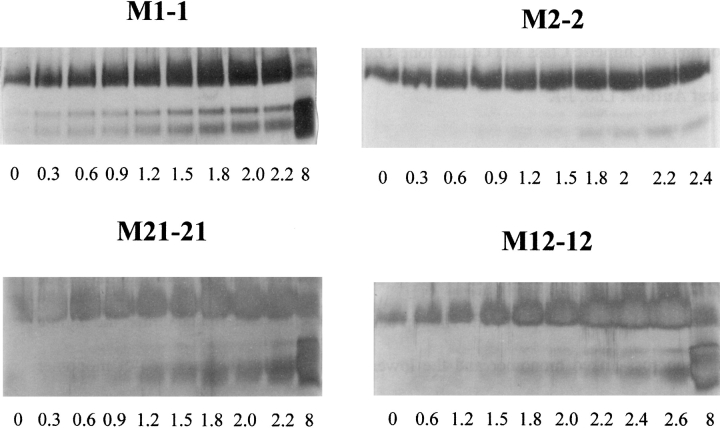
Fig. 6.
SDS-PAGE analysis of wild-type and chimeric enzymes cross-linked with glutaraldehyde. A total of 2 μM enzyme incubated in various concentrations of urea (numbers below the gels denote the molar concentration of urea used) were reacted with glutaraldehyde and resolved on 15% SDS-PAGE. The top band represents cross-linked dimer, the middle band uncross-linked monomer, and the bottom band indicates the presence of monomer with intramolecular cross-links.
Table 1.
Thermodynamic parameters of unfolding of native and chimeric class μ glutathione transferases
| Fluorescencea | CDb | |||||
| Transition 1 | Transition 2 | |||||
| Protein | ΔG(H2O)1 (kcal • mol−1) | m1 (kcal • mol−1 • M1) | ΔG(H2O)2 (kcal • mol−1) | m2 (kcal • mol−1 • M1) | ΔG(H2O) (kcal • mol−1) | m (kcal • mol−1 • M1) |
| M1–1c | 10.8 ± 1.8 | 1.0 ± 0.2 | 16.5 ± 0.1 | 3.5 ± 0.3 | 19.6 ± 0.2 | 3.38 ± 0.02 |
| M2–2c | 12.4 ± 0.9 | 1.8 ± 0.1 | 14.8 ± 0.8 | 3.1 ± 1.1 | 17.1 ± 1.1 | 3.7 ± 0.03 |
| M12–12 | 11.7 ± 0.8 | 1.0 ± 0.2 | 14.2 ± 1.9 | 3.1 ± 0.3 | 16.5 ± 0.4 | 3.5 ± 0.1 |
| M21–21 | 10.2 ± 0.8 | 0.8 ± 0.2 | 11.8 ± 1.4 | 2.5 ± 0.2 | 9.9 ± 0.3 | 1.99 ± 0.05 |
a Fluorescence data were fitted to a three-state unfolding mechanism (N2 ↔ 2I ↔ 2U).
b The CD data were fit to a two-state mechanism.
c Data for the native enzymes are from Hornby et al. (2000).
Glutaraldehyde cross-linking
The fact that the first fluorescence-unfolding phase observed for both chimeras corresponds to dimer dissociation was confirmed by glutaraldehyde cross-linking experiments (Fig. 6 ▶). Similar experiments were reported for the wild-type proteins (Hornby et al. 2000). M1-1 and M2-2 begin to dissociate at ∼0.3 to 0.6 M urea and at ∼1.8–2 M urea, respectively, confirming our recent finding that the M2-2 dimer is more stable than the M1-1 dimer (Hornby et al. 2000). The data in Figure 6 ▶ indicate the M(12)-(12) chimera shares a similar dimer stability to M2-2, with monomers appearing at ∼1.5–1.8 M urea. The stability of the M(21)-(21) dimer, however, is similar to that of M1-1, with monomers appearing at ∼0.6 M urea.
Discussion
Interdomain contacts are involved in the assembly of GST subunits and dimers. The crystal structures of M1-1 and M(12)-(12) show that the exchange of entire domains between class μ isozymes does not impact significantly on overall protein architecture. This is also evident from fluorescence and far-UV CD-spectral data for the wild-type and chimeric proteins. Furthermore, exchanging domains 1 and 2 between M1-1 and M2-2 does not alter their three-state equilibrium unfolding mechanism (N2 ↔ 2I ↔ 2U), as shown for the M(12)-(12) and M(21)-(21) chimeras. Although dissociation of the dimers does not significantly affect the secondary structure, it results in changes in the tertiary structure of the subunits, as indicated by the large increases in tryptophan fluorescence and increased solvent exposure of tryptophans. Interactions across the dimer interface, therefore, play an important role in stabilizing not only the dimeric structure but also the native tertiary structure of subunits in class μ (this work; Hornby et al. 2000) and other gene classes (for review, see Dirr 2001). Dissociation results in catalytically inactive monomers. The small values for m1 (Table 1) are consistent with the dissociation of the dimers to structured monomers. They are, however, larger than the expected value of ∼0.5 kcal mole−1M−1 urea based on the surface areas that are buried at the dimer interfaces in the crystal structures of M1-1 and M(12)-(12). The monomeric intermediates, therefore, have a native-like secondary structure but display less compact tertiary structures (this work; Hornby et al. 2000).
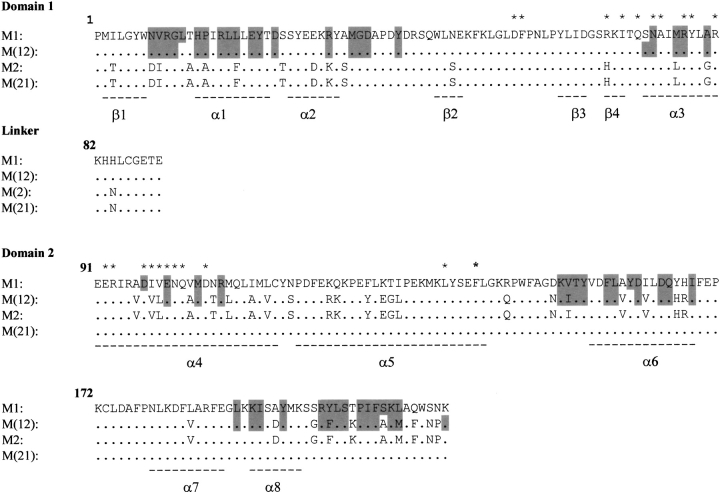
Spectroscopic and cross-linking data for the equilibrium unfolding of the wild-type and chimeric class μ GSTs show that the order of dimer stability is M2-2 > M(12)-(12) > M1-1 ∼ M(21)-(21) (this work; Hornby et al. 2000). The ranking suggests that the interacting surfaces in M2-2 have the most favorable geometric and chemical complementarity, and that domain 2 plays an important role in determining dimer stability. Hydrophobic and electrostatic interactions are the major forces stabilizing GST dimers. The amino acid residues involved in intersubunit contacts in the crystal structures of M1-1 and M(12)-(12) together with the corresponding residues in the sequences of M2-2 and M(21)-(21) are largely conserved in the four proteins (Fig. 7 ▶). Although many of the interactions between domain 1 of one subunit and domain 2 of the other subunit are conserved in the two crystal structures, there are significant differences in charge-cluster interactions at the dimer twofold axis (Fig. 8 ▶). The buried mixed-charge cluster is proposed to stabilize the quaternary structure of GSTs (Zhu and Karlin 1996). At the dimer interfaces of both M1-1 and M(12)-(12), Arg 81 in domain 1 forms salt bridges to Glu 90 and Asp 97 in domain 2 of the neighboring subunit (Fig. 8 ▶). A major difference in the charge cluster of M1-1 and M(12)-(12) lies in the orientation of the side chain of Arg 77 (Fig. 8A,B ▶). In M1-1, there are five water molecules within 4Å of Arg 77, at least two of which are able to hydrogen bond with the Arg 77 guanidino group. Furthermore, Arg 77 forms salt-bridges with Asp 97 and Glu 100 in domain 2 of the same M1 subunit (Fig. 8A ▶). In M(12)-(12), however, the orientation of the Arg 77 side-chain is different from that in M1-1 (Fig. 8B ▶), with only one water molecule within 4Å. A WHATIF (Vriend 1990) calculation of the loss of solvent-accessible surface area upon dimerization for Arg 77 in M(12)-(12) indicates that it is almost twice as buried as Arg 77 in M1-1. The orientation of Arg 77 in M12-12 causes it to lose its intrasubunit salt bridge to Asp 97 but, instead, it forms salt bridges to Glu 100 and Asp 97 in the neighboring subunit (Fig. 8B ▶). These additional interactions might explain the greater stability of the M(12)-(12) dimer relative to the M1-1 dimer. Calculated solvent-accessible surface areas (Vriend 1990) indicate a loss of ∼1700 Å2 nonpolar surface area and ∼980 Å2 polar surface area upon dimerization of M1-1. The corresponding values for M(12)-(12) are 1630 Å2 and 1050 Å2, respectively. The ratio of nonpolar-to-polar surface areas indicates a burial of more polar surface in M(12)-(12) (ratio = 1.55) than in M1-1 (ratio = 1.73) upon dimerization, suggestive of the importance of polar interactions in enhancing the stability of M(12)-(12). Arg 77 is topologically conserved in GSTs. Due to the absence of crystal structures for rat M2-2 and M(21)-(21), details of the interactions at their dimer interfaces are not known.
Fig. 7.
Aligned sequences for M1, M2, M(12), and M(21). (*) Residues involved in intersubunit interactions. Shaded residues indicate those involved in interdomain contacts. Positions of helices (α) and strands (β) are indicated by broken lines under the sequences.
Fig. 8.
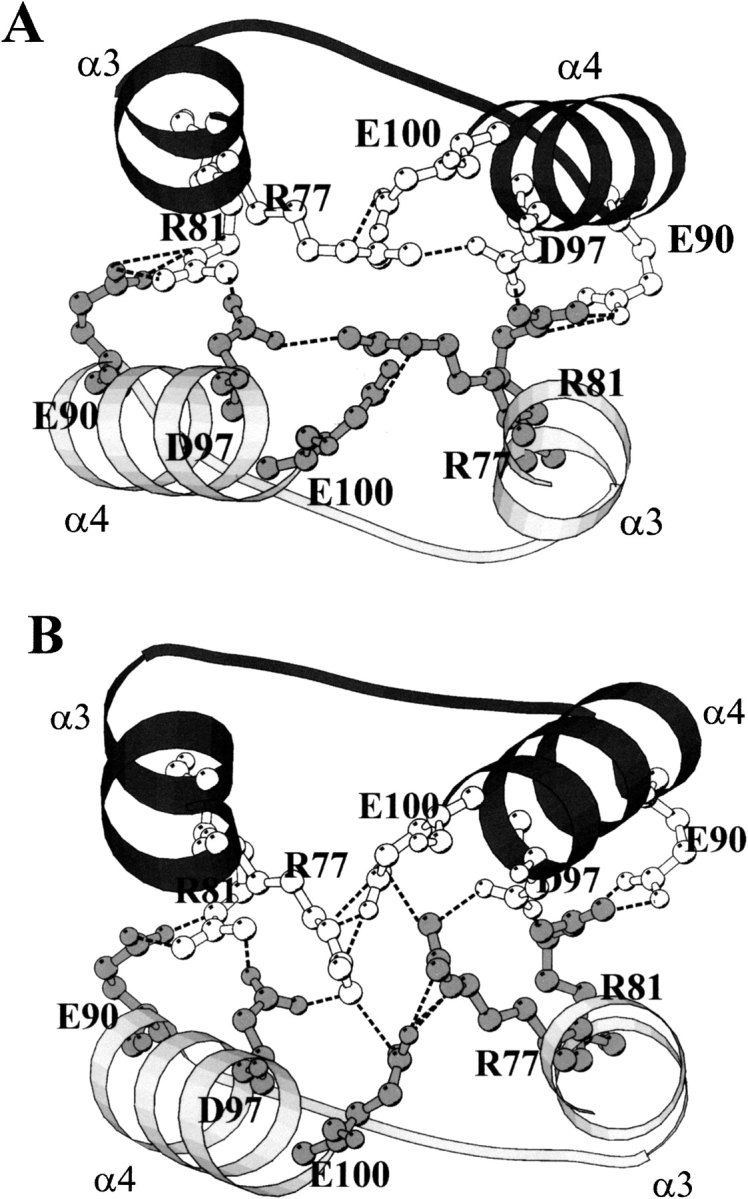
Ball-and-stick representation of the amino acid residues in the charge cluster at the subunit interface of (A) M1-1 (Ji et al. 1992), and (B) M(12)-(12)2.
Another difference observed at the dimer interface of M1-1 and M(12)-(12) involves a conserved hydrophobic motif (Fig. 1 ▶). In this motif, the side-chain of Phe 56 in domain 1 of one subunit fits into a hydrophobic pocket in domain 2 of the neighboring subunit. In the structure of M(12)-(12) (and by analogy M2-2), the hydrophobic pocket includes the side chain of Leu 99 that makes van der Waals contact with the phenyl ring of Phe 56 and could increase dimer stability. On the other hand, the corresponding residue (Val 99) in M1-1 [and by analogy M(21)-(21)], is too short to interact with Phe 56. The intersubunit hydrophobic motif, although not essential for dimerization, has been shown to contribute significantly toward stabilizing GST dimers from class α (Sayed et al. 2000), class π (Stenberg et al. 2000) and class μ (J.A.T. Hornby, S. Codreanu, R.N. Armstrong, and H.W. Dirr, in prep.).
Equilibrium unfolding of the wild-type (Hornby et al. 2000) and chimeric proteins (this work) show that the order of stability of the monomeric folding intermediate is M1 > M2 ∼ M(12) > M(21). Because structural details of the monomers and how their domain interfaces might be affected by dimer dissociation are unknown, the contributions of the intrinsic stability of domains 1 and 2 and domain–domain interactions toward the overall stability of the monomers are not known. Partial dissociation of the domains during the unfolding of class α GSTA1-1 has been reported to occur in the region of helix 1 in domain 1 and helix 8 in domain 2 (Wallace et al. 2000). Amino acid residues that make interdomain contacts in the native subunit crystal structures of M1 and M(12) are shown in Figure 7 ▶. Three of them are variable but represent conservative mutations [i.e., V152I, Y202F, and L211M in M(12) relative to M1], and probably would not affect stability. Calculated solvent-accessible surface areas (Vriend 1990) indicate a loss of ∼900 Å2 in nonpolar surface area and 700 Å2 in polar surface area for domain 1 of both M1 and M(12) upon association with domain 2. The corresponding values for domain 2 upon association with domain 1 are 1020 Å2 and 510 Å2 for nonpolar and polar surface areas, respectively. The ratios of nonpolar-to-polar buried surface areas indicate very little differences between M1 and M(12). It is interesting to note that Arg 77, which is involved in intersubunit contacts (see Fig. 8 ▶), also forms interdomain contacts in the M1 and M(12) subunit structures. In the M1 subunit, Arg 77 in helix 3 forms interdomain hydrogen bonds with Asp 97 and Glu 100 in helix 4 and with Tyr 154 in helix 6. In the M(12) subunit, Arg 77 hydrogen bonds only to Glu 100 in helix 4. Should these interactions persevere in the structures of the folding monomers, they might explain, in part, the greater stability of the M1 monomer. Contacts between helix 3 in domain 1 and helices 4 and 6, which, with helix 5, form the core of domain 2, might play an important role in stabilizing the monomers (see below).
It was originally thought that the forces driving protein oligomerization would be similar to that of protein folding and so the interface would bear some of the characteristics of folded proteins, such as a central hydrophobic core surrounded by polar or charged residues (Chothia and Janin 1975; Miller 1989). However, a more recent survey of 136 homodimeric proteins showed that only 43 displayed a recognizable hydrophobic core (Larsen et. al. 1998). The majority of the proteins showed a patchy character, with hydrophobic and hydrophilic interactions interspersed throughout the interface. The dimer interface of the GSTs investigated in this study fall within this classification. Interfaces, in general, show intermediate hydrophobicity between the compact protein core and the protein surface (Jones and Thornton 1996). Therefore, the energy cost of burying an ion-pair within the interface is not as high as the equivalent burial in folding and leads to the greater contribution of ion pairs, hydrogen bonding, and charge clusters in stabilizing the interface between protein subunits (Zhu and Karlin 1996; Xu et. al. 1997).
The class of interfaces with a hydrophobic core correspond to proteins that are proposed to associate by a two-state mechanism, whereas the interfaces with mixed hydrophobicity belong to the proteins that associate by a three-state process. This is supported by the unfolding mechanism of the class μ glutathione transferases, in which the mix of hydrophobic and hydrophilic interactions at the dimer interface allow for the existence of a stable monomeric intermediate. Folded monomeric intermediates have also been reported for desulfoferrodoxin, which shows an open, polar dimer interface (Apiyo et. al. 2001), and for tertrameric peanut agglutinin, which possesses a similar open oligomeric interface (Reddy et. al. 1999). In contrast, the Escherichia coli Trp repressor, a dimeric protein with an interdigitated hydrophobic dimer interface, unfolds via a two-state mechanism (Gittelman and Matthews 1990) However, other experimental studies have shown that the folding mechanism does not always follow from the nature of the interface. The arc repressor protein, for example, which also possesses a highly interdigitated hydrophobic interface, shows a stable folding intermediate (Silva et. al. 1992).
There is no evidence to suggest that the individual domains in the class μ chimeric (this work) and wild-type (Hornby et al. 2000) monomers unfold independently. Their tryptophan fluorescence and far-UV CD unfolding transitions are coincident and monophasic. The cooperative unfolding of domain 1 and domain 2 is shown by the steep unfolding transitions and their corresponding m2-values; the latter being consistent with the surface area that becomes exposed to solvent during unfolding of structured monomers. Cooperativity between domains 1 and 2 has also been observed for GSTs from class α (Wallace et al. 1998), class σ (Stevens et al. 1998), class π (Erhardt and Dirr 1995), and Sj26GST (Kaplan et al. 1997). However, a recent thermal irreversible denaturation study with hGSTP1-1 suggests that domain 1 is less stable than domain 2 and that an unfolding monomeric intermediate might exist consisting of a structured domain 2 and partially unfolded domain 1 (Dragani et al. 1998). It should be noted that one of the two tryptophans in the class π GST is located in the highly dynamic helix 2 (Stella et al. 1998; Vega et al. 1998), and that the fluorescence probe most likely monitors local rather than global conformational changes in domain 1. Local unfolding has been reported for the highly flexible helix 2 in hGSTP1-1 (Hitchens et al. 2001) and a class σ GST (Stevens et al. 1998). The corresponding helical region in class μ (including Sj26GST) and α GSTs is more stable (Sinning et al. 1993; McCallum et al. 2000).
Traditionally, a domain is defined as a compact, self-contained structural region having more contacts with itself than with the rest of the protein and that a minimum amount of surface area becomes newly exposed upon dissection of the protein into individual domains (Jaenicke 1999; Peng and Wu 2000). The interface between the domains in the native class μ subunits is extensive; the surface area that becomes buried when domain 1 and domain 2 associate is calculated to be ∼1500 Å, which is slightly more than that buried at the dimer interface (1200 Å). This most likely explains why rGSTM1-1 is predicted to have one rather than two domains (Siddiqui and Barton 1995). The entropic penalty due to folding of the subunit would be paid in part by a gain in enthalpy arising from extensive interdomain contacts. Domain 1 in GSTs was recruited from a thioredoxin-like ancestral protein (Martin 1995; Rossjohn et al. 1998). Limited proteolysis studies suggest that domain 1 is comprised of two subdomains, a β1α1β2α2 N-subdomain and a β3β4α3 C-subdomain (Martini et al. 1993; Aceto et al. 1995a). These subdomains are similar to those found in thioredoxin (Tasayco et al. 2000). It is proposed that the folding of thioredoxin-like proteins/domains is initiated by interactions between hydrophobic regions corresponding to the neighboring β strands in the N and C subdomains (e.g., β1 and β3 in GST) (Tasayco et al. 2000). The cooperativity between the domains in the GST subunit appear to be maintained primarily by interactions between helix 3 in the C-subdomain and helices 4 and 6 in domain 2 (Gulick et al. 1992; Martini et al. 1993; Aceto et al. 1995b; Dragani et al. 1998). Removal of the N-subdomain (Martini et al. 1993; Aceto et al. 1995b) or truncating a key N-subdomain-domain 2 contact (Wallace et al. 2000) does not impact significantly on the structure of domain 2.
In summary, exchanging entire domains between class mu GSTs is accommodated by the GST fold. Although the equilibrium folding mechanism is not altered, the domain interchange has a significant impact on the conformational stability of the native dimers and monomeric folding intermediates. Interactions involving Arg 77, which is topologically conserved in GSTs, appears to play an important role in the stability of both the native dimeric and the folded monomeric structures.
Materials and methods
Materials
Ultrapure urea was from Merck, glutathione was from ICN Biomedicals Inc., and all other reagents were of analytical grade. Silver staining was performed with the Amersham Quicksilver kit. Wild-type M1-1 and M2-2 were expressed and purified as described previously (Hornby et al. 2000).
Chimera expression and purification
The M(12)-(12) and M(21)-(21) chimeric proteins were overexpressed in Escherichia coli M5219 as described (Zhang and Armstrong 1990). They were purified either by CM-Sephadex ion-exchange chromatography (Hornby et al. 2000) or by S-hexylglutathione affinity chromatography (Stenberg et al. 1992) and subsequently buffer exchanged into Buffer 1 (20 mM sodium phosphate at pH 6.5, 1 mM EDTA, 0.1 M NaCl, 0.02% NaN3). Protein concentration was determined using a ɛ280 of 82 820 M−1 • cm−1 for M1-1 and M(21)-(21), and a ɛ280 of 80 140 M−1 • cm−1 for M2-2 and M(12)-(12), as calculated by the method of Perkins (1986).
Spectroscopic studies
Fluorescence emission spectra and other fluorescence measurements were made in Buffer 1 at 20°C with a Hitachi model 850 spectrofluorimeter. Excitation was at 295 nm. Far-UV CD spectra were determined using a 2-mm pathlength cuvette in a Jasco model 810 spectropolarimeter. Mean residue ellipticity (θ) (deg • cm2 • dmol−1) was calculated as [θ] = 100(signal)/Cnl, in which (signal) denotes the ellipticity signal after subtraction of the solvent baseline, C is the millimolar concentration of protein, n is the number of residues, and l is the pathlength in centimeters.
Equilibrium unfolding
Equilibrium unfolding was performed in Buffer 1 containing various concentrations of urea as described (Hornby et al. 2000). Structural changes in the proteins during unfolding were monitored by tryptophan fluorescence and far-UV CD spectroscopy. Unfolding curves were fitted using SigmaPlot 5.0 (Hornby et al. 2000). Reversibility of unfolding was measured following a 10-fold dilution of denatured protein into nondenaturing Buffer 1. Samples (100 μL) of protein denatured in various concentrations of urea were also subjected to glutaraldehyde cross-linking by adding 20 μL of 25% glutaraldehyde to each sample and allowing the cross-linking reaction to proceed for 20 min. SDS-PAGE in 15% gels was performed on cross-linked samples, followed by silver staining.
Acknowledgments
This work was supported by the University of the Witwatersrand, the South African Foundation for Research and Development, the Wellcome Trust, the Forgarty International Collaboration Award TW00779, and Grant GM30910 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
CD, circular dichroism
CDNB, 1-chloro-2,4-dinitrobenzene
GSH, reduced glutathione
GST(s), glutathione S-transferase(s)
rGSTM1-1, μ class GST from rat, a dimer of two type-1 subunits
rGSTM2-2, as rGSTM1-1, but a dimer of two type-2 subunits
SDS-PAGE, sodium dodecyl sulphate polyacrylamide gel electrophoresis
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0208002.
References
- Aceto, A., Dragani, B., Allocati, N., Angelucci, S., Bucciarelli, T., Sacchetta, P., Di Ilio, C.D., and Martini, F. 1995a. Analysis by limited proteolysis of domain organization and GSH-site arrangement of bacterial glutathione transferase B1-1. Int. J. Biochem. Cell Biol. 27 1033–1041. [DOI] [PubMed] [Google Scholar]
- Aceto, A., Sacchetta, P., Bucciarelli, T., Dragani, B., Angelucci, S., Radatti, G.L., and Di Ilio, C. 1995b. Structural and functional properties of the 34-kDa fragment produced by the N-terminal chymotryptic cleavage of glutathione transferase P1-1. Arch. Biochem. Biophys. 316 873–878. [DOI] [PubMed] [Google Scholar]
- Apiyo, D., Jones, K., Guidry, J., and Wittung-Stafshede, P. 2001. Equilibrium unfolding of dimeric desulfoferrodoxin involves a monomeric intermediate: Iron cofactors dissociate after polypeptide unfolding. Biochemistry 40 4940–4948. [DOI] [PubMed] [Google Scholar]
- Armstrong, R.N. 1997. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 10 2–18. [DOI] [PubMed] [Google Scholar]
- Chothia, C. and Janin, J. 1975. Principles of protein-protein recognition. Nature 256 705–708. [DOI] [PubMed] [Google Scholar]
- Dirr, H., Reinemer, P., and Huber, R. 1994. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur. J. Biochem. 220 645–661. [DOI] [PubMed] [Google Scholar]
- Dirr, H.W. 2001. Folding and assembly of glutathione transferases. Chem. Biol. Interact. 133 19–23. [Google Scholar]
- Dragani, B., Iannarelli, V., Allocati, N., Masulli, M., Cicconetti, M., and Aceto, A. 1998. Irreversible thermal denaturation of glutathione transferase P1-1. Evidence for varying structural stability of different domains. Int. J. Biochem. Cell Biol. 30 155–163. [Google Scholar]
- Erhardt, J. and Dirr, H. 1995. Native dimer stabilizes the subunit tertiary structure of porcine class pi glutathione S-transferase. Eur. J. Biochem. 230 614–620. [PubMed] [Google Scholar]
- Fan, Y.X., Zhou, J.M., Kihara, H., and Tsou, C.L. 1998. Unfolding and refolding of dimeric creatine kinase equilibrium and kinetic studies. Protein Sci. 7 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittelman, M.S. and Matthews, C.R. 1990. Folding and stability of trp aporepressor from Escherichia coli. Biochemistry 29 7011–7020. [DOI] [PubMed] [Google Scholar]
- Gulick, A.M., Goihl, A.L., and Fahl, W.E. 1992. Structural studies on human glutathione S-transferase pi. Family of native-specific monoclonal antibodies used to block catalysis. J. Biol. Chem. 267 18946–18952. [PubMed] [Google Scholar]
- Hitchens, T.K., Mannervik, B., and Rule, G.S. 2001. Disorder-to-order transition of the active site of human class Pi glutathione transferase, GST P1-1. Biochemistry 40 11660–11669. [DOI] [PubMed] [Google Scholar]
- Hornby, J.A., Luo, J.K., Stevens, J.M., Wallace, L.A., Kaplan, W., Armstrong, R.N., and Dirr, H.W. 2000. Equilibrium folding of dimeric class mu glutathione transferases involves a stable monomeric intermediate. Biochemistry 39 12336–12344. [DOI] [PubMed] [Google Scholar]
- Jaenicke, R. 1999. Stability and folding of domain proteins. Prog. Biophys. Mol. Biol. 71 155–241. [DOI] [PubMed] [Google Scholar]
- Jaenicke, R. and Lilie, H. 2000. Folding and association of oligomeric and multimeric proteins. Adv. Protein Chem. 53 329–401. [DOI] [PubMed] [Google Scholar]
- Ji, X., Zhang, P., Armstrong, R.N., and Gilliland, G.L. 1992. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3–3 and glutathione at 2.2-A resolution. Biochemistry 31 10169–10184. [DOI] [PubMed] [Google Scholar]
- Jones, S. and Thornton, J.M. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. 93 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, W., Husler, P., Klump, H., Erhardt, J., Sluis-Cremer, N., and Dirr, H. 1997. Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase: A detoxification enzyme and fusion-protein affinity tag. Protein Sci. 6 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, T.A., Olson, A.J., and Goodsell, D.S. 1998. Morphology of protein-protein interfaces. Structure. 6 421–427. [DOI] [PubMed] [Google Scholar]
- Martin, J.L. 1995. Thioredoxin–a fold for all reasons. Structure 3 245–250. [DOI] [PubMed] [Google Scholar]
- Martini, F., Aceto, A., Sacchetta, P., Bucciarelli, T., Dragani, B., and Di Ilio, C. 1993. Investigation of intra-domain and inter-domain interactions of glutathione transferase P1-1 by limited chymotryptic cleavage. Eur. J. Biochem. 218 845–851. [DOI] [PubMed] [Google Scholar]
- McCallum, S.A., Hitchens, T.K., Torborg, C., and Rule, G.S. 2000. Ligand-induced changes in the structure and dynamics of a human class Mu glutathione S-transferase. Biochemistry 39 7343–7356. [DOI] [PubMed] [Google Scholar]
- Miller, S. 1989. The structure of interfaces between subunits of dimeric and tetrameric proteins. Protein Eng. 3 77–83. [DOI] [PubMed] [Google Scholar]
- Peng, Z.Y. and Wu, L.C. 2000. Autonomous protein folding units. Adv. Protein Chem. 53 1–47. [DOI] [PubMed] [Google Scholar]
- Perkins, S.J. 1986. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur. J. Biochem. 157 169–180. [DOI] [PubMed] [Google Scholar]
- Perrett, S., Freeman, S.J., Butler, P.J., and Fersht, A.R. 1999. Equilibrium folding properties of the yeast prion protein determinant Ure2. J. Mol. Biol. 290 331–345. [DOI] [PubMed] [Google Scholar]
- Reddy, G.B., Bharadwaj, S., and Surolia, A. 1999. Thermal stability and mode of oligomerization of the tetrameric peanut agglutinin: A differential scanning calorimetry study. Biochemistry 38 4464–4470. [DOI] [PubMed] [Google Scholar]
- Rossjohn, J., Polekhina, G., Feil, S.C., Allocati, N., Masulli, M., De Illio, C., and Parker, M.W. 1998. A mixed disulfide bond in bacterial glutathione transferase: Functional and evolutionary implications. Structure 6 721–734. [DOI] [PubMed] [Google Scholar]
- Rossmann, M.G. and Argos, P. 1981. Protein folding. Annu. Rev. Biochem. 50 497–532. [DOI] [PubMed] [Google Scholar]
- Sayed, Y., Wallace, L.A., and Dirr, H.W. 2000. The hydrophobic lock-and-key intersubunit motif of glutathione transferase A1-1: Implications for catalysis, ligandin function and stability. FEBS Lett. 465 169–172. [DOI] [PubMed] [Google Scholar]
- Siddiqui, A.S. and Barton, G.J. 1995. Continuous and discontinuous domains: An algorithm for the automatic generation of reliable protein domain definitions. Protein Sci. 4 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, J.L., Silveira, C.F., Correia, J.A., and Pontes, L. 1992. Dissociation of a native dimer to a molten globule monomer. Effects of pressure and dilution on the association equilibrium of arc repressor. J. Mol. Biol. 223 545–555. [DOI] [PubMed] [Google Scholar]
- Sinning, I., Kleywegt, G.J., Cowan, S.W., Reinemer, P., Dirr, H.W., Huber, R., Gilliland, G.L., Armstrong, R.N., Ji, X., and Board, P.G. 1993. Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes. J. Mol. Biol. 232 192–212. [DOI] [PubMed] [Google Scholar]
- Stella, L., Caccuri, A.M., Rosato, N., Nicotra, M., Lo, B.M., De Matteis, F., Mazzetti, A.P., Federici, G., and Ricci, G. 1998. Flexibility of helix 2 in the human glutathione transferase P1-1. time-resolved fluorescence spectroscopy. J. Biol. Chem. 273 23267–23273. [DOI] [PubMed] [Google Scholar]
- Stenberg, G., Bjornestedt, R., and Mannervik, B. 1992. Heterologous expression of recombinant human glutathione transferase A1-1 from a hepatoma cell line. Protein Expr. Purif. 3 80–84. [DOI] [PubMed] [Google Scholar]
- Stenberg, G., Abdalla, A.M., and Mannervik, B. 2000. Tyrosine 50 at the subunit interface of dimeric human glutathione transferase P1-1 is a structural key residue for modulating protein stability and catalytic function. Biochem. Biophys. Res. Commun. 271 59–63. [DOI] [PubMed] [Google Scholar]
- Stevens, J.M., Hornby, J.A., Armstrong, R.N., and Dirr, H.W. 1998. Class σ glutathione transferase unfolds via a dimeric and a monomeric intermediate: Impact of subunit interface on conformational stability in the superfamily. Biochemistry 37 15534–15541. [DOI] [PubMed] [Google Scholar]
- Tasayco, M.L., Fuchs, J., Yang, X.M., Dyalram, D., and Georgescu, R.E. 2000. Interaction between two discontiguous chain segments from the β-sheet of Escherichia coli thioredoxin suggests an initiation site for folding. Biochemistry 39 10613–10618. [DOI] [PubMed] [Google Scholar]
- Vega, M.C., Walsh, S.B., Mantle, T.J., and Coll, M. 1998. The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the αB-310B region. J. Biol. Chem. 273 2844–2850. [DOI] [PubMed] [Google Scholar]
- Vriend, G. 1990. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 8 52–56, 29. [DOI] [PubMed] [Google Scholar]
- Wallace, L.A., Sluis-Cremer, N., and Dirr, H.W. 1998. Equilibrium and kinetic unfolding properties of dimeric human glutathione transferase A1-1. Biochemistry 37 5320–5328. [DOI] [PubMed] [Google Scholar]
- Wallace, L.A., Burke, J., and Dirr, H.W. 2000. Domain-domain interface packing at conserved Trp-20 in class α glutathione transferase impacts on protein stability. Biochim. Biophys. Acta 1478 325–332. [DOI] [PubMed] [Google Scholar]
- Wetlaufer, D.B. 1973. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl. Acad. Sci. 70 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilce, M.C. and Parker, M.W. 1994. Structure and function of glutathione S-transferases. Biochim. Biophys. Acta 1205 1–18. [DOI] [PubMed] [Google Scholar]
- Xu, D., Lin, S.L., and Nussinov, R. 1997. Protein binding versus protein folding: The role of hydrophilic bridges in protein associations. J. Mol. Biol. 265 68–84. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Liu, S., Shan, S.O., Ji, X., Gilliland, G.L., and Armstrong, R.N. 1992. Modular mutagenesis of exons 1, 2, and 8 of a glutathione S-transferase from the mu class. Mechanistic and structural consequences for chimeras of isoenzyme 3-3. Biochemistry 31 10185–10193. [DOI] [PubMed] [Google Scholar]
- Zhang, P.H. and Armstrong, R.N. 1990. Construction, expression, and preliminary characterization of chimeric class mu glutathione S-transferases with altered catalytic properties. Biopolymers 29 159–169. [DOI] [PubMed] [Google Scholar]
- Zhu, Z.Y. and Karlin, S. 1996. Clusters of charged residues in protein three-dimensional structures. Proc. Natl. Acad. Sci. 93 8350–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]