Abstract
By means of profile-matching procedures, conservation of functionally important residues, and fold-recognition techniques, we show that two distinct families of lipopolysaccharide kinases encoded in the genomes of Gram-negative bacteria are related to each other and to two distinct classes of proteins, namely eukaryotic protein kinases and right open reading frame (RIO1). Members of one of the lipopolysaccharide kinase families are identified only in pathogenic bacteria. Phosphorylation by these enzymes is relevant in the construction of outer membrane, immune response, and pathogenic virulence. The class of proteins called RIO1, also related to eukaryotic protein kinases and previously known to occur only in archaea and eukaryotes, are now identified in eubacteria as well. It has been suggested here that RIO1 proteins are intermediately related to lipopolysaccharide kinases and eukaryotic protein kinases implying an evolutionary relationship between the three classes of proteins.
Keywords: Lipopolysaccharide, outer-cell membrane, phosphorylation, profile matching, protein kinases, sequence comparison
Lipopolysaccharides (LPSs), a major constituent of the outer membrane of all Gram-negative bacteria, play a critical role in bacterial viability and virulence (White et al. 1997). They act as a permeability barrier to certain antibacterial agents and host defense factors. The outer membrane has three conserved components, namely, lipid A, core (comprised of phosphoryl derivatives of sugar residues) and the O-antigen. The phosphoryl substituents of the core serve to stabilize the outer membrane by means of cross-linking to adjacent LPS molecules via divalent cations (Yethon and Whitfield 2001).
Lack of phosphorylation in the core has been shown to result in strains that are hypersensitive to detergents and hydrophobic antibiotics, and also being less virulent (Yethon and Whitfield 2001). Two key enzymes involved in the core phosphorylation, namely WaaP gene product (heptose kinase) and the Kdo (3-Deoxy-d-manno-octulosonic acid) kinase, have been well characterized (White et al. 1999; Yethon and Whitfield 2001). These two LPS kinases differ in their substrate specificity, despite their involvement in the same lipopolysaccharide biosynthesis pathway. As can be seen later in this paper, these two classes of LPS phosphorylating enzymes are related to each other and occur only in Gram-negative bacteria. Occurrence of Kdo kinases has been restricted to Gram-negative pathogens. Earlier studies have shown that the Kdo kinase is responsible for the virulence in Haemophilus influenzae (White et al. 1999). Here, we discuss the results of our analysis indicating significant similarity between amino-acid sequences of these LPS phosphorylating enzymes and eukaryotic protein kinases.
A class of kinase-like sequences, right open reading frame (RIO1), previously has been shown to be coded in the genomes of archaea and eukaryotes (Plowman et al. 1999). Here, we report the occurrence of RIO1 in eubacteria as well, in addition to its occurrence in archaea and eukaryotes, suggesting RIO1 as an evolutionary link between eukaryotic protein kinase-like sequences in prokaryotes and eukaryotic protein kinases. Further, we suggest that RIO1 sequences are intermediately related to LPS phosphorylating enzymes and eukaryotic protein kinases.
Protein kinases encoded in the eukaryotic genomes form one of the largest families of proteins (Hanks and Hunter 1995). The catalytic domain of these kinases share a common three-dimensional (3-D) fold irrespective of their association with diverse domains and subunits required for their regulation (Johnson et al. 1996). The key sequence motifs associated with functional properties and conserved among most protein kinases (Hanks et al. 1988) include the Gly-rich loop (sub-domain I), which occurs close to ATP; the invariant lysine (sub-domain II), which is hydrogen bonded to ATP; and a glutamate (sub-domain III), which anchors the phosphates of the ATP through the invariant lysine. Further functional residues include the catalytic aspartate and invariant asparagine (both in sub-domain VIb) and another aspartate (sub-domain VII), which are required for the chelation of the Mg2+ ions. An invariant aspartate (sub-domain IX) helps in the stabilization of the catalytic loop by interacting with the arginine in the catalytic loop. An arginine preceding the catalytic Asp interacts with the phosphorylated Ser/Thr/Tyr residue to enable correct disposition of the various catalytic residues in those kinases that are regulated by phosphorylation in their activation segment (Johnson et al. 1996). The occurrences of the eukaryotic protein kinase-like sequences in the prokaryotes are becoming increasingly evident with the completion of genome sequencing of many archeaebacterial and eubacterial species (Leonard et al. 1998; Kennelly 2002). However, the most well-studied protein kinases of the prokaryotes belong to the histidine kinase family, which does not share similarity in terms of structure, sequence, and modes of regulation (Dutta et al. 1999) with Ser/Thr/Tyr kinases.
A relationship has been established in the current study between LPS phosphorylating enzymes in Gram-negative bacteria and eukaryotic protein kinases using a match of sequences with profiles of protein families. The compatibility of the sequences of these bacterial proteins to the eukaryotic kinase fold, as deduced from different inverse folding procedures, also is shown to be excellent. These sequences also show a high degree of conservation of functionally critical residues. This suggests that the eukaryotic protein kinases are related to lipopolysaccharide kinases in addition to the lipid kinases (Walker et al. 1999) and aminoglycoside kinases (Hon et al. 1997).
Results and Discussion
A large number of searches has been performed using the powerful sequence search method, PSI-BLAST (Altschul et al. 1997), in the nonredundant database (NRDB) of protein sequences and in a data-set containing the predicted amino-acid sequences of proteins in 41 complete bacterial genomes to identify the homologs of Kdo kinase (KdoK), WaaP gene product, and the kinase-like sequences, RIO1. A search for KdoK of H. influenzae in NRDB is able to identify other homologs in Vibrio cholerae, Xylella fastidiosa, and Pastuerella multocida with very significant E-values (10−72 to 10−29) and sharing >33% sequence identity with the KdoK of H. influenzae. All these gene products are <260 amino acids long and no domain, other than kinase domain, has been identified in these gene products. Interestingly, all the sources of KdoK correspond to Gram-negative pathogens. While V. cholerae and H. influenzae are human pathogens and P. multocida causes cholera in poultry animals, X. fastidiosa is known to be a plant pathogen. It is not obvious why Kdo kinase occurs only in a subset of Gram-negative bacteria that happens to be pathogens. Identification of WaaP gene products from Escherichia coli, Pseudomonas aeruginosa, and Salmonella typhimurium (E = 6 × 10−5), as well as eukaryotic protein kinases (E = 2 × 10−5) in the subsequent cycles of PSI-BLAST with KdoK as the query, suggests that the two families of LPS kinases (KdoK and WaaP gene products) are related to the eukaryotic protein kinases.
A search for the WaaP homologs in the NRDB identified various isoforms of WaaP gene products in E. coli and variants in P. aeruginosa and S. typhimurium. All the WaaP gene products contained kinase-like domain only. The subsequent cycles of PSI-BLAST also picked up KdoK variants followed by eukaryotic protein kinases in support of the relationship between the LPS kinases and the eukaryotic protein kinases.
A search for the eukaryotic protein kinase-like sequences in the database containing the 41 bacterial genomes detected the RIO1 gene products followed by the Kdo kinases in the subsequent cycles. This suggests that the eukaryotic protein kinases are more closely related to the Kdo kinase than to the WaaP gene product. The sequence identity between WaaP and Kdo kinases is low (<14%), while the sequence identities among WaaP proteins and among Kdo kinases are high (>53% and >33%, respectively), suggesting that WaaP and Kdo kinases form different families of LPS kinases.
The biological function of RIO1 is unclear, and it was previously identified only in archaea and in the genomes of Saccharomyces cerevisiae and Caenorhabditis elegans (Angenmayr and Bandlow 1997; Leonard et al. 1998; Plowman et al. 1999). In this study, RIO1 homologs have been further identified in two species of eubacteria, P. aeruginosa and Deinococcus radians (E<10−12). This suggests the RIO1 sequences of bacteria could possibly be an intermediate link in the evolution of the eukaryotic protein kinase-like sequences in bacteria and protein kinases in eukaryotes. The results of the PSI-BLAST search with human ERK and RIO1 sequences as queries also suggests that RIO1 sequences are intermediately linked to the KdoK and the eukaryotic protein kinases.
To further ensure that the Kdo kinases are related to the eukaryotic protein kinase family, a profile-based search method IMPALA (Schaffer et al. 1999) was employed. The protein sequence of each variant of KdoK was searched in a profile database comprising 2764 profiles corresponding to the 2697 families of proteins in Pfam (Bateman et al. 2000) and 67 subfamily profiles of the eukaryotic protein kinases. The search picked up about 50 hits below the E-value of 5 × 10−4 and all the hits corresponded to the profiles of various subfamilies of eukaryotic protein kinases. The top-most hits corresponded to extracellularly regulated kinase (ERK) variants from yeast, worm, and human with the E-values of the order of 5 × 10−6. The above results therefore confirm that the KdoK is related to the eukaryotic protein kinase family. Results of the IMPALA search with WaaP gene products are similar to those obtained for Kdo kinases.
Fold recognition of the Kdo kinase sequence has been performed using GENTHREADER (Jones 1999) and 3D-PSSM (Kelley et al. 2000). These procedures work on different principles. GENTHREADER identified 10 hits of known 3-D structure, all of which correspond to the eukaryotic protein kinases, which share the common fold, although the details of the structures vary to some extent. The probability of the hits being the correct fold as suggested by GENTHREADER is one for all of the 10 hits. This corresponds to a GENTHREADER confidence level "Certain; >99%." Interestingly, the sequence identity between Kdo kinase and protein kinases of known structure is very low, ranging between 9% and 15%. The only lipid kinase (PI3 kinase γ) structure (Walker et al. 1999) available is not picked up as one of the hits and its fold involves several long insertions compared to Ser/Thr and Tyr kinases. It would have been interesting to explore if the Kdo kinase sequence fits better with the structure of antibiotics phosphorylating enzyme (APH) from Enterococcus (Hon et al. 1997), as the fold of APH is the same as that of Ser/Thr protein kinases. However, the structure of APH is not yet available in the protein data bank, hence this analysis could not be made by us. Result of fold recognition using 3D-PSSM is very similar to that of GENTHREADER. With a KdoK as the query, all of the 20 hits from 3D-PSSM corresponded to Ser/Thr or Tyr kinases with an E-value of the order of 8 × 10−2, which suggests highly reliable hits according to 3D-PSSM.
A multiple sequence alignment of Kdo kinases, WaaP variants, and RIO1 sequences has been generated (Fig. 1 ▶) to analyze the extent of conservation of the functionally critical residues with respect to eukaryotic cyclic AMP-dependent protein kinase (cAPK) (Zheng et al. 1993). The catalytic aspartate (D166) conserved in all known eukaryotic protein kinases; a critical lysine (K72) involved in ATP anchoring; an aspargine and aspartate (N171 and D184) involved in chelation of Mg2+; and aspartates (D208, D220) involved in the stabilization of the catalytic loop are all highly conserved in all the bacterial proteins under current investigation. Various buried polar and nonpolar residues (Fig. 1 ▶) occurring in α-helices and β-strands of cAPK are well conserved in these sequences. The arginine preceding D166 is identified only in the WaaP variants, an indicator of phosphorylation in the activation segment, as seen in the eukaryotic protein kinases (Johnson et al. 1996). Indeed, very recently it has been shown that a site-directed mutant of E. coli WaaP wherein the critical aspartate is replaced by alanine showed almost no phosphorylation on LPS (Yethon and Whitfield 2001). The glycine-rich region is known to occur close to the ATP binding site of eukaryotic protein kinases. While three glycines are present in the "equivalent" region in the bacterial proteins, the sequence pattern deviates from that of eukaryotic protein kinases. However, the only eukaryotic protein kinase-like bacterial kinase of known structure (Hon et al. 1997) that is known to phosphorylate a carbohydrate (aminoglycoside [APH]) lacks a glycine-rich region, although the crystal structure shows ATP binding and remarkable similarity of the overall fold to cAPK (Hon et al. 1997). Based on the conservation of various residues critical for the function in LPS kinases, it is proposed that the mechanism of action LPS kinases, in employing ATP as the phosphate donor and in catalyzing phosphorylation, is similar to that of eukaryotic protein kinases although the substrate specificity of LPS kinases and eukaryotic proteins are very different.
Fig. 1.
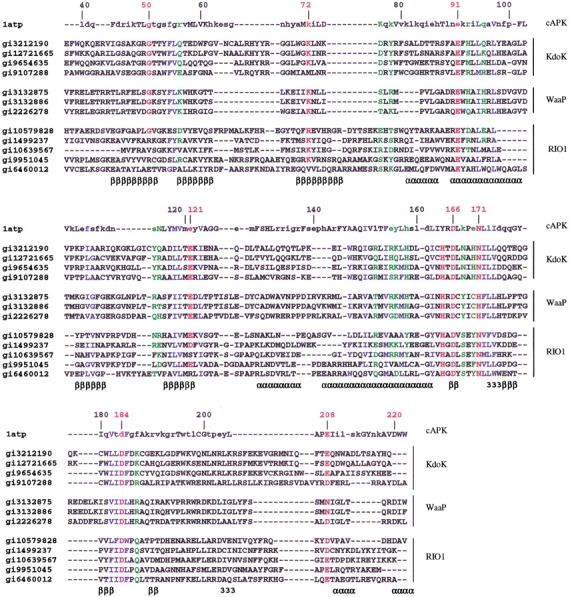
Multiple sequence alignment of cyclic-AMP–dependent protein kinase (cAPK-1atp) with the KDO kinase from Haemophilus influenzae (gi3212190, 29–241), Pasteurella multocida (gi12721665, 29–241), Vibrio cholerae (gi9654635, 46–251), Xylella fastidiosa (gi9107288, 48–254), WaaP homologs from Escherichia coli (gi3132875, 29–225), Salmonella typhimurium (gi3132886, 29–225), Pseudomonas aeruginosa (gi226278, 27–225) and RIO1 homologs of Halobacterium sp. NRC-1 (gi10579828, 75–289), Methanococcus janaschii (gi1499237, 68–277), Thermoplasma acidophilum (gi10639567, 1–186), Pseudomonas aeruginosa (gi9951045, 18–246), and Deinococcus radiodurans (gi6460012, 70–263). The secondary structure of cAPK is indicated in the last line of each alignment block. α, β, and 3 represent α-helical, β-strand, and 310-helical regions, respectively. Residues of cAPK that are shown in lower case and in upper case represent the solvent accessible and buried residues (defined by <7% of residue accessibility), respectively. The residue numbering shown above each sequence block corresponds to that used in the crystal structure of cAPK (1atp [Zheng et al. 1993]). The functionally critical residues of cAPK that are conserved in the various sequences are marked red. Residues with green and blue colors represent the polar and nonpolar residues, respectively, conserved across the KDO, WaaP, and RIO1 members. As can be seen from the alignment, the functional residues of cAPK are well conserved in most of the sequences. The conservation of the arginine of the `RD' is, however, seen only in the WaaP variants, indicating that they could probably undergo phosphorylation in the activation segment as a key regulating mechanism as seen in cAPK. Further, it is interesting to note that the D220 that is known to interact with the R of the `RD' to stabilize the catalytic loop also is conserved in all the WaaP variants. The conservation of catalytic aspartate (D166) of cAPK in all the sequences indicate a mechanism of phosphorylation similar to that of the eukaryotic protein kinases in KDO kinases and WaaP variants and probably the RIO1 variants whose function is not known. The residues involved in the anchoring of ATP in cAPK, namely G50, K72, E91, and the residues involved in chelation of Mg2+ (N171, D184) are also well conserved in most of the sequences.
Conclusions
The current analysis therefore reveals that the substrate specificity of the eukaryotic protein kinase superfamily is more diverse and now includes LPSs apart from serine/threonine/tyrosine-containing sequences, lipids and aminoglycosides. The conservation of the catalytic residues of the eukaryotic protein kinases also suggests the cofactor requirement and the catalytic mechanism of the LPS kinases to be similar to the eukaryotic protein kinases. The current study also suggests potential similarity between the three classes of proteins, namely the LPS kinases, eukaryotic protein kinases, and similar sequences in bacteria. The identification of RIO1-like proteins in eubacteria that have been previously known to be restricted to archaea and eukaryotes indicates a possible evolutionary relationship between RIO1 and the three classes of proteins mentioned above. The low sequence similarity between the RIO1, the two classes of LPSK, and the ePKs, but a significant similarity in the 3-D–fold and catalytic residues, suggests an independent as well as divergent evolution of these three classes of proteins from a common ancestor. Based on the current study, we also suggest an evolutionary relationship between the WaaP gene products and Kdo kinases with a higher likelihood of the latter being evolved divergently from WaaP gene product to enable different substrate specificity and hence for the evolution of virulence in the organisms that encode them. Potential similarity of the structures, ATP binding, and the catalytic residues of the cAPK in the Kdo kinase, therefore indicates an evolutionary relation between the two. This similarity therefore could be investigated experimentally further to test the effectiveness of eukaryotic kinase inhibitors and their variants on Kdo kinase and hence guides in the design of an inhibitor against Kdo kinase, which plays a significant role in the virulence of pathogens.
Materials and methods
Data set
A database of 41 complete sets of predicted protein sequences of bacteria and the NRDB of proteins have been obtained from the ftp web site ncbi.nlm.nih.gov.
Profile matching
Iterative searches have been made using PSI-BLAST (Altschul et al. 1997) in a linux platform using a stand-alone version, on the bacterial genomes and the NRDB with an E-value cut-off of 0.0005. Every PSI-BLAST output has been examined manually to ensure that there is no obvious "drift" as the cycles of the search progressed, which could lead to false positives. A series of searches has been made, using IMPALA (Schaffer et al, 1999), with each one of the Kdo kinase sequences and WaaP gene products as query in a database of 2764 profiles (position-specific score matrices [PSSMs]) corresponding to 2697 protein families in Pfam database (Bateman et al. 2000) and 67 profiles corresponding to various subfamilies of protein kinases. The PSSMs of families in Pfam have been constructed, using the "seed alignments" given in Pfam, to explore using the powers of IMPALA, which uses profile matching in addition to using Hidden Markov Model-based searches in Pfam. The subfamily profiles of eukaryotic protein kinases have been constructed by using the sequences and alignment corresponding to the various subfamilies of eukaryotic protein kinases available at the Protein Kinase Resource (http://www.sdsc.edu/Kinases) (Smith et al. 1997). The multiple sequence alignment shown in the Figure 1 ▶ initially was generated by MALIGN (Johnson et al. 1993).
Fold recognition
Two methods, GENTHREADER (Jones 1999) and 3D-PSSM (Kelly et al. 2000), which use very different principles in fold recognition, have been used to predict the fold of KdoK and WaaP gene products.
Acknowledgments
We thank Ms. S. Sujatha for generating the profiles of families in Pfam database. A.K. is supported by a fellowship from C.S.I.R, India. This research is supported by a Senior Fellowship to N.S. from the Wellcome Trust, U.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.3560102.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids. Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenmayr, M. and Bandlow, W. 1997. The type of basal promoter determines the regulated or constitutive mode of transcription in the common control region of the yeast gene pair GCY/RIO1. J. Biol. Chem. 272 31360–31635. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L.L. 2000. The Pfam protein families database. Nucl. Acids. Res. 28 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, R., Qin, L., and Inouye, M. 1999. Histidine kinases: Diversity of domain organization. Mol. Microbiol. 34 633–640. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., Quin, A.M., Hunter, T. 1988. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241 42–52. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K. and Hunter, T. 1995. The eukaryotic protein kinase family: Kinase (catalytic) domain structure and classification. FASEB J. 9 576–596. [PubMed] [Google Scholar]
- Hon, W.C., McKay, G.A., Thompson, P.R., Sweet, R.M., Yang, D.S.C., Wright, G.D., and Berghuis, A.M. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89 887–895. [DOI] [PubMed] [Google Scholar]
- Johnson, M.S., Overington, J.P., and Blundell, T.L. 1993. Alignment and searching for common protein folds using a data bank of structural templates. J. Mol. Biol. 231 735–752. [DOI] [PubMed] [Google Scholar]
- Johnson, L.N., Noble, M.E.M., and Owen, D.J. 1996. Active and inactive protein kinases: Structural basis for regulation. Cell 85 149–158. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. 1999. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287 797–815. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J.E. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299 499–526. [DOI] [PubMed] [Google Scholar]
- Kennelly, P.J. 2002. Protein kinases and protein phosphatases in prokaryotes: A genomic perspective. FEMS Microbiol. Lett. 206 1–8. [DOI] [PubMed] [Google Scholar]
- Leonard, C.J., Aravind, L., and Koonin, E.V. 1998. Novel families of putative protein kinases in bacteria and archaea: Evolution of the "Eukaryotic" protein kinase superfamily. Genome Res. 8 1038–1047. [DOI] [PubMed] [Google Scholar]
- Plowman, G.D., Sudarsanam, S., Bingham, J., Whyte, D., and Hunter, T. 1999. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. 96 13603–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, A.A., Wolf, Y.I., Ponting, C.P., Koonin, E.V., Aravind, L., and Altschul, S.F. 1999. IMPALA: Matching a protein sequence against a collection of PSI-BLAST–constructed position specific score matrices. Bioinformatics 15 1000–1011. [DOI] [PubMed] [Google Scholar]
- Smith, C.M., Shindyalov, I.N., Veretnik, S., Gribskov, M., Taylor, S.S., TenEyck, L.F., and Bourne, P.E. 1997. The protein kinase resource. Trends Biochem. Sci. 22 444–446. [DOI] [PubMed] [Google Scholar]
- Walker, E.H., Perisic, O., Ried, C., Stephens, L., and Williams, R.L. 1999. Structural insights into PI3K catalysis and signalling. Nature 402 313–320. [DOI] [PubMed] [Google Scholar]
- White, K.A., Kaltashov, I.A., Cotter, R.J., and Raetz, C.R.H. 1997. A monofunctional 3-deoxy-D-manno-octusulonic acid (Kdo) transferase and a Kdo kinase in extracts of Haemophilus influenzae. J. Biol. Chem. 272 16555–16563. [DOI] [PubMed] [Google Scholar]
- White, K.A., Lin, S.H., Cotter, R.J., and Raetz, C.R.H. 1999. A Haemophilus influenzae that encodes a membrane bound 3-deoxy-D-manno-octulosonic acid (Kdo) kinase. Possible involvement of Kdo phosphorylation in bacterial virulence. J. Biol. Chem. 274 31391–31400. [DOI] [PubMed] [Google Scholar]
- Yethon, J.A. and Whitfield, C. 2001. Purification and characterization of WaaP from Escherichia coli, a lipopolysaccharide kinase essential for outer membrane stability. J. Biol. Chem. 276 5498–5504. [DOI] [PubMed] [Google Scholar]
- Zheng, J.H., Knighton, D.R., Teneyck, L.F., Karlsson, R., Xuong, N.H., Taylor, S.S., and Sowadski, J.M. 1993. Crystal structure of the catalytic sub-unit of cAMP dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry 32 2154–2161. [DOI] [PubMed] [Google Scholar]


