Abstract
To further investigate the mechanism and function of allosteric activation by chloride in some α-amylases, the structure of the bacterial α-amylase from the psychrophilic micro-organism Pseudoalteromonas haloplanktis in complex with nitrate has been solved at 2.1 Å, as well as the structure of the mutants Lys300Gln (2.5 Å) and Lys300Arg (2.25 Å). Nitrate binds strongly to α-amylase but is a weak activator. Mutation of the critical chloride ligand Lys300 into Gln results in a chloride-independent enzyme, whereas the mutation into Arg mimics the binding site as is found in animal α-amylases with, however, a lower affinity for chloride. These structures reveal that the triangular conformation of the chloride ligands and the nearly equatorial coordination allow the perfect accommodation of planar trigonal monovalent anions such as NO3−, explaining their unusual strong binding. It is also shown that a localized negative charge such as that of Cl−, rather than a delocalized charge as in the case of nitrate, is essential for maximal activation. The chloride-free mutant Lys300Gln indicates that chloride is not mandatory for the catalytic mechanism but strongly increases the reactivity at the active site. Disappearance of the putative catalytic water molecule in this weakly active mutant supports the view that chloride helps to polarize the hydrolytic water molecule and enhances the rate of the second step in the catalytic reaction.
Keywords: Allosteric activation, family 13 glycosyl hydrolases, α-amylase, psychrophilic, crystal structures, chloride ion, monovalent anions, catalysis
α-Amylases (α-1,4-glucan-4-glucanohydrolase, EC 3.2.1.1) are ubiquitous enzymes synthesized in all life genera (Janecek 1994, 1997), which hydrolyze starch and related α-1,4–linked glycosyl polysaccharides. These enzymes of ∼50 kD share a similar molecular architecture made of a central (β/α)8 barrel or domain A, a small β-pleated domain B protruding between β3 and α3, and a C-terminal globular domain C consisting of a Greek key motif differing in size according to the organism. All α-amylases bind at least one strongly conserved Ca2+ ion that is required for structural integrity and for enzymatic activity.
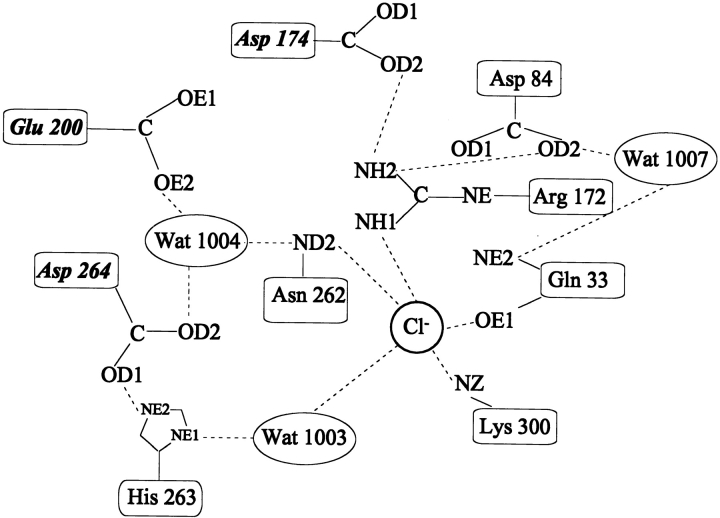
Among this large enzyme family, one group is allosterically activated by the binding of a chloride ion. These chloride-dependent α-amylases are specific to all animals (vertebrates and invertebrates) but are also found in some gram-negative bacteria such as Pseudoalteromonas haloplanktis, formerly Alteromonas haloplanctis (D'Amico et al. 2000). Both human salivary and pancreatic α-amylases belong to this group, which is subjected to intensive investigations for fundamental, therapeutic, or diagnostic purposes. Although significant advances in the elucidation of the reaction mechanism have been made recently (McCarter and Withers 1996; Uitdehaag et al. 1999; Brayer et al. 2000; Aghajari et al. 2002), the function of the chloride ion remains unclear. In all chloride-dependent crystal structures solved so far—namely, α-amylases from pig pancreas (Qian et al. 1993; Larson et al. 1994), human salivary (Ramasubbu et al. 1996) and pancreatic (Brayer et al. 1995) glands, the beetle Tenebrio molitor (Strobl et al. 1998), and the bacteria P. haloplanktis (Aghajari et al. 1998a,b)—the anion binds to a common site, close to the center of the barrel and in the near vicinity of the active site. However, three additional chloride ions were observed near the active site in a high-resolution structure of pig pancreatic α-amylase (Qian et al. 2001). The protein ligands for the conserved chloride ion and the network of interactions within the active site are depicted in Figure 1 ▶. The chloride ligands Arg195 and Asn298 are conserved in most chloride-independent α-amylases, whereas the bidentate coordination by the basic side-chain of Arg337 is specific to all animal chloride-dependent α-amylases and is therefore the key feature for anion binding (Feller, et al. 1996; D'Amico, et al. 2000). This latter arginine is substituted by a lysine in bacterial α-amylases (Lys300 in P. haloplanktis α-amylase [AHA]), providing a monodentate coordination through NZ, which seems responsible for the lower binding constant (Ka ∼ 102 M−1 in bacteria; Ka ∼ 103 M−1 in vertebrates). The fourth Cl− ligand is a conserved water molecule (Wat1003 in AHA).
Fig. 1.
Schematic representation of the chloride binding site and of the interaction network with active site residues (adapted from Qian et al. 1994). The essential catalytic residues are in italics. Chloride ligands are Lys300(337), Arg172(195), Asn262(298), and H2O1003(525); active site residues are Glu200(233), Asp174(197), and Asp264(300) the putative catalytic water molecule H2O1004(cat) and His263(299). Numbers in parentheses refer to pig pancreas α-amylase.
Chloride-free α-amylases display a low level of basal activity, typically <0.5%, and activation by chloride follows a simple binding isotherm (Levitzki and Steer 1974; Feller et al. 1996). However, activation is also provided, but to a lower extent, by other small monovalent anions and notably by NO3− or ClO3−, which bind even more strongly to α-amylases than does Cl−. This indicates that a negative charge, not restricted to that of Cl−, is essential for the amylolytic reaction and, furthermore, that the anion binding strength and the activation capacity are not correlated.
To get better insights into the mechanism and function of allosteric activation by chloride, the complex of AHA with NO3− has been crystallized, as well as the chloride-free mutant Lys300Gln and the mutant Lys300Arg in complex with chloride, and the corresponding three-dimensional structures were determined by X-ray crystallography.
Results
Replacement of the chloride activator by a NO3− ion
As shown in Table 1, the binding of NO3− to AHA is 3 times stronger than that of Cl−, but only 45% of the maximal enzyme activity is restored. The stronger binding of NO3− compared with Cl− can be explained from the three-dimensional structure of the complex in terms of binding distances and geometry. The nitrate ion has a trigonal planar configuration that fits ideally into the chloride binding site, where it has optimal interactions with Arg172 NH2 and NE, each performing two hydrogen bonds to this NO3− ion (Table 2; Figs. 2, 3 ▶ ▶). The crucial difference between chloride and nitrate binding modes is found in the coordination by Lys300 and Asn262. Both residues, via their side-chain nitrogen atoms, perform two hydrogen bonds or electrostatic interactions instead of one, as in the case of chloride binding. In addition, the binding distances (Table 2) indicate that interactions of the respective amino acids with NO3− are much stronger (Asn262 ND2 excepted) than the corresponding ones with Cl−.
Table 1.
Main biochemical parameters for the AIIA/NO3−, complex and for the mutants K300Q and K300R
| Enzyme | Anion bound | Kd | kcal |
| mM | s−1 | ||
| AHA | Cl− | 5.9 | 609 |
| AHA | NO3− | 1.9 | 274 |
| K300Q | none | — | 77 |
| K300R | Cl− | 44 | 338 |
Activation kinetics and activities using 4-nitrophenyl-α-d-maltoheptaoside-4, 6-O-ethylidene as substrate (Feller et al. 1996).
Table 2.
Binding distances between the nitrate ion and its ligands
| Amino acid/water | NO3−-ion | Distance (Å) | Distance (Å) Cl− |
| Arg 172 NH2 | O2 | 2.98 | 3.42 |
| Arg 172 NE | O1 | 2.82 | 3.36 |
| (Arg 172 NH2 | O1 | 3.55) | |
| (Arg 172 NE | O2 | 3.62) | |
| Asn 262 ND2 | O2 | 3.36 | 3.33 |
| Asn 262 ND2 | O3 | 2.91 | |
| Lys 300 NZ | O1 | 2.69 | 3.26 |
| Lys 300 NZ | O3 | 2.95 | |
| Water 1003 | 3.26 |
The distances between chloride (Aghajari et al. 1998a) and these ligands in native AHA are given for comparison.
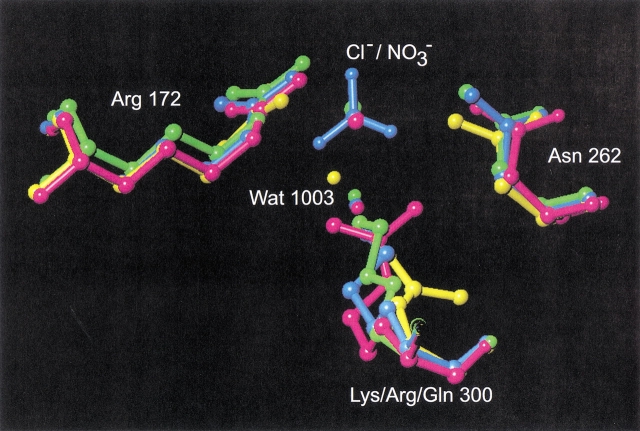
Fig. 2.
Superposition of the chloride binding site in P. haloplanktis α-amylase (AHA), in green, complex AHA/nitrate in blue, mutant K300Q in yellow, and mutant K300R in pink.
Fig. 3.
Nitrate replacing chloride in the complex P. haloplanktis α-amylase (AHA)/NO3−. Stereo view of the 2Fo − Fc electron density (in blue) after the insertion of the NO3− ion, showing its interactions with Arg172, Asn262, and Lys300.
The mutation Lys300Gln
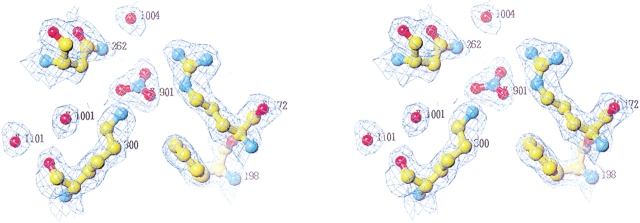
The mutation of Lys300 to a glutamine results in an active but chloride-independent enzyme (Table 1), thus mimicking the residue organization found in Cl−-independent bacterial α-amylases such as those from Bacillus amyloliquefaciens and B. licheniformis (Machius et al. 1998). The absence of chloride in the Lys300Gln mutant structure is clearly seen from the difference Fourier density maps calculated on the basis of phases from the native enzyme with chloride included (Fig. 4 ▶). Compared to the three-dimensional structure of the native enzyme, the side-chain of Gln300 now replaces a water molecule (Wat1001), being the third water molecule (when starting counting from the active site) in the chloride water pocket described earlier (Aghajari et al. 1998a). Furthermore, Arg172 turns its guanidinium group in another fashion than the one found in the chloride binding site in the native enzyme (Fig. 2 ▶), toward the active site, where it is located between Asp174 and Glu200. In addition to these changes, Wat1003 (first water molecule in the chloride water pocket) has moved into the site where chloride was previously bound, and very interestingly, the putative catalytic water molecule, Wat1004 (Aghajari et al. 1998a,b), has disappeared. In other α-amylase three-dimensional structures, this conserved water molecule is in a perfect position to attack the α-1,4 glycosidic bond in the substrate. Both aspects, Wat1003 replacing the Cl− ion as well as the absence of Wat1004, are probably correlated with the eightfold lower activity of this mutant compared with the native enzyme (Table 1). One could argue that the missing Wat1004 is just not visible in the electron density because of the rather poor data (Table 3), but in this particular case, which concerns a water molecule that is found to be conserved in all α-amylase three-dimensional structures, in which it is firmly hydrogen bonded to the catalytic residues Glu200 and Asp264, it obviously does not seem fortuitous. It should be noticed that this inactive enzyme is quite difficult to crystallize and that the few crystals obtained are very small and unstable. This indicates that this mutant enzyme probably is quite labile.
Fig. 4.
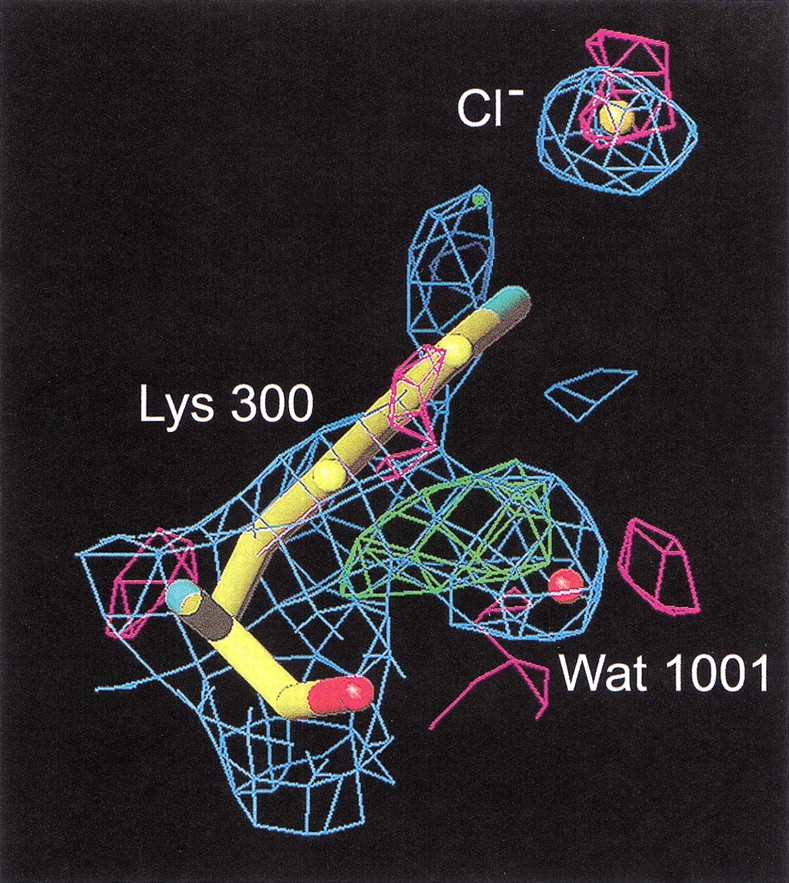
Chloride binding site in the inactive mutant K300Q. 2Fo − Fc density, negative Fo − Fc density, and positive Fo − Fc density are shown in blue, green, and pink, respectively. Here phases have been generated before the replacement of Lys300 by Gln. It is clearly seen that the obtained structure is devoid of chloride by the positive Fo − Fc density and that the glutamine residue that replaces lysine at position 300 points away from the former binding site. Please notice that the glutamine replaces a water molecule (Wat1001) in the water pocket earlier described (Aghajari et al. 1998a).
Table 3.
Data collection statistics for the AHA/NO3− complex and the mutants K300Q and K300R/Cl−
| K300R/Cl− | K300Q | AHA/NO3− | |
| Space group | C2221 | C2221 | C2221 |
| Unit cell (Å) | a = 70.3, b = 136.4, c = 113.5 | a = 71.5, b = 138.4, c = 115.4 | a = 71.3, b = 138.4, c = 114.9 |
| Resolution (Å) | 2.25 | 2.5 | 2.1 |
| Completeness | 95.8% | 81.8% | 89.3% |
| Completeness outermost shell | 53.9% | 39.2% | 90.5% |
| Number of reflections | 147154 | 57811 | 149687 |
| Number of reflections (unique) | 25155 | 16759 | 29641 |
| RI (all reflections, no cut-off) | 13.3% | 21.6% | 15.0% |
| I/σ(I) > 2 | 61.5% | 68.8% | 83.2% |
| I/σ(I) > 2 (outermost) | 15.0% | 34.0% | 65.9% |
| Rfactor | 17.88% | 18.04% | 14.61% |
| Rfree | 21.46% | 24.47% | 18.01% |
| No. amino acids | 448 | 448 | 448 |
| No. water molecules | 384 | 168 | 216 |
| No. Ca2+ ions | 1 | 1 | 1 |
| No. Cl− ions | 2 | 0 | 0 |
| No. NO3− ions | 0 | 0 | 1 |
| Cryo | yes | no | no |
RI = Σhkl Σi|I(hkl)i − 〈I(hkl)〉|/Σhkl Σi I(hkl)i
When comparing with the corresponding site in α-amylase from B. licheniformis (BLA; Machius et al. 1998), which is constituted of Arg229 (Arg172, AHA), Asn326 (Asn262, AHA), and Gln360 (Gln300, AHA), it is seen that although these residues are identical in nature, the sites are quite different spatially. The Asn (262, AHA; 326, BLA) side-chains are the only residues which display exactly the same conformation in the two enzymes, whereas the arginine (172, AHA; 229, BLA) side-chains adopt diverging positions in the respective enzymes. Considering a sphere with a radius of ∼20 Å centered on the chloride binding site, it appears that a significant number of residues, present in the first and second shell, are not conserved. Among the most important differences, we find that in the B. licheniformis enzyme, Phe284 (Thr221, AHA) and Trp41 (Gln33, AHA), as well as the proton donor in the hydrolytic reaction, Glu261 (Glu200 AHA), point directly into the binding site. Within this sphere, the number of aromatic residues in BLA is higher than in the psychrophilic amylase, and this site is clearly more positively charged in AHA than in BLA, even with the mutation Lys300Gln. In addition, the water molecule, which seems to have moved from the water pocket (Aghajari et al. 1998a), Wat1003, to the chloride binding site is found in BLA as well and corresponds to Wat2007.
The mutation Lys300Arg
The low binding constant of bacterial α-amylases for chloride ions has been related to the unidentate ligation of chloride by a lysine, whereas this ligand in animal α-amylases is replaced by an arginine, providing a bidentate coordination (Feller et al. 1996). However, as shown in Table 1, the replacement of Lys300 by Arg in AHA strikingly failed to restore the high affinity of mammalian α-amylases for chloride, with Kd ∼ 0.3 mM (Levitzki and Steer 1974). The X-ray structure of the mutant Lys300Arg shows that the Arg side-chain perfectly superimposes onto Lys in wild-type AHA (Fig. 2 ▶) and to the corresponding Arg in animal α-amylases. The reason for the lower affinity must therefore be found elsewhere than in the ligation mode of the chloride ion. The charge distribution within a sphere of 20 Å around the chloride binding site does not seem to differ significantly between AHA, pig pancreas α-amylase (PPA), or human pancreas α-amylase (HPA). Within this sphere, the only mutation in the first shell of amino acid residues surrounding the chloride binding site is Phe198 (AHA), which is a Tyr231 in HPA but a Phe in PPA. Other mutations within this sphere, between the bacterial and animal enzymes, are Phe5 (AHA) to Ile13 (animals), Thr85 to Ala97, Leu86 to Val98, Phe173 to Ile196 (PPA)/Leu196 (HPA), Val196 to Phe229, Ser226 to Gly259, Val257 to Leu293, Tyr298 to Phe335, and Pro299 to Thr336. This indicates that residues in the second shell also modulate the ion binding strength.
A probable second chloride binding site at the surface of the enzyme has been identified. Ligands binding to this chloride ion (Cl− 901 in AHA) are Arg73, Asn18, and Wat1461. This Cl−, which replaces Wat1158 in other AHA structures, however, seems to be loosely bound, as judged from the temperature factor (49 Å2 compared with 15 Å2 for the "catalytic" chloride ion) and from binding distances, which are 3.8 Å to Asn18NE2, 3.5 Å to Arg73, and 3.2 Å to Wat1461O. The data collection and refinement statistics for the three-dimensional structures are given in Table 3, and the main biochemical parameters are summarized in Table 1.
Chloride-free α-amylase
Despite numerous attempts, it was not possible to obtain crystals suitable for X-ray studies of the Cl−-free AHA. Micro crystals grown under the same conditions as the holo-form (Aghajari et al. 1996) disappeared after 3 weeks from appearance. One should note that AHA is a heat-labile and marginally stable enzyme. It has been shown by differential scanning calorimetry that chloride provides a positive contribution to the enthalpy of unfolding of 10 kcal/mole, probably by bridging the secondary structures bearing the chloride ligands (Feller et al. 1999). It is likely that the apo-form, although properly folded (Feller et al. 1996), is too unstable to maintain an ordered conformation required for crystallization.
Discussion
The structures described here allow us to not only explain the binding mode and the activation capacity of chloride but also provide new insights into the contribution of the effector to the catalytic mechanism. The anion binding site can accommodate ions of various size with different affinities (Levitzki and Steer 1974; Feller et al. 1996). As shown by the crystal structures of all chloride-dependent α-amylases (Qian et al. 1993; Larson et al. 1994; Brayer et al. 1995; Ramasubbu et al. 1996; Aghajari et al. 1998a,b; Strobl et al. 1998), the three protein ligands adopt a triangular and nearly equatorial conformation around the chloride ion. This conformation probably explains the previously reported relation between the anion binding constants and their ionic radius (Levitzki and Steer 1974; Feller et al. 1996). As an additional consequence, planar monovalent anions having a trigonal geometry, such as NO3− or ClO3−, bind optimally to α-amylases through stronger and extra electrostatic interactions with the usual chloride ligands (Tables 1, 2; Fig. 3 ▶). It can be inferred that other monovalent anions lacking this optimized geometry, such as NO2−, HCOO−, CNO−, or N3−, bind weakly to α-amylases. Incidentally, nitrate in polluted waters could be an unsuspected weak inhibitor of human and animal α-amylases as a result of its high affinity but low activation of these enzymes.
The complex of AHA with nitrate is also instrumental in explaining the difference in activation capacity displayed by the various monovalent anions. Indeed, when the chloride ion is bound to its site, the negative charge is localized. In the case of NO3−, the charge is delocalized on the three oxygen atoms and therefore is more efficiently counterbalanced as a consequence of the increased number of interactions performed with the protein ligands compared with the chloride ion. Because the charge is now delocalized (Fig. 3 ▶), it may influence the environment of catalytic residues and of water molecules (Fig. 5 ▶) in a less efficient way, as indicated by a loss of half of the maximal activity (Table 1). One should, however, not exclude dynamics correlated with structural features. As mentioned above, an increased number of interactions that are stronger (Table 2) are performed between protein ligands and the nitrate ion compared to those of the chloride ion. The entire hydrogen bonding network in and around the active site is consequently altered, and hydrogen bonds to the nitrate ion lock down ligating residues, which in turn may reduce the flexibility of the active site residues and thus the catalytic activity.
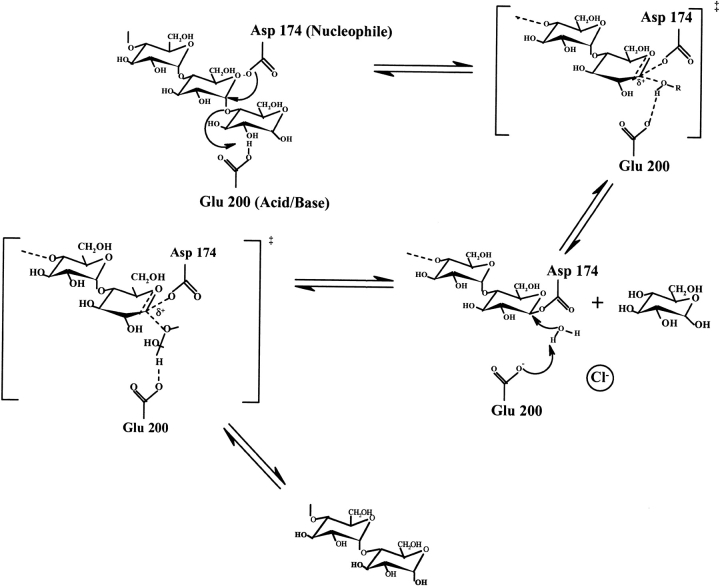
Fig. 5.
Reaction mechanism for P. haloplanktis α-amylase (AHA) and chloride-dependent α-amylases in general. By analogy with other studies of glycosidases, Asp 174 has been proven to be the catalytic nucleophile (McCarter and Withers 1996), whereas Glu200 is the best candidate for being the proton donor (Svensson and Søgaard 1993). The role of the third catalytic residue, Asp264, is more unclear, but it has been proposed that it stabilizes the protonated state of the glutamic side-chain (Strokopytov et al. 1995) and that it may contribute to control and/or maintain an elevated pKa value of the nearby proton donor (Brzozowski and Davies 1997). These studies propose another function of Asp264: In conjunction with the chloride ion, it creates a negative field that forces the water molecules implicated in hydrolysis (after being deprotonated by Glu200) to move, because of electrostatic repulsion, toward the anomeric C1 atom, where it attacks the substrate.
The crystal structure of the chloride-free mutant Lys300Gln indicates that Cl− is not mandatory for the amylolytic activity or for proper enzyme folding but that the anion dramatically enhances the enzyme reactivity and structural stability. This is possibly the evolutionary advantage of the chloride-dependent α-amylases found in all animals, whereas plant, yeast and most microbial α-amylases have to perform catalysis in chloride-poor or variable environments. There is strong experimental evidence that the chloride ion in AHA allows the acid/base catalyst Glu200 to be protonated at the pH of maximal activity (around neutrality) either through a charge-relay system (Fig. 1 ▶) or through a direct effect on the carboxylate (Qian et al. 1994; Feller et al. 1996). In addition, the electric field created by the chloride ion would help to polarize the putative catalytic water molecule Wat1004, which after its deprotonation assisted by Glu200 would be in a very unfavorable position close to the chloride ion (Fig. 5 ▶). Repulsion of the HO− moiety toward the anomeric carbon of the cleaved substrate can drastically enhance the second hydrolytic step of the reaction pathway (Fig. 5 ▶), leading to the formation of a new reducing end. The low activity of the chloride-free mutant Lys300Gln, in which the putative catalytic water molecule has disappeared after the mutation, supports a close relation between Cl− and Wat1004. Nevertheless, one should not exclude that both possible functions of chloride, namely, the pKa shift of Glu200 and the polarization of the catalytic Wat1004, are not mutually incompatible.
Materials and methods
Production of enzyme and crystal growth
Substitution of Cl− by NO3−was achieved by dialyzing the purified wild-type AHA (Feller et al. 1992) against 5 mM NH4HCO3, and lyophilization of the resulting chloride-free enzyme and solubilization in 10 mM Tris-HNO3, 25 mM NaNO3, 1 mM CaCl2, and 1 mM NaN3 (pH 8.0) at a final concentration of 18 mg/mL. The mutations Lys300Gln and Lys300Arg were introduced in the α-amylase gene by inverse PCR, and the mutant enzymes were expressed in Escherichia coli and purified as described (Feller et al. 1996).
Crystals of AHA mutants Lys300Gln and Lys300Arg, as well as the complex between native wild-type AHA and NO3−, were grown under conditions as described elsewhere (Aghajari et al. 1996) for the native enzyme and using crystals of wild-type AHA for cross-seeding. Small crystals grown to a size of ∼0.1 × 0.2 × 0.3 mm3 appeared after ∼4 weeks for Lys300Arg and after 3 weeks for the nitrate complex. As concerns mutant Lys300Gln, crystals grow to a size of 0.1 × 0.1 × 0.15 mm3 after ∼4 weeks.
X-ray data collections, structure determination, and refinement
Data for the mutant Lys300Arg and for the complex AHA/NO3− were collected on an in house MARresearch 345-mm image plate system, using CuKα radiation generated from a Nonius FR 591 rotating anode operating at 40 kV and 80 mA, with a graphite monochromator in the case of the AHA/NO3− complex and osmic mirrors in the case of the mutant K300R. Data for the mutant Lys300Arg were collected at cryogenic temperature (100 K), and crystals were flash-frozen in liquid nitrogen directly from the drop without the need for additional cryo-protectants.
Data for the mutant Lys300Gln were collected at 15°C on a MARresearch 300-mm image plate system with a graphite monochromator, with CuKα X-rays generated from a Rigaku RU200 rotating anode operating at 40 kV and 80 mA. Diffraction data were integrated and scaled using DENZO (Otwinowski and Minor 1997) and SCALA (Collaborative Computing Project Number 4 1994) for mutant Lys300Gln and the complex with the nitrate ion, whereas SCALEPACK (Otwinowski and Minor 1997) was used for scaling cryo mutant data. Initial phases came from the native structure of AHA (Aghajari et al. 1998a), and refinements were performed using the program X-PLOR (Brünger et al. 1987; Brünger 1992) followed by a final cycle of refinement by CNS (Brünger et al. 1998) in the case of the nitrate complex and the mutant Lys300Gln, whereas CNS was used throughout for refinement of Lys300Arg.
The atomic coordinates and structure factors (codes 1JD7 [K300R], 1JD9 [K300Q], and 1L0P [complex with NO3−]) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Acknowledgments
This work was supported by the European Human Capital and Mobility Network CHRX-CT-94-0521 and the BIOTECH programme COLDZYME. Support from the Centre National de la Recherche Scientifique is greatfully acknowledged as well.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0202602.
References
- Aghajari, N., Feller, G., Gerday, C. and Haser, R. 1996. Crystallization and preliminary X-ray diffraction studies of α-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. Protein Sci. 5 2128–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998a. Crystal structures of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 7 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998b. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure 6 1503–1516. [DOI] [PubMed] [Google Scholar]
- Aghajari, N., Roth, M., and Haser, R. 2002. Crystallographic evidence of a transglycosylation reaction: Ternary complexes of a psychrophilic α-amylase. Biochemistry 41 4273–4280. [DOI] [PubMed] [Google Scholar]
- Brayer, G.D., Luo, Y., and Withers, S.G. 1995. The structure of human pancreatic α-amylase at 1.8 Å resolution and comparisons with related enzymes. Protein Sci. 4 1730–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayer, G.D., Sidhu, G., Maurus, R., Rydberg, E.H., Braun, C., Wang, Y., Nguyen, N.T., Overall, C.M., and Withers, S.G. 2000. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry 39 4778–4791. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1992. The free R-value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355 472–474. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Kuriyan, J., and Karplus, M. 1987. Crystallographic R-factor refinement by molecular dynamics. Science 235 458–460. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T., and Warren, G.L. 1998. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Brzozowski, A.M. and Davies, G.J. 1997. Structure of Aspergillus oryzae α-amylase complexed with the inhibitor acarbose at 2.0 Å resolution. Biochemistry 36 10837–10845. [DOI] [PubMed] [Google Scholar]
- Collaborative Computing Project Number 4. 1994. The CCP4 Suite: Programs for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- D'Amico, S., Gerday, C., and Feller, G. 2000. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene 253 95–105. [DOI] [PubMed] [Google Scholar]
- Feller, G., Lonhienne, T., Deroanne, C., Libioulle, C., Van Beeumen, J., and Gerday, C. 1992. Purification, characterization, and nucleotide sequence of the thermolabile α-amylase from the Antarctic psychrotroph Alteromonas haloplanctis A23. J. Biol. Chem. 267 5217–5221. [PubMed] [Google Scholar]
- Feller, G., Bussy, O., Houssier, C., and Gerday, C. 1996. Structural and functional aspects of chloride binding to Alteromonas haloplanctis α-amylase. J. Biol. Chem. 271 23836–23841. [DOI] [PubMed] [Google Scholar]
- Feller, G., D'Amico, S., and Gerday, C. 1999. Thermodynamic stability of a cold-active α-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38 4613–4619. [DOI] [PubMed] [Google Scholar]
- Janecek, S. 1994. Sequence similarities and evolutionary relationships of microbial, plant and animal α-amylases. Eur. J. Biochem. 224 519–524. [DOI] [PubMed] [Google Scholar]
- ———. 1997. α-Amylase family: Molecular biology and evolution. Prog. Biophys. Mol. Biol. 67 67–97. [DOI] [PubMed] [Google Scholar]
- Larson, S.B., Greenwood, A., Cascio, D., Day, J., and McPherson, A. 1994. Refined molecular structure of pig pancreatic α-amylase at 2.1 Å resolution. J. Mol. Biol. 235 1560–1584. [DOI] [PubMed] [Google Scholar]
- Levitzki, A. and Steer, M. L. 1974. The allosteric activation of mammalian α-amylase by chloride. Eur. J. Biochem. 41 171–180. [DOI] [PubMed] [Google Scholar]
- Machius, M., Declerck, N., Huber, R., and Wiegand, G. 1998. Activation of Bacillus licheniformis α-amylase through a disorder→order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad. Structure 6 281–292. [DOI] [PubMed] [Google Scholar]
- McCarter, J.D. and Withers, S.G. 1996. Unequivocal identification of Asp-214 as the catalytic nucleophile of Saccharomyces cerevisiae α-glucosidase using 5-fluoro glycosyl fluorides. J. Biol. Chem. 271 6889–6894. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Qian, M., Haser, R., and Payan, F. 1993. Structure and molecular model refinement of pig pancreatic α-amylase at 2.1 Å resolution. J. Mol. Biol. 231 785–799. [DOI] [PubMed] [Google Scholar]
- Qian, M., Haser, R., Buisson, G., Duee, E., and Payan, F. 1994. The active center of a mammalian α-amylase: Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2-Å resolution. Biochemistry 33 6284–6294. [DOI] [PubMed] [Google Scholar]
- Qian, M., Nahoum, V., Bonicel, J., Bischoff, H., Henrissat, B., and Payan, F. 2001. Enzyme-catalyzed condesation reaction in a mammalian α-amylase: High resolution structure analysis of an enzyme inhibitor complex. Biochemistry 40 7700–7709. [DOI] [PubMed] [Google Scholar]
- Ramasubbu, N., Paloth, V., Luo, Y., Brayer, G.D., and Levine, M.J. 1996. Structure of human salivary α-amylase at 1.6 Å resolution: Implication for its role in the oral cavity. Acta Crystallogr. D Biol. Crystallogr. 52 435–446. [DOI] [PubMed] [Google Scholar]
- Strobl, S., Maskos, K., Betz, M., Wiegand, G., Huber, R., Gomis-Ruth, F.X., and Glockshuber, R. 1998. Crystal structure of yellow meal worm α-amylase at 1.64 Å resolution. J. Mol. Biol. 278 617–628. [DOI] [PubMed] [Google Scholar]
- Strokopytov, B., Penninga, D. Rozeboom, H.J., Kalk, K.H., Dijkhuizen, L., and Dijkstra, B.W. 1995. X-ray structure of cyclodextrin glycosyl transferase complexed with acarbose: Implications for the catalytic mechanism of glycosidases. Biochemistry 34 2234–2240. [DOI] [PubMed] [Google Scholar]
- Svensson, B. and Søgaard, M. 1993. Mutational analysis of glycosilase function. J. Biotechnol. 29 1–37. [Google Scholar]
- Uitdehaag, J.C., Mosi, R., Kalk, K.H., van der Veen, B.A., Dijkhuizen, L., Withers, S.G., and Dijkstra, B.W. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 6 432–436. [DOI] [PubMed] [Google Scholar]