Abstract
Many outer membrane proteins (OMPs) in Gram-negative bacteria possess known β-barrel three-dimensional (3D) structures. These proteins, including channel-forming transmembrane porins, are diverse in sequence but exhibit common structural features. We here report computational analyses of six outer membrane proteins of known 3D structures with respect to (1) secondary structure, (2) hydropathy, and (3) amphipathicity. Using these characteristics, as well as the presence of an N-terminal targeting sequence, a program was developed allowing prediction of integral membrane β-barrel proteins encoded within any completely sequenced prokaryotic genome. This program, termed the β-barrel finder (BBF) program, was used to analyze the proteins encoded within the Escherichia coli genome. Out of 4290 sequences examined, 118 (2.8%) were retrieved. Of these, almost all known outer membrane proteins with established β-barrel structures as well as many probable outer membrane proteins were identified. This program should be useful for predicting the occurrence of outer membrane proteins in bacteria with completely sequenced genomes.
Keywords: Computer program, bacteria, outer membranes, β-barrel porins, genome sequences, hydropathy, amphipathicity, protein structure
Gram-negative bacteria are surrounded by two concentric lipid bilayer membranes. Both membranes contain proteins that facilitate the transport of nutrients, end products of metabolism, toxic substances, ionic species, macromolecules, and other molecular species of biological importance (aSaier 2000a). Although integral membrane transporters of the inner membrane are generally of α-structures, traversing the membrane as α-helices, those of the outer membranes consist largely of β-structures, forming β-barrels (Koebnik et al. 2000; Saier 2000a; Schulz 2000). Structural features may provide targeting signals for these two membranes (Hancock 1991; Buchanan 1999).
Among the outer membrane proteins (OMPs) of Gram-negative bacteria are the oligomeric, often trimeric channel-forming porins, several of which have been structurally characterized by X-ray crystallography (Hancock et al. 1990; Jeanteur et al. 1991; Meyer et al. 1997). These proteins can transport small molecules nonselectively, or they can be highly selective for a single class of molecules (Nieweg and Bremer 1997; Wang et al. 1997; Buchanan 1999). Similar proteins are found in outer membranes of mitochondria and plant plastids (Blachly-Dyson et al. 1990; Fischer et al. 1994; Bathori et al. 2000). They may also be present in the outer mycolic acid-containing membranes of acid-fast Gram-positive bacteria such as species of mycobacteria, corynebacteria, and Nocardia (Riess et al. 1998; Senaratne et al. 1998; Kartmann et al. 1999).
Because of their unique structures and subcellular locations, outer membrane β-barrel porins are classified in their own category in our transporter classification (TC) system (Category 1.B) separately from the α-type cytoplasmic membrane channel proteins (Category 1.A) and from the pore-forming toxins, which are synthesized in cells other than the ones in which they exert their toxic effects (Category 1.C) (Saier 1999a, 1999b, 1999c, 2000a, 2000b; Saier and Tseng 1999). There are currently 35 families classified as integral β-barrel porins in our TC system under Category 1.B (aSaier 2000a; see our transporter classification database, TCDB). Twenty-nine of these protein families are derived from Gram-negative bacteria with one from mitochondria, three from chloroplasts, and two from acid fast Gram-positive bacteria. No significant sequence similarity between members of different families can be detected (see Saier 1994, for consideration of significance levels), and in at least some of these families, known structural differences suggest independent evolutionary origins. Because many families of β-barrel porins are not yet recognized, and the sequences of these kind of proteins are very diverse, recognition and characterization of new members of β-barrel porin families is a challenging and interesting task (Achouak et al. 2001).
In this article, we report an analysis of the sequences and available structural information of all β-barrel porins for which 3D structures had been determined when we initiated these studies. Using this information, we developed an algorithm that can be used to judge if a protein has a high probability of having an integral membrane β-barrel structure. This method can be used to screen whole genomes for proteins of predominantly β-structure and to select candidates that may be outer membrane β-barrel porin proteins. Such studies, applied to Escherichia coli, the organism with the most functionally characterized OMPs, are reported here. The method reported complements an early method described by Neuwald et al. (1995) and applied to mitochondrial proteins by Mannella et al. (1996), as well as a neural network-based approach reported by Jacoboni et al. (2001) and a distinct approach based on amino acid composition and protein architecture reported by Wimley (2002). The latter two reports were published after completion of the work reported here. The BBF program is freely available to academic users upon request to the corresponding author (M.H.S.).
Results
Transmembrane signal sequence analysis
Systematic hydropathy analyses of all protein sequences encoded within completely sequenced genomes enabled us to rule out most of the cytoplasmic and integral inner membrane proteins because most outer membrane protein precursors have N-terminal hydrophobic transmembrane signal sequences. Because of the inaccuracies of the hydropathy analyses, there is a possibility that more than one transmembrane hydrophobic peak will be identified for a given sequence. We have used this program to analyze all proteins classified as β-barrel proteins in the TC system (aSaier 2000a) as reported on our Web site. Of the 80 outer membrane proteins with (putative) β-barrel structures thus identified, 49 had only one predicted TMS, 22 had two predicted TMSs, and 9 proteins had three or four predicted TMSs. None of these proteins displayed more than four putative TMSs. Eighty-three percent of the proteins analyzed had transmembrane segments near their N-termini (first 50 residues). Applying the BBF program to 4290 sequences encoded within the E. coli genome, the MEMSAT program predicted 1730 sequences to have at least one transmembrane segment within their N-terminal 50 residues. Among them, 1073 sequences had only this one TMS, 266 sequences had two putative TMSs, and 191 sequences had three or four predicted TMSs. All of these 1730 sequences were candidates for further analysis.
Outer membrane protein screen program
All identified candidate protein sequences were analyzed for potential β-barrel structures by combining three programs. A hydrophobic peak should coincide with an amphipathic peak in a region predicted by Jnet to be a β-strand. Of the 1730 sequences screened, our program retrieved 70 sequences with one predicted TMS, 35 sequences with two predicted TMSs, 9 sequences with three predicted TMSs, and 4 sequences with four predicted TMSs. The total number of sequences is 118. Thus, 2.8% of the sequences in E. coli were selected by our method as outer membrane protein candidates.
Family identification using BLAST
Using the protein sequences obtained, we performed a systematic BLAST search against the databases for sequences exhibiting clear similarity. We then identified known or putative outer membrane proteins based on the database annotations. The results are presented in Table 1. As the results show, 47 sequences (40%) are either known outer membrane proteins with β-barrel structures, or are putative outer membrane proteins that exhibit sequence similarity with known outer membrane proteins. Other proteins retrieved by the program include 12 extracytoplasmic fimbrial chaperone proteins and 7 extracytoplasmic lipoproteins, both probably largely of β-structure (Choudhury et al. 1999) (Table 2). Fifty-two additional proteins, most of unknown structure and function, are also tabulated. Several of these are known to be extracytoplasmic.
Table 1.
(Putative) E. coli OMPs retrieved by the BBF program
| Family | |||
| TC # | Abbreviation | Known OMPs and proteins with high similarity to known OMPs | Database description |
| — | — | PldA | Phospholipase A1 precursor (EC 3.I.1.32) |
| 1.B.1 | GBP | OmpF | Outer membrane protein F precursor |
| 1.B.1 | GMP | PhoE | Outer membrane pore protein E (phosphoporin) |
| 1.B.1 | GMP | NmpC | Outer membrane porin protein NmpC precursor |
| 1.B.1 | GMP | OmpC | Outer membrane protein C precursor |
| 1.B.1 | GMP | b1377 | Outer membrane protein C precursor homolog |
| 1.B.3 | LamB | LamB | Maltoporin precursor (lambda receptor protein) |
| 1.B.6 | OOP | OmpA | Outer membrane protein A precursor |
| 1.B.6 | OOP | YiaD | Hypothetical lipoprotein |
| 1.B.9 | FadL | FadL | Long-chain fatty acid transport protein precursor |
| 1.B.10 | Tsx | Tsx | Nucleoside-specific channel-forming protein precursor |
| 1.B.11 | FUP | b0532 | Outer membrane usher protein SfmD precursor |
| 1.B.11 | FUP | b0718 | Hypothetical outer membrane usher protein (YbgQ) |
| 1.B.11 | FUP | b0940 | Hypothetical outer membrane usher protein (YcbS) |
| 1.B.11 | FUP | FimD | FimD protein |
| 1.B.11 | FUP | YehB | Hypothetical outer membrane usher protein |
| 1.B.11 | FUP | b3046 | Hypothetical outer membrane usher protein (YqiG) |
| 1.B.12 | AT | b1169 | Hypothetical protein |
| 1.B.12 | AT | b1202 | Hypothetical protein |
| 1.B.12 | AT | b1170 | (AidA) Adhesin AidA-I precursor |
| 1.B.12 | AT | YdbA | Hypothetical protein |
| 1.B.14 | OMR | FhuA | Ferrichrome-iron receptor precursor |
| 1.B.14 | OMR | FepA | Ferrienterochelin receptor precursor |
| 1.B.14 | OMR | FhuE | Ferric-coprogen receptor protein precursor |
| 1.B.14 | OMR | b1451 | Probable TonB-dependent receptor (YncD) |
| 1.B.14 | OMR | CirA | Colicin I receptor precursor |
| 1.B.14 | OMR | b0805 | Probable TonB-dependent receptor (YbiL) |
| 1.B.25 | Opr | b0681 | Hypothetical protein (YbfM) |
| Proteins with low similarity to known OMPs | |||
| 1.B.6 | OOP | OmpW | |
| 1.B.6 | OOP | YciD | |
| 1.B.9 | FadL | b1834 | |
| 1.B.12 | AT | YehA | |
| 1.B.12 | AT | b1381 | |
| 1.B.12 | AT | b2974 | |
| 1.B.14 | OMR | b1722 | |
| 1.B.14 | OMR | Imp | |
| 1.B.17 | OMF | b2332 | |
| 1.B.21 | OmpG | YjhA | |
| 1.B.25 | Opr | b0371 | |
| 1.B.25 | Opr | YbhC | |
| 1.B.26 | CDP | b3875 | |
| — | — | b0964 | Homolog of OmpH from Photobacterium SS9 |
| — | — | b1428 | Homolog of probable outer membrane protein PMP6 in Chlamydophila pneumoniae |
| — | — | b1782 (YeaF) | Homolog of porin-like membrane protein Omp26La of Listonella anguillarum |
| — | — | YfeN | Homolog of outer membrane protein OmpK in Vibrio parahaemolyticus |
| — | — | YiaT | Homolog of porin-like membrane protein Omp26La of Listonella anguillarum |
| — | — | YtfM | Homolog of putative outer membrane proteins in several Gram-negative bacteria |
Table 2.
E. coli proteins retrieved by the BBF program that lack sequence similarity to known OMPs
| (Probable) extracytoplasmic fimbrial chaperones | |||
| b0531 | Fimbrial chaperone precursor (SmfC) | b0717 | Hypothetical fimbrial chaperone (YbgP) |
| b0939 | Hypothetical fimbrial chaperone (YcbR) | YehC | Hypothetical fimbrial chaperone |
| YcbF | Hypothetical fimbrial chaperone | YraI | Hypothetical fimbrial chaperone |
| b2336 | Hypothetical fimbrial chaperone (YfcS) | YhcA | Hypothetical fimbrial chaperone |
| FimC | Type 1 fimbrial chaperone | b3047 | Hypothetical fimbrial chaperone (YqiH) |
| b0716 | Hypothetical protein (YbgO) | EcpD | Fimbriae biogenesis protein homologue |
| (Probable) lipoproteins | |||
| b0986 | YmcC, hypothetical lipoprotein | NlpA | Lipoprotein-28 precursor |
| YhiU | Hypothetical protein | YjbF | Hypothetical lipoprotein |
| LolB/HemM | Outer membrane lipoprotein | YjbH | Hypothetical protein |
| YaeF | Hypothetical lipoprotein | b2512 | Homologue of probably lipoprotein in Ralstonia solanacearum |
| Others | |||
| YacK | Probable blue-copper protein | GalM | Aldose 1-epimerase (EC 5.1.3.3) (mutarotase) |
| b0574 | Hypothetical protein (YlcD) | b0837 | Hypothetical protein (YliI) |
| b0819 | Hypothetical protein | b1424 | Hypothetical protein |
| b0947 | Hypothetical protein | b1487 | Periplasmic dipeptide transport protein (dipeptide) binding protein (Dbp) (DppA) |
| FlgA | Flagellar basal body p-ring formation protein precursor | b1588 | Dimethylsulfoxide reductase chain A (DmsA) |
| b1113 | Hypothetical protein | b2043 | Colananic acid biosynthesis glycosyl transferse WcaA |
| b1440 | Hypothetical protein | NapG | Ferredoxin-type protein NapG |
| b1452 | Hypothetical protein | MepA | Penicillin-insensitive murein endopeptidase precursor |
| b1598 | Hypothetical protein | YffE | Hydrogenase-4 component A (HyfA) |
| b1678 | Hypothetical protein | YraK | Hypothetical protein |
| b1780 | Unknown protein from 2D-page (YzzQ) | b3219 | Hypothetical protein (YhcF) |
| ErfK | Protein ErfK/SrfK precursor | YihV | Hypothetical sugar kinase |
| AtoA | Acetyl CoA-transferase β subunit | b0941 | Hypothetical protein (YcbT) |
| SufI | SufI protein precursor | b1834 | Hypothetical protein |
| AcrE | Acriflavin resistance protein E precursor (EnvC) (Membrane Fusion Protein TC #8.A.1) | b3524 | Hypothetical ABC transporter ATP-binding protein (YhiG) |
| YiaS | Putative hexulose-6-phosphate isomerase | YihR | Hypothetical protein |
| YjbP | Acid phosphatase (AphA/NapA) | YjhT | Hypothetical protein |
| CpdB | 2`,3`-cyclic-nucleotide 2`-phosphodiesterase precursor | FimH | FimH protein |
| YagV | Hypothetical protein | b2225 | Hypothetical protein |
| UshA | UDP-sugar hydrolase precursor | b2466 | Hypothetical protein |
| YeiP | Hypothetical protein | YicH | Hypothetical protein |
| EndA | Endonuclease | YijF | Hypothetical protein |
| NrfC | NrfC protein | YjaH | Hypothetical protein |
| YadE | Hypothetical protein (same as YadJ) | YagW | Hypothetical protein |
| YadC | Hypothetical fimbrial-like protein | b1585 | Hypothetical protein (YnfC) |
| TolB | TolB protein | ||
Crossreferencing the TC system
The final step was to screen the E. coli database for members of recognized outer membrane barrel protein families tabulated in the TC system (aSaier 2000a) using the BLAST search tool. Thirteen out of the 28 Gram-negative bacterial families were found to be represented in E. coli. Among them, 10 were recognized by the BBF program. The results are presented in Table 3. Column A tabulates the Gram-negative bacterial porin families that are listed in TC category 1.B; column B reveals families in E. coli that were obtained by our program (marked by ×); column C lists the families that are represented in E. coli but were not retrieved by our program (marked with a star) and column D tabulates those families that have no recognized or predicted homologues in E. coli (marked with an open circle).
Table 3.
Retrieval of E. coli proteins included within 28 Gram-negative bacterial outer membrane protein (OMP) families (TC Category 1.B)
| Family | A | B | C | D | Family | A | B | C | D | Family | A | B | C | D | Family | A | B | C | D |
| GBP | 1 | × | FadL | 9 | × | SAP | 16 | ○ | CBP | 23 | ○ | ||||||||
| CP | 2 | ○ | Tsx | 10 | × | OMF | 17 | ★ | OPr | 25 | × | ||||||||
| SP | 3 | × | FUP | 11 | × | OMA | 18 | ★ | CDP | 26 | ○ | ||||||||
| BRP | 4 | ○ | AT | 12 | × | OprB | 19 | ○ | HOP | 27 | ○ | ||||||||
| POP | 5 | ○ | AEP | 13 | ○ | TPS | 20 | ○ | MomP | 31 | ○ | ||||||||
| OOP | 6 | × | OMR | 14 | × | OmpG | 21 | ★ | FomP | 32 | ○ | ||||||||
| RPP | 7 | ○ | RafY | 15 | ○ | Secretin | 22 | ★ | VC/NP | 33 | ○ |
A: OMP family TC number in category 1.B (from our Web site: www-biology.ucsd.edu/~msaier/transport/).
B: OMP families represented in E. coli based on the results obtained with the BBF program (×).
C: OMP families represented in E. coli but not recognized by the BBF program (★).
D: OMP families that have no predicted or known homologs in E. coli (○).
Known OMPs not retrieved by the BBF program
Family 17, the Outer Membrane Factor (OMF) family (TC #1.B.17) is one of the families not identified by our program. The 3D structure of one member of this family, TolC of E. coli, has been solved (Koronakis et al. 2000). TolC has a distinctive and previously unknown fold with three TolC protomers assembled to form a continuous channel tunnel over 140 Å long that spans both the outer membrane and the periplasmic space. Export of a transported substrate is brought about by the reversible interaction of the substrate-specific inner membrane protein with an outer membrane protein, thus bypassing the intervening periplasm. Only a small part of each chain has an antiparallel β-sheet structure, and each chain contributes only one-third of the β-barrel in the outer membrane. A large part of the protein has an α-helical structure. Because the β- and α-structures are interspersed, the BBF program did not identify it. Another two families, the Outer Membrane Auxillary (OMA family; TC #1.B.18) (Paulsen et al. 1997; Arrecubieta et al. 2001) and the Outer Membrane Secretin (Secretin) family (TC #1.B.22) (Nguyen et al. 2000) were also not retrieved, possibly for similar reasons. Predictions of the Jnet program suggest that these proteins have relatively large percentages of α-structure, higher than the preselected threshold values used in the BBF program. The OmpG porin (OmpG) family (TC #1.B.21) is the fourth family that was not identified by our program. The single functionally characterized protein in E. coli was suggested to have a structure with a 16 β-stranded barrel lacking the large external loop, L3, that constricts the pores in other porins (Conlan et al. 2000). This protein was not retrieved by our program because its hydrophobic signal sequence begins beyond the first 50 N-terminal residues, at position 55.
Uncharacterized proteins retrieved by the BBF program
The BBF program identified many proteins not known to be β-barrel outer membrane proteins. The functions of most of these proteins are unknown, and they are not classified in the TC system. BLAST searches with these proteins gave either just one hit (to themselves) or multiple hits. The BBF program suggests that these proteins contain large proportions of β-structure, and most of the predicted β-sheet regions contain regions with corresponding hydrophobic and amphipathic peaks. For example, YfeN, a hypothetical 29.2 KD protein, like many other proteins retrieved, displays no functionally characterized homologs when screened with Ψ-BLAST (Altschul et al. 1997). Nevertheless, the secondary structure predictions suggest a large percentage of β-structure, and most of the β-structural regions reveal amphipathic peaks corresponding to hydrophobic peaks. Although the function is unknown, the annotation in Genport notes that it exhibits similarity to an outer membrane protein in V. parahaemolyticus, OmpK. This protein may function as a receptor for the broad host-range vibriophage KVP40 (Inoue et al. 1995). All of the information available suggests that this protein is an outer membrane β-barrel protein. Similarly, a TC-BLAST search (Zhai et al. 2002) revealed that YfeN shows greatest similarity (24% identity; 36% similarity in 150 residue positions) to OmpK of Vibrio parahaemolyticus (Table 1, bottom).
YeaF (Itoh et al. 1996) and YiaT (Sofia et al. 1994) represent two additional proteins retrieved. They have few homologs as indicated using the Ψ-BLAST program. These two proteins are homologs of each other and the outer membrane protein OmpV in V. cholerae (Pohlner et al. 1986). Ψ-BLAST also reveals that these proteins are homologous to a porin-like outer membrane protein Omp26La in L. anguillarum (Table 2; Suzuki et al. 1998). The functions of YiaT and YeaF are currently unknown, but the analyses of our program show that they may be of β-barrel structure. TC-BLAST also revealed that these proteins exhibit short stretches with significant similarity to proteins of the outer membrane receptor (OMR) family (TC #1.B.14).
The outer membrane protein W precursor, OmpW (Stoltzfus et al. 1988), has many homologs. A TC-BLAST search revealed that it exhibits 25% identity and 42% similarity with PorF of Pseudomonas aeruginosa, a member of the OOP family (TC #1.B.6) (Table 1, bottom).
YtfM (Burland et al. 1995) is a hypothetical 64.8 kD protein. Ψ-BLAST results showed that it has similarity with other potential outer membrane proteins from various Gram-negative bacteria (Parkhill et al. 2000a, 2000b). By sequence analysis, it appears that these proteins all have β-barrel structures.
Discussion
In this article we summarize analyses of six families of outer membrane proteins with known β-barrel structures, thereby deriving parameters that delineate individual transmembrane β-strands. We show that most of these β-strands are characterized by peaks of both hydrophobicity and amphipathicity when the angle is set at 180°. This results because of the occurrence of an increased proportion of hydrophobic residues and of alternating hydrophilic and hydrophobic residues in the transmembrane region, respectively. Based on these characteristics of outer membrane barrel proteins, we designed a program that combined three preexisting programs. After screening for an N-terminal signal sequence, we first predict the secondary structure of a protein sequence, and then we calculate the hydropathy and amphipathicity values for the regions predicted to form β-strands. Proteins that meet the assigned criteria are selected as candidates for outer membrane proteins with β-barrel structures. All such proteins are then examined for homologs, one or more of which may have been functionally or structurally characterized, or which may have been localized to the outer membrane of the Gram-negative bacterial envelope.
This method has been designed to automatically screen whole genome protein sequence databases. In this report, we use the method to systematically screen the protein sequences encoded within the E. coli genome. Members of nine of the 13 outer membrane protein families known to be represented in E. coli that had been classified in the TC system were retrieved. The program still has limitations resulting from predictive inaccuracies and unusual positions of transmembrane signal segments as well as inadequate secondary structure predictions. Improvement of these methods should increase the accuracy of the predictions.
We use a sliding window size of seven residues to calculate hydropathy and amphipathicity values, and these values are plotted as a function of protein length. This relatively small window size creates a substantial amount of noise. This problem can be minimized by calculating average hydropathy and amphipathicity values for several aligned sequences (Zhai and Saier 2001b). Using this method together with other bioinformatic tools, we may be able to retrieve a greater proportion of the proteins that are likely to have β-barrel structures. These studies provide a guide to further analyses into the structure/function relationships of outer membrane proteins.
Materials and methods
Sequence analyses of β-barrel proteins with known 3D structures
Three-dimensional structures of several outer-membrane β-barrel proteins, belonging to seven families, had been solved using the technique of X-ray crystallography when we initiated these studies. Five families include proteins that are known to form channels across the outer membranes of Gram-negative bacteria. According to our TC system (aSaier 2000a), these families are as follows: (1) the general bacterial porin (GBP) family (TC# 1.B.1) including OmpF (Cowan et al. 1995) and phoE (Peuptit et al. 1991) of E. coli; (2) the Rhodobacter PorCa Porin (RPP) family (TC #1.B.7) (Kreusch et al. 1994; Kreusch and Schulz 1994; Schulz 2000); (3) the OmpA-OmpF Porin (OOP) family (TC# 1.B.6 ) including OmpA of E. coli (Movva et al. 1980; Pautsch and Schulz 2000); (4) the Sugar Porin (SP) family (TC# 1.B.3) including maltoporin (LamB) of E. coli (Schirmer et al. 1995) and the sucrose porin, ScrY of Salmonella typhimurium (Forst et al. 1998); and (5) the Outer Membrane Receptor (OMR) family (TC# 1.B.14) including FhuA of E. coli (Locher et al. 1998; Koronakis et al. 2000). Two other families that include members that have not been shown to be channels but for which the 3D structures have been solved and shown to be β-barrels include phospholipase A (Snijder et al. 1999) and the OmpX protein (Vogt and Schulz 1999), both of E. coli. We analyzed the sequences and structures of these proteins, and from the results derived a program for identifying candidates with an increased probability of being outer membrane proteins with β-barrel structures.
The GBP family (TC# 1.B.1) and the RPP family (TC #1.B.7)
The general PorCa porin from Rhodobacter capsulatus was the first outer membrane porin for which a 3D X-ray structure was solved (Weiss et al. 1991). Soon thereafter, two other porins, OmpF and PhoE from E. coli, were obtained (Cowan et al. 1992). These porins form homotrimeric structures in the outer bacterial membrane with 16 β-strands spanning the membrane in each monomer. Figure 1 ▶ shows the hydropathy and amphipathicity plots for the mature OmpF protein of E. coli (lacking its hydrophobic leader) using a sliding window of seven residues and an angle of 180° as is appropriate for assessing amphipathicity for β-strands (Kyte and Doolittle 1982; Le et al. 1999). The solid curve is the hydropathy plot, while the dotted curve is the amphipathicity plot (Zhai and Saier 2001a). The lines at the bottom of the figure reveal the positions of established transmembrane β-strands. As can be seen from the figure, the hydropathy values of these strands are generally less than those for a transmembrane α-helical segment (TMS) in a strongly hydrophobic cytoplasmic integral membrane protein, as expected. Outer membrane proteins would probably not cross the cytoplasmic membrane if they exhibited regions other than the cleavable signal segments capable of inserting permanently in the cytoplasmic membrane.
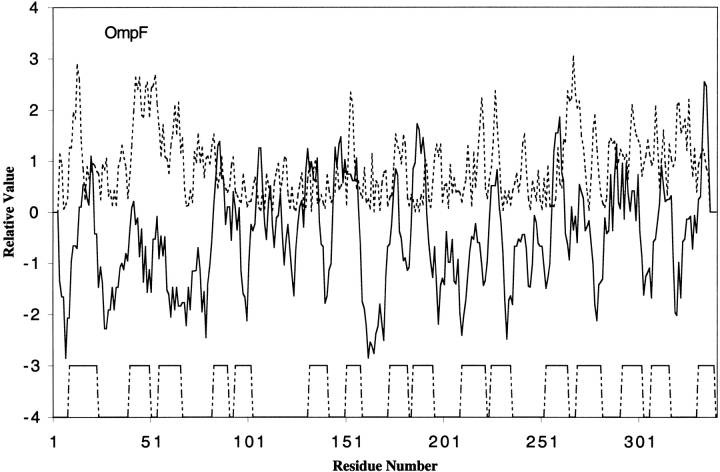
Fig. 1.
Hydropathy and amphipathicity plots for OmpF of the GBP family. The PDB code for OmpF is 2OMF. Solid lines indicate hydropathy; dotted lines indicate amphipathicity; dashed lines indicate predicted β-structures. The format of presentation used here is also used in Figures 2–6 ▶ ▶ ▶ ▶ ▶.
Almost every transmembrane (TM) β-strand corresponds in position to a peak of hydrophobicity and one of amphipathicity. This fact is in agreement with the property that OmpF and other proteins of this family form aqueous transmembrane channels, allowing the diffusion of small hydrophilic molecules across the membrane. A peak of amphipathicity results from side chains in opposite orientation alternately exhibiting hydrophilic and hydrophobic character (Eisenberg et al. 1982).
In the OmpF protein, the first hydrophobic/amphipathic peak corresponds to the first transmembrane β-strand. The high peak of amphipathicity just preceding the peak of hydrophobicity is due to the side chain orientations of hydrophilic residues K10, D12, and K16 versus hydrophobic residues V11, L13, and G15. The second transmembrane β-strand also corresponds to hydrophobic and amphipathicity peaks, generated by hydrophilic residues R42, K46, E48, and Q50 and hydrophobic residues A41, L43, F45, and G47. The third hydrophobic/amphipathic peak occurs at the edge of the third TM β-strand. The amphipathicity is caused by hydrophilic residues Q60, E62, N64, and Q66 and hydrophobic residues G59, W61, Y63, and F65. All remaining pairs of β-strands exhibit hydrophobic peaks in which the peak of the second β-strand merges with or is immediately adjacent to that of the first.
As we compare the hydrophobic and amphipathic peaks with the 3D structure of the protein, we note that the side chains of the hydrophobic residues in TM β-strands all point toward the lipid bilayer while hydrophilic side chains point towards the interior of the pore, as expected. From the figure, we notice that the amphipathicity peaks of transmembrane β-strands 5 and 6 are not obvious. This is because loop L3, connecting strands 5 and 6, is folded back in the barrel, allowing these two strands to be shorter than normal. The short β-sheet in this region and the effect of a coiled structure near them diminishes the amphipathic peaks. Nevertheless, hydrophilic residues K89 in β-strand 4, D97 in β-strand 5, and R140 in β-strand 6 all face toward the inner channel of the barrel.
The OmpA-OmpF porin (OOP) family (TC# 1.B.6)
The large OOP family includes the functionally well-characterized OmpA porin of E. coli (Sugawara and Nikaido 1994) as well as the OmpF porin of Pseudomonas aeruginosa (Sugawara et al. 1996). These proteins and their many homologs are believed to form structures consisting of eight transmembrane β-strands (Baldermann et al. 1998). OmpA provides a model system for studying the mechanism of insertion, folding and assembly of constitutive integral membrane proteins both in vivo and in vitro. The function of OmpA is not currently well understood, but channel formation has been demonstrated (Arora et al. 2000).
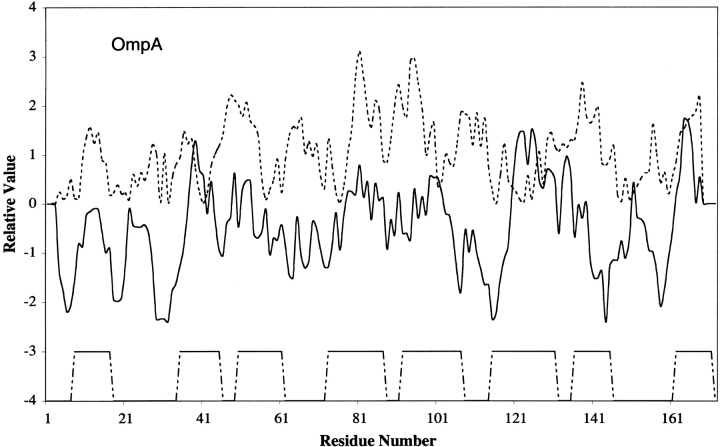
Figure 2 ▶ shows the hydropathy and amphipathicity plots for the mature form of OmpA. Secondary structural information, based on the 3D structure (Pautsch and Schulz 1998, 2000) is shown at the bottom of the figure. As for the results presented in Figure 1 ▶, each TM β-strand generally corresponds to one hydrophobic peak and one amphipathic peak. The hydrophilic residues that contribute to each of these amphipathic peaks are K13 in β-strand 1, K35 in β-strand 2, E53 and D57 in β-strand 3, Q79 and K83 in β-strand 4, D93 and Q97 in β-strand 5, E129 in β-strand 6, R139, E141, Q143, and N147 in β-strand 7, and R170 in β-strand 8. They all face inwards, and are important for formation of the hydrophilic ion channel.
Fig. 2.
Hydropathy and amphipathicity plots for OmpA of the OOP family. The PDB code for OmpA is 1BXW.
The sugar porin (SP) family (TC #1.B.3)
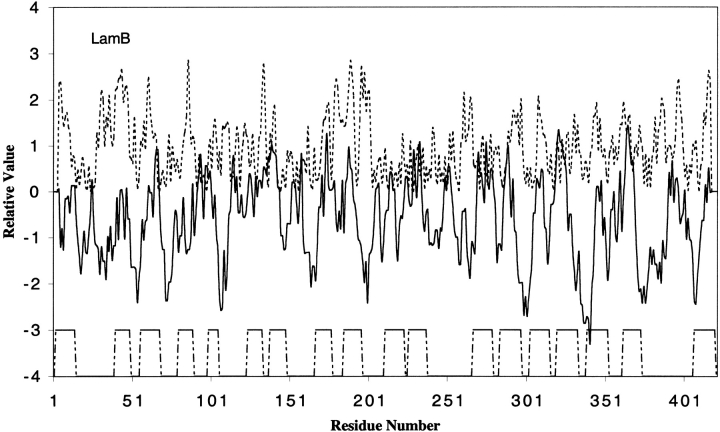
The SP family includes the well-characterized maltooligosaccharide-specific maltoporin of E. coli, LamB, and the sucrose-specific porin, ScrY, of S. typhimurium. The 3D structures of both of these two proteins have been solved (Schirmer et al. 1995; Forst et al. 1998). Figure 3 ▶ shows our amphipathicity/hydrophobicity analysis for the mature form of the E. coli maltoporin. The 18 transmembrane β-strands are shown at the bottom of the figure. Most of the hydrophilic side chains contributing to the amphipathic peaks point inwards, lining the channel. These residues are, for example, R8 in β-strand 1, E43, K45, and E49 in β-strand 2, E61 and N63 in β-strand 3, E83, N85, Q87, and K89 in β-strand 4, R105 in β-strand 5, and so on. These residues are important for the channel characteristics as they form a hydrogen bonded network with the hydroxyl groups of the substrate sugar molecules.
Fig. 3.
Hydropathy and amphipathicity plots for LamB of the SP family. The PDB code for LamB is 1MAL.
The outer membrane receptor (OMR) family (TC #1.B.14)
The OMR family includes a large number of sequenced Gram-negative bacterial outer membrane proteins that form transmembrane pores and transport relatively large molecules from the external milieu to the periplasm in an energized process. As for the other families discussed above, no OMR member has been identified in a Gram-positive bacterium, an archaeon or a eukaryote. Energization of transport across the outer membrane requires a heterotrimeric complex of proteins, the TonB-ExbB-ExbD complex (Moeck and Coulton 1998) or in some cases, the TolA-TolQ-TolR complex (Locher et al. 1998). Energization requires the proton motive force (pmf) across the cytoplasmic membrane. In the absence of a pmf or one of the three energy-coupling proteins of the complex, the receptor binds its substrate, but transport does not occur.
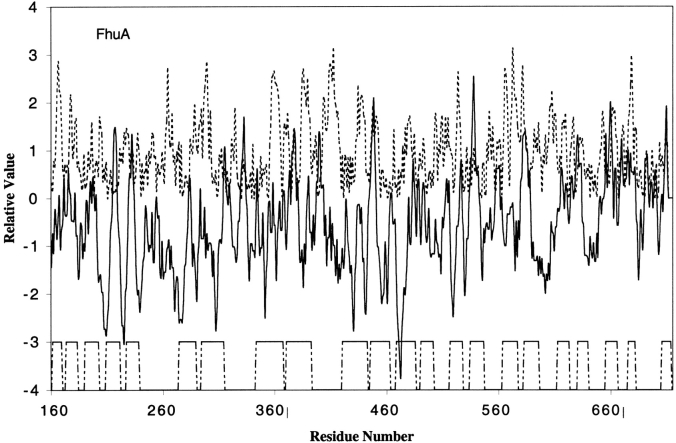
The proteins in this family form a C-terminal 22-stranded β-barrel and an N-terminal plug domain. The plug is located inside the barrel and thus obstructs the channel interior. This domain tightly binds the barrel by more than 60 hydrogen bonds and nine salt bridges (Locher et al. 1998). Figure 4 ▶ shows the sequence analysis of the β-barrel domain of the TonB-dependent receptor, TolC. Similar to Figures 1–3 ▶ ▶ ▶, most of the transmembrane β-strands correspond to hydrophobic/amphipathic peaks, and most of the hydrophilic residues that contribute to the amphipathic peak face inwards to assist in the formation of hydrogen bonds and salt bridges. For example, E163, Q165, and K167 in β-strand 1, Q175 and D179 in β-strand 2, and R199 in β-strand 3 all line the channel.
Fig. 4.
Hydropathy and amphipathicity plots for FhuA of the OMR family. The PDB code for FhuA is 1BY3.
OmpX and phospholipase A
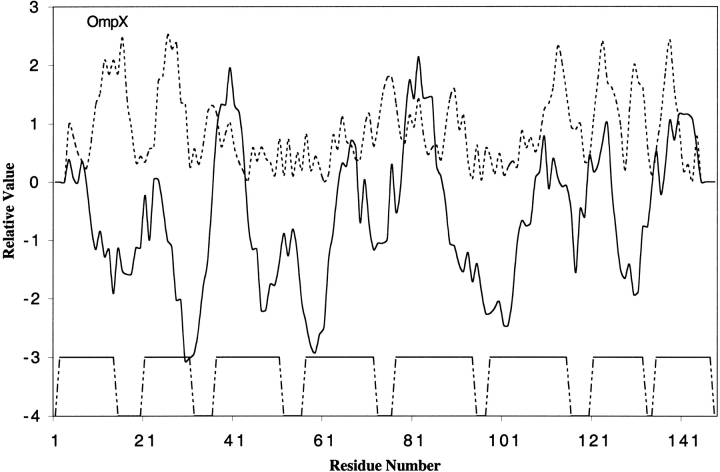
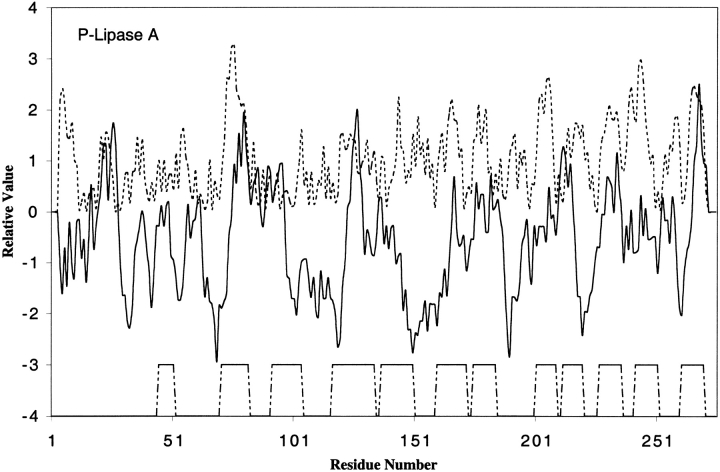
Two other outer membrane β-barrel proteins for which 3D structures are available are OmpX (Vogt and Schulz 1999) and phospholipase A (Snijder et al. 1999). OmpX belongs to a family of proteins that are important for virulence. They are believed to neutralize host defense mechanisms. Similar to OmpA, OmpX has eight transmembrane β-strands but they are different in shear number (a measure of the stages of the strands in the β-sheet (Koebnik et al. 2000), one of the important parameters of β-barrel structures). Phospholipase A, an enzyme that hydrolyzes phospholipids, has 12 transmembrane β-strands arranged in a barrel. These two proteins are similar to outer membrane β-barrel porins with hydrophilic residues facing inwards to form a hydrogen-bonded network. When analyzed for hydrophobicity and amphipathicity, most transmembrane β-strands in these proteins yield hydrophobic/amphipathic peaks (Figs. 5 and 6 ▶ ▶). It is not known if these β-barrels form transmembrane aqueous channels.
Fig. 5.
Hydropathy and amphipathicity plots for OmpX. The PDB code for OmpX is 1QJ8.
Fig. 6.
Hydropathy and amphipathicity plots for phospholipase A. The PDB code for Phospholipase A is 1QD5.
β-Barrel structural analyses: conclusions
From the hydropathy and amphipathicity analyses of the outer membrane β-barrel proteins of known 3D structure discussed above, we found that (1) most of the transmembrane strands correspond to a peak of hydrophobicity, although the hydrophobic values of these peaks are generally not as high as those of the transmembrane α-helices of cytoplasmic integral membrane proteins as expected; and (2) most of the transmembrane β-strands exhibit peaks of amphipathicity caused by the alternating hydrophilic residues located inside the barrel and the hydrophobic residues located outside the barrel. These properties of outer membrane β-barrel proteins have been noted previously by several investigators including our study of the proteins of the autotransporter (AT) family (TC #1.B.12) (Loveless and Saier 1997).
Whole genome sequence screening
Based on the sequence analyses described above, we identified criteria for searching for outer membrane β-barrel proteins in any protein sequence database, and based on these criteria, we developed a program for identifying such proteins. We combine three sequence analysis methods. The first is secondary structure prediction; the second is hydropathy analysis; and the third is amphipathicity analysis. For secondary structure prediction, we use the program Jnet, developed by the Barton group (Cuff et al. 1998; Cuff and Barton 2000). This program uses a two-level neutral network algorithm and gives better predictive results than the other programs we have examined. After obtaining the secondary structure results, we calculate the hydropathy and amphipathicity values using a window size of 7, which we found to be optimal for transmembrane β-sheets. Each predicted β-strand that also exhibits a peak of hydrophobicity and a peak of amphipathicity is recorded as a transmembrane β-strand. We can define an overall value based on these three parameters. The higher the value, the higher the probability that the region is in a true transmembrane β-strand, and the presence of multiple such regions increases the probability that the protein has a β-barrel structure. A specified threshold value can arbitrarily be assigned to allow the program to count potential TM β-strands.
Calculation method
We have used our program to screen proteins encoded within the E. coli genome (Blattner et al. 1997) for predicted transmembrane β-barrel structures. The whole genome sequence analysis includes three steps as follows: (1) automated hydropathy analysis of the protein sequences. Because there should be a transmembrane signal sequence for any protein precursor located in the outer membrane or periplasm, this step is used to rule out most cytoplasmic and integral inner membrane proteins. The program we used is a modified version of MEMSAT using default values for prediction of transmembrane α-helices (Jones et al. 1994, modified by us). Due to the inaccuracies of the program, we selected proteins that have at least one but no more than four putative TMSs with the first TMS within the N-terminal 50 residues. (2) Automated secondary structure, hydropathy, and amphipathicity predictions for each sequence in the genome database. Proteins that are selected exhibit 70% or greater of predicted transmembrane β-strands either for the whole protein if the full-length protein comprises the β-barrel, or for that domain that does comprise the β-barrel in the case of multidomain proteins. The cutoff point hydropathy value for β-strands is 1.5, and the amphipathicity value must be ≥1.0. However, if the hydropathy value is less than 1.5 but greater than 0.5, an amphipathicity value of ≥2.0 will compensate for the lower hydropathy value and the strand will be counted. (3) Automated similarity search. This third step is performed for each sequence to find homologs, some of which may be known as outer membrane proteins. Database searches are performed using the BLAST search tool (Altschul et al. 1990).
Acknowledgments
Work in our laboratory was supported by NIH Grants RR07861 and GM64368. We thank Mary Beth Hiller for her assistance in the preparation of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0209002.
References
- Achouak, W., Heulin, T., and Pagés, J.-M. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199 1–7. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, A., Rinehart, D., Szabo, G., and Tamm, L.K. 2000. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 275 1594–1600. [DOI] [PubMed] [Google Scholar]
- Arrecubieta, C., Hammarton, T.C., Barrett, B., Chareonsudjai, S., Hodson, N., Rainey, D., and Roberts, I.S. 2001. The transport of group 2 capsular polysaccharides across the periplasmic space in Escherichia coli. J. Biol. Chem. 276 4245–4250. [DOI] [PubMed] [Google Scholar]
- Baldermann, C., Lupas, A., Lubieniecki, J., and Engelhardt, H. 1998. The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J. Bacteriol. 180 3741–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathori, G., Parolini, I., Szabo, I., Tombola, F., Messina, A., Oliva, M., Sargiacomo, M., De Pinto, V., and Zoratti, M. 2000. Extramitochondrial porin: Facts and hypotheses. J. Bioenerg. Biomembr. 32 79–89. [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson, E., Peng, S., Colombini, M., and Forte, M. 1990. Selectivity changes in site-directed mutants of the VDAC ion channel: Structural implications. Science 247 1233–1236. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., Gregor, J., Davis, N.W., Kirkpatrick, H.A., Goeden, M.A., Rose, D.J., Mau, B., and Shao, Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277 1453–1474. [DOI] [PubMed] [Google Scholar]
- Buchanan, S.K. 1999. Beta-barrel proteins from bacterial outer membranes: Structure, function and refolding. Curr. Opin. Struct. Biol. 9 455–461. [DOI] [PubMed] [Google Scholar]
- Burland, V., Plunkett III, G., Sofia, H.J., Daniels, D.L., and Blattner, F.R. 1995. Analysis of the Escherichia coli genome. VI. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 23 2105–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, D., Thompson, A., Stojanoff, V., Langermann, S., Pinkner, J., Hultgren, S.J., and Knight, S.D. 1999. X-ray structure of the FimC-FimH chaperone–adhesin complex from uropathogenic Escherichia coli. Science 285 1061–1066. [DOI] [PubMed] [Google Scholar]
- Conlan, S., Zhang, Y., Cheley, S., and Bayley, H. 2000. Biochemical and biophysical characterization of OmpG: A monomeric porin. Biochemistry 39 11845–11854. [DOI] [PubMed] [Google Scholar]
- Cowan, S.W., Garavito, R.M., Jansonius, J.N., Jenkins, J.A., Karlsson, R., Konig, N., Pai, E.F., Pauptit, R.A., Rizkallah, P.J., Rosenbusch, J.P., Rummel, G., and Schirmer, T. 1995. The structure of OmpF porin in a tetragonal crystal form. Structure 3 1041–1050. [DOI] [PubMed] [Google Scholar]
- Cowan, S.W., Schirmer, T., Rummel, G., Steiert, M., Ghosh, R., Pauptit, R.A., Jansonius, J.N., and Rosenbusch, J.P. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358 727–733. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A. and Barton, G.J. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40 502–511. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M., and Barton, G.J. 1998. JPred: A consensus secondary structure prediction server. Bioinformatics 14 892–893. [DOI] [PubMed] [Google Scholar]
- Eisenberg, D., Weiss, R.M., and Terwilliger, T.C. 1982. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 299 371–374. [DOI] [PubMed] [Google Scholar]
- Fischer, K., Weber, A., Brink, S., Arbinger, B., Schunemann, D., Borchert, S., Heldt, H.W., Popp, B., Benz, R., and Link, T.A. 1994. Molecular cloning and functional characterization of two new members of the porin family. J. Biol. Chem. 269 25754–25760. [PubMed] [Google Scholar]
- Forst, D., Welte, W., Wacker, T., and Diederichs, K. 1998. Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose. Nat. Struct. Biol. 5 37–46. [DOI] [PubMed] [Google Scholar]
- Hancock, R.E. 1991. Bacterial outer membranes: Evolving concepts. Specific structures provide Gram-negative bacteria with several unique advantages. ASM News 57 175–182. [Google Scholar]
- Hancock, R.E., Siehnel, R., and Martin, N. 1990. Outer membrane proteins of Pseudomonas. Mol. Microbiol. 4 1069–1075. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Matsuzaki, S., and Tanaka, S. 1995. Cloning and sequence analysis of Vibrio parahaemolyticus ompK gene encoding a 26-kDa outer membrane protein, OmpK, that serves as receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett. 134 245–249. [DOI] [PubMed] [Google Scholar]
- Itoh, T., Aiba, H., Baba, T., Hayashi, K., Inada, T., Isono, K., Kasai, H., Kimura, S., Kitakawa, M., Kitagawa, M., Makino, K., Miki, T., Mizobuchi, K., Mori, H., Mori, T., Motomura, K., Nakade, S., Nakamura, Y., Nashimoto, H., Nishio, Y., Oshima, T., Saito, N., Sampei, G., Seki, Y., and Horiuchi, T. 1996. A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1–50.0 min region on the linkage map. DNA Res. 3 379–392. [DOI] [PubMed] [Google Scholar]
- Jacoboni, I., Martelli, P.L., Fariselli, P., De Pinto, V., and Casadio, R. 2001. Prediction of the transmembrane regions of β-barrel membrane proteins with a neural network-based predictor. Protein Sci. 10 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur, D., Lakey, J.H., and Pattus, F. 1991. The bacterial porin superfamily: Sequence alignment and structure prediction. Mol. Microbiol. 5 2153–2164. [DOI] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33 3038–3049. [DOI] [PubMed] [Google Scholar]
- Kartmann, B., Stenger, S., Niederweis, M., and Stengler, S. 1999. Porins in the cell wall of Mycobacterium tuberculosis. J. Bacteriol. 181 6543–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik, R., Locher, K.P., and Van Gelder, P. 2000. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 37 239–253. [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., and Hughes, C. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405 914–919. [DOI] [PubMed] [Google Scholar]
- Kreusch, A. and Schulz, G.E. 1994. Refined structure of the porin from Rhodopseudomonas blastica. Comparison with the porin from Rhodobacter capsulatus. J. Mol. Biol. 243 891–905. [DOI] [PubMed] [Google Scholar]
- Kreusch, A., Neubuser, A., Schiltz, E., Weckesser, J., and Schulz, G.E. 1994. Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 Å resolution. Protein Sci. 3 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J. and Doolittle, R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157 105–132. [DOI] [PubMed] [Google Scholar]
- Le, T., Tseng, T.-T., and Saier, M.H., Jr. 1999. Flexible programs for the prediction of average amphipathicity of multiply aligned homologous proteins: Application to integral membrane transport proteins. Mol. Mem. Biol. 16 173–179. [DOI] [PubMed] [Google Scholar]
- Locher, K.P., Rees, B., Koebnik, R., Mitschler, A., Moulinier, L., Rosenbusch, J.P., and Moras, D. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: Crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95 771–778. [DOI] [PubMed] [Google Scholar]
- Loveless, B.J. and Saier, M.H., Jr. 1997. A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol. Membr. Biol. 14 113–123. [DOI] [PubMed] [Google Scholar]
- Mannella, C.A., Neuwald, A.F., and Lawrence, C.E. 1996. Detection of likely transmembrane beta strand regions in sequences of mitochondrial pore proteins using the Gibbs sampler. J. Bioenerg. Biomembr. 28 163–169. [DOI] [PubMed] [Google Scholar]
- Meyer, J.E., Hofnung, M., and Schulz, G.E. 1997. Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside. J. Mol. Biol. 266 761–775. [DOI] [PubMed] [Google Scholar]
- Moeck, G.S. and Coulton, J.W. 1998. TonB-dependent iron acquisition: Mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28 675–681. [DOI] [PubMed] [Google Scholar]
- Movva, N.R., Nakamura, K., and Inouye, M. 1980. OmpA gene structure of the OmpA protein, a major surface protein of Escherichia coli required for cell-cell interaction. J. Mol. Biol. 143 317–328. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., Liu, J.S., and Lawrence, C.E. 1995. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 4 1618–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L., Paulsen, I.T., Tchieu, J., Hueck, C.J., and Saier, M.H., Jr. 2000. Phylogenetic analyses of the constituents of type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2 125–144. [PubMed] [Google Scholar]
- Nieweg, A. and Bremer, E. 1997. The nucleoside-specific Tsx channel from the outer membrane of Salmonella typhimurium, Klebsiella pneumoniae and Enterobacter aerogenes: Functional characterization and DNA sequence analysis of the tsx genes. Microbiology 143 603–615. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Achtman, M., James, K.D., Bentley, S.D., Churcher, C., Klee, S.R., Morelli, G., Basham, D., Brown, D., Chillingworth, T., Davies, R.M., Davis, P., Devlin, K., Feltwell, T., Hamlin, N., Holroyd, S., Jagels, K., Leather, S., Moule, S., Mungall, K., Quail, M.A., Rajandream, M.A., Rutherford, K.M., Simmonds, M., Skelton, J., Whitehead, S., Spratt, B.G., and Barrell, B.G. 2000a. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404 502–506. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B.W., Mungall, K., Ketley, J.M., Churcher, C., Basham, D., Chillingworth, T., Davies, R.M., Feltwell, T., Holroyd, S., Jagels, K., Karlyshev, A.V., Moule, S., Pallen, M.J., Penn, C.W., Quail, M.A., Rajandream, M.A., Rutherford, K.M., van Vliet, A.H., Whitehead, S., and Barrell, B.G. 2000b. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403 665–668. [DOI] [PubMed] [Google Scholar]
- Paulsen, I.T., Beness, A.M., and Saier, M.H., Jr. 1997. Computer-based analyses of the protein constituents of transport systems catalyzing export of complex carbohydrates in bacteria. Microbiology 143 2685–2699. [DOI] [PubMed] [Google Scholar]
- Pauptit, R.A., Schirmer, T., Jansonius, J.N., Rosenbusch, J.P., Parker, M.W., Tucker, A.D., Tsernoglou, D., Weiss, M.S., and Schulz, G.E. 1991. A common channel-forming motif in evolutionarily distant porins. J. Struct. Biol. 107 136–145. [DOI] [PubMed] [Google Scholar]
- Pautsch, A. and Schulz, G.E. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5 1013–1017. [DOI] [PubMed] [Google Scholar]
- ———. 2000. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298 273–282. [DOI] [PubMed] [Google Scholar]
- Pohlner, J., Meyer, T.F., Jalajakumari, M.B., and Manning, P.A. 1986. Nucleotide sequence of ompV, the gene for a major Vibrio cholerae outer membrane protein. Mol. Gen. Genet. 205 494–500. [DOI] [PubMed] [Google Scholar]
- Riess, F.G., Lichtinger, T., Cseh, R., Yassin, A.F., Schaal, K.P., and Benz, R. 1998. The cell wall porin of Nocardia farcinica: Biochemical identification of the channel-forming protein and biophysical characterization of the channel properties. Mol. Microbiol. 29 139–150. [DOI] [PubMed] [Google Scholar]
- Saier, M.H., Jr. 1994. Computer aided analysis of transport protein sequences: Gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol. Rev. 58 71–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1999a. A proposal for classification of transmembrane transport proteins in living organisms. In Biomembrane transport (ed. L. Van Winkle), pp. 265–276. Academic Press, San Diego, CA.
- ———. 1999b. Eukaryotic transmembrane solute transport systems. In International review of cytology: A survey of cell biology (ed. K.W. Jeon), pp. 61–136. Academic Press, San Diego, CA. [DOI] [PubMed]
- ———. 1999c. Genome archeology leading to the characterization and classification of transport proteins. Curr. Opin. Microbiol. 2 555–561. [DOI] [PubMed] [Google Scholar]
- ———. 2000a. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64 354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000b. Families of proteins forming transmembrane channels. Membr. Biol. 175 165–180. [DOI] [PubMed] [Google Scholar]
- Saier, M.H., Jr. and Tseng, T.-T. 1999. Evolutionary origins of transmembrane transport systems. In Transport of molecules across microbial membranes (Society for General Microbiology Symposium) (eds. J.K. Broome-Smith, S. Baumberg, C.J. Stirling, and F.B. Ward), pp. 252–274. Cambridge University Press, Cambridge, UK.
- Senaratne, R.H., Mobasheri, H., Papavinasasundaram, K.G., Jenner, P., Lea, E.J., and Draper, P. (1998). Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J. Bacteriol. 180 3541–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, T., Keller, T.A., Wang, Y.F., and Rosenbusch, J.P. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267 512–514. [DOI] [PubMed] [Google Scholar]
- Schulz, G.E. 2000. β-Barrel membrane proteins. Curr. Opin. Struct. Biol. 10 443–447. [DOI] [PubMed] [Google Scholar]
- Snijder, H.J., Ubarretxena-Belandia, I., Blaauw, M., Kalk, K.H., Verheij, H.M., Egmond, M.R., Dekker, N., and Dijkstra, B.W. 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401 717–721. [DOI] [PubMed] [Google Scholar]
- Sofia, H.J., Burland, V., Daniels, D.L., Plunkett III, G., and Blattner, F.R. 1994. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 22 2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, A., Leslie, J.F., and Milkman, R. 1988. Molecular evolution of the Escherichia coli chromosome. I. Analysis of structure and natural variation in a previously uncharacterized region between trp and tonB. Genetics 120 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, E. and Nikaido, H. 1994. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269 17981–17987. [PubMed] [Google Scholar]
- Sugawara, E., Steiert, M., Rouhani, S., and Nikaido, H. 1996. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J. Bacteriol. 178 6067–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, S., Yasue, K., and Kusuda, R. 1998. A porin like outer membrane protein (Omp26La) appears to increase in an oxytetracycline resistant strain of marine fish pathogen Vibrio (Listonella) anguillarum. Microbes Environ. 13 197–202. [Google Scholar]
- Vogt, J. and Schulz, G.E. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Struct. Fold Des. 7 1301–1309. [DOI] [PubMed] [Google Scholar]
- Wang, Y.F., Dutzler, R., Rizkallah, P.J., Rosenbusch, J.P., and Schirmer, T. 1997. Channel specificity: Structural basis for sugar discrimination and differential flux rates in maltoporin. J. Mol. Biol. 272 56–63. [DOI] [PubMed] [Google Scholar]
- Weiss, M.S., Abele, U., Weckesser, J., Welte, W., Schiltz, E., and Schulz, G.E. 1991. Molecular architecture and electrostatic properties of a bacterial porin. Science 254 1627–1630. [DOI] [PubMed] [Google Scholar]
- Wimley, W.C. 2002. Toward genomic identification of β-barrel membrane proteins: Composition and architecture of known structures. Protein Sci. 11 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Y. and Saier, M.H., Jr. 2001a. A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J. Mol. Microbiol. Biotechnol. 3 501–502. [PubMed] [Google Scholar]
- ———. 2001b. A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J. Mol. Microbiol. Biotechnol. 3 285–286. [PubMed] [Google Scholar]
- Zhai, Y., Tchieu, J., and Saier, M.H., Jr. 2002. A web-based Tree View (TV) program for the visualization of phylogenetic trees. J. Mol. Microbiol. Biotechnol. 4 69–70. [PubMed] [Google Scholar]