Abstract
BetaCore is a designed ∼50-residue protein in which two BPTI-derived core modules, CM I and CM II, are connected by a 22-atom cross-link. At low temperature and pH 3, homo- and heteronuclear NMR data report a dominant folded (`f') conformation with well-dispersed chemical shifts, i, i+1 periodicity, numerous long-range NOEs, and slowed amide hydrogen isotope exchange patterns that is a four-stranded antiparallel β-sheet with nonsymmetrical and specific association of CM I and CM II. BetaCore `f' conformations undergo reversible, global, moderately cooperative, non-two-state thermal transitions to an equilibrium ensemble of unfolded `u' conformations. There is a significant energy barrier between `f' and `u' conformations. This is the first designed four-stranded antiparallel β-sheet that folds in water.
Keywords: Protein design, protein folding, β-sheet protein, core modules, cross-link
The active field of `protein design' focuses on generating sequences that can adopt a unique ensemble of closely related three-dimensional structures under conditions approaching physiological, and as such, provides an opportunity to apply and test ideas about protein folding and stability (Richardson et al. 1992; Hodges 1996; Beasley and Hecht 1997; Tuchscherer et al. 1998; DeGrado et al. 1999). De novo design involves the construction of sequences that are not directly related to those of any natural protein (DeGrado et al. 1999). Alternatively, naturally occurring proteins can be redesigned to simpler `minimalist' sequences (Vita et al. 1998; Imperiali and Ottesen 1999). Automated, structure-based strategies begin with an experimentally determined architecture of a natural protein and are used to produce new proteins with enhanced stabilities or novel functions (Dahidat and Mayo 1997). These approaches can extend the repertoire of 20 proteinogenic L-amino acid residues linked in a polypeptide chain(s) with additional building blocks (including D-stereoisomers) and/or templates, and are generally based on the premise that incipient secondary structural elements such as α-helices, β-sheets, β-turns, etc. are further stabilized in the final designed proteins by spontaneous self-assemblies (monomeric intramolecular or multimeric such as coiled-coil or bundles), covalent cross-links (e.g., disulfides), and/or metal ligations.
Considerable successes in the design of α-helical proteins (Hill et al. 2000) are in contrast to the myriad challenges of creating β-sheet proteins (Smith and Regan 1997). Specific intramolecular long-range backbone and side chain interstrand interactions required to achieve these structures are countered by related intermolecular interactions; the latter are manifested by a pronounced tendency to aggregate in aqueous media. Nevertheless, building on information about conformational propensities of residues in β-turns and β-sheets revealed in protein structural databases as well as through experimental studies of model β-hairpins (Blanco et al. 1998; Gellman 1998), de novo design has been carried out for three-stranded antiparallel β-sheets that fold in water (Kortemme et al. 1998; Schenck and Gellman 1998; De Alba et al. 1999) and in organic solvents (Das et al. 1998; Sharman and Searle 1998), and a four-stranded β-sheet has been reported to be stable in MeOH and 50% MeOH/H2O (Das et al. 2000). Moreover, a three-stranded antiparallel β-sheet protein was constructed by redesign of naturally occurring toxin hand motifs (Ottesen and Imperiali 2001), and three-stranded antiparallel β-sheets derived from the WW domain motif have been studied extensively (Koepf et al. 1999; Jiang et al. 2001).
Here we report the characterization and structural analysis of the first water-soluble designed four-stranded antiparallel β-sheet, called BetaCore, that has properties approaching those of native proteins. Because of the importance of β-sheets in protein dimerization, protein-protein recognition, and protein aggregation (Maitra and Nowick 2000), including a likely role in the progression of neurodegenerative diseases (e.g., Alzheimer's, Creutzfeldt-Jakob, and Huntington's), this work may be of interest beyond the initial protein design emphasis.
Results and Discussion
Protein design
We recently proposed a novel strategy to identify domains favoring native-like secondary structure, and developed a relevant experimental model system based on bovine pancreatic trypsin inhibitor (BPTI; 58 residues and three disulfides) (Carulla et al. 2000). Protein `core elements,' that is, stretches of residues containing the slowest exchanging NH protons in the native protein (Li and Woodward 1999), are identified, and combined into a single polypeptide, termed `core module' (CM), composed of these core elements (Woodward et al. 2001). We expect that by incorporating into a core module a cross-link, for example, a disulfide bridge, more extended and flexible (high entropy) conformations will be excluded while more collapsed, native-like core structure will be favored in the ensemble of still-accessible conformations. Accordingly, the 25-residue spanning disulfide-cross-linked `oxidized core module,' patterned on BPTI residues 14–38, favors native-like β-sheet structure in rapid equilibrium with populations of nonnative β-sheet (Carulla et al. 2000); similar equilibria but with much smaller populations are observed with the corresponding `reduced core module' (Carulla et al. 2000).
We hypothesize further that covalent linkage of two CM units will enhance self-association and mutual stabilization (Carulla et al. 2000; Woodward et al. 2001), and result in an ensemble that favors more compact conformations resembling a protein native state. A dimeric form of BPTI exists in solution and undergoes rapid association–dissociation (Gallagher and Woodward 1989; Ilyina et al. 1997), and molecular modeling studies on dimeric BPTI (assumed to be antiparallel) suggest that residues at the hydrophobic monomer-monomer interface are primarily in CM (Zielenkiewicz et al. 1991). Using oxime-forming ligation chemistry to create covalent CM dimers, six candidates each of ∼50 residues were produced with variations in position and length of the cross-link (Carulla et al. 2001). One-dimensional proton (1D 1H) NMR at pH 2 was used as a qualitative criterion to gauge ordered structure in the conformational ensembles of each CM dimer, and the candidate with the narrowest line widths and highest degree of chemical shift dispersion, that is, BetaCore, was chosen for the more detailed investigations reported herein.
Note that although the CM building blocks of BetaCore have identical residues except for the ones in the cross-link, combining two CMs results in a formal loss of symmetry. In CM I, Arg17 is replaced to form the cross-link; in CM II, Leu29 is replaced. Previous synthetic protocols (Carulla et al. 2001) were readily adapted to prepare CM I and CM II, and the resultant BetaCore constructs, with site-specific 15N labels (in individual CMs or in both), hence facilitating heteronuclear NMR experiments.
BetaCore is monomeric at pH 3 in the absence of salt
Since most β-sheet forming peptides have a strong tendency to aggregate, it is imperative to confirm that BetaCore is monomeric under conditions of the NMR structural studies. Size-exclusion chromatography (SEC) at 3°C with 0.3 mM protein in CH3CN–10 mM aqueous NaCl (1:19), pH 2, shows an elution volume consistent with monomeric molecular weight (calcd 5919.9 Da) and the absence of noncovalent associations (Carulla et al. 2001). Elution profiles were calibrated with standards of known size; salt and organic solvent were included to suppress, respectively, ionic and hydrophobic interactions between protein and cross-linked agarose/dextran column material.
A clearer gauge of aggregation states was provided by analytical ultracentrifuge sedimentation equilibrium experiments; these were conducted at 5°C, with protein loading concentrations ranging from 2.5 μM to 0.05 mM in water at pH 3, and two rotor speeds. Data analysis gave average molecular weights (i.e., ∼4468 Da, with considerable scatter) somewhat lower than the molecular weight calculated from the monomeric amino acid sequence; this is a commonly observed result attributable to the opposition of protein electrostatic effects (note that at pH 3, BetaCore has seven positive charges) and ultracentrifugal forces (Schenck and Gellman 1998; De Alba et al. 1999). Therefore, a second virial coefficient, B, was included to account for nonideality effects, resulting in a good global fit of the data to a single species of average mass (Mav) 5878 Da [95% confidence 5800 to 5945 Da]. These results are consistent with the monomer mass and the absence of significant self-association over the concentration range examined. The fit value of B is 0.691 mL/mg [95% confidence 0.651 to 0.725], consistent with strong repulsion.
Given the plan to characterize BetaCore by NMR at concentrations higher than those used in the sedimentation analysis, 1D 1H NMR spectra were recorded in water at pH 3 and 5°C for samples at 0.05 mM and 0.4 mM. The observation that line widths as well as chemical shifts are indistinguishable is good evidence that the protein is monomeric into the 0.4 mM concentration range.
One means to suppress nonideal sedimentation behavior is to add salt, for example, 20 mM NaCl. Under these conditions at pH 3, the average molecular weights [i.e., ∼7400 Da, with a range from ∼5500 to ∼9000 depending on concentration] were higher than the calculated monomeric weight. The data are best fit to a monomer-dimer equilibrium, dissociation constant 87 μM, with the monomer Mav of 5911 Da [95% confidence from 5731 to 6033 Da]; any higher stoichiometries (trimer, tetramer, etc.) do not give reasonable fits or estimated monomer masses. Inclusion of a second virial coefficient in the monomer-dimer fit does not provide significant improvement and returns a negative (attractive) value for B.
Based on these controls, the conditions used for NMR experiments are 0.4 mM in water at pH 3 with no added salt. A pH of 3 was chosen because line widths and chemical shift dispersions are optimal (with respect to higher pH), solubility is acceptable, and amide proton-solvent exchange is near the minimum rate.
Assignments of folded conformations of BetaCore
BetaCore samples were prepared containing 15N labels at selected positions in either CM I or CM II (see Fig. 1 ▶ and accompanying text for nomenclature). In 15N HSQC spectra recorded at 1°C, the number of peaks is equal to the number of labeled residues in each molecule, with chemical shifts well resolved from the random coil envelope (Fig. 2a ▶). Moreover, the ten peaks from the BetaCore sample in which CM I is labeled are all different from the nine peaks from the sample in which CM II is labeled at the same sequence positions (note that CM II has a cross-link in place of Leu at position 29). This result clearly indicates asymmetry between the two core modules comprising BetaCore, which can be correlated directly to the presence of the cross-link, and suggests that the observed peaks correspond to folded (`f') conformations.
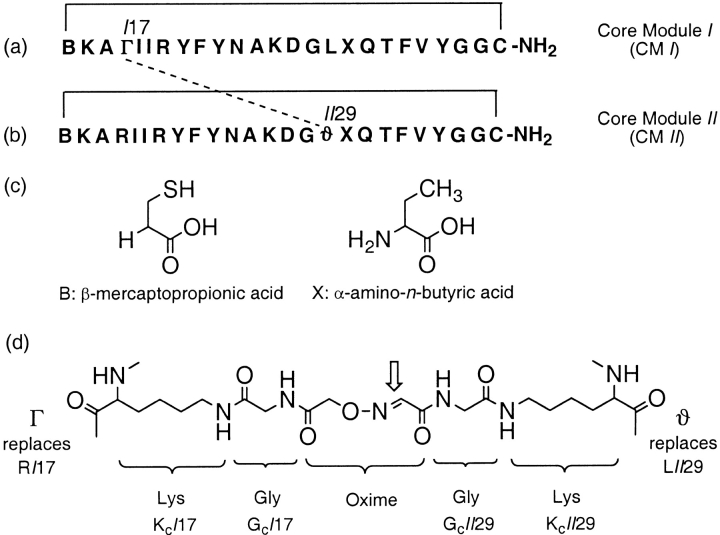
Fig. 1.
Chemical construction of BetaCore, based on an `oxidized core module' (CM) structure (Carulla et al. 2000). (a) Amino acid sequence of CM I, in which Arg17 of CM is formally replaced by residue Γ, which consists of Lys17 the side chain of which has been extended with a Gly and an (aminooxy)acetyl moiety. (b) Sequence of CM II, in which Leu29 of CM is formally replaced by residue ϑ, which consists of Lys29 the side chain of which has been extended with Gly and Ser, with subsequent NaIO4 oxidation providing a glyoxylyl moiety. (c) Chemical structures of B, β-mercaptopropionic acid, and X, α-amino-n-butyric acid, which are incorporated into the CMs. (d) Detailed chemical structure of final cross-link, formed at position indicated by thick arrow. The dotted line between Γ in (a) and ϑ in (b) also depicts the cross-link, and the solid lines in both (a) and (b) each indicate spanning disulfide bridges in CM. Residues are designated by the same numbering used for the corresponding residues in native BPTI; for example, the N-terminal Mpa and C-terminal Cys are respectively 14 and 38. A `I' or `II' before the residue number indicates the appropriate one of the two asymmetric core modules. Assemblies of CM I and CM II by solid-phase methods, cleavage from the support, disulfide creation, and the formation of the oxime bond to connect them to each other have been described (Carulla et al. 2001). Designations and abbreviations of CMs and BetaCore focus on the specific polypeptides of the present studies, and supersede the necessarily more detailed and general nomenclature of two earlier works in which additional molecules were evaluated (Carulla et al. 2000, 2001).
Fig. 2.
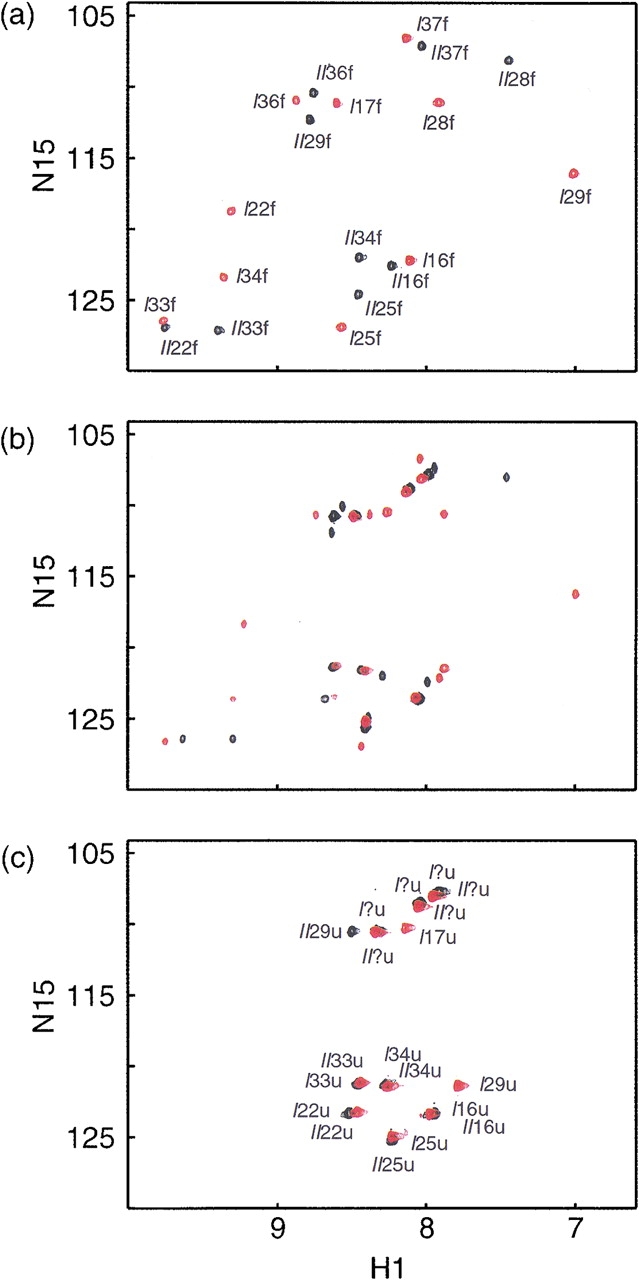
BetaCore 15N-1H HSQC spectra taken at 800 MHz in H2O:D2O (9:1 by vol) at pH 3 and (a) 1°C, (b) 35°C, and (c) 55°C. In one series of experiments, ten positions of CM I were 15N-labeled (cross-peaks shown in red), and in the other series, nine positions of CM II were 15N-labeled (cross-peaks shown in black). Assignments were carried out in other experiments (see text); each cross-peak in parts (a) and (c) in this Figure is designated by CM (I or II)-residue number (between 14 and 38)-conformation (`f' or `u'), except for six glycine residues in the `u' conformation for which the position in the sequence could not be determined unambiguously. Cross-peaks in part (b) of this Figure are a superposition of those assigned in parts (a) and (c).
TOCSY spectra of unlabeled BetaCore were obtained at pH 3, and 5° or 15°C. At 5°C, many of the peaks in the NH–CαH region are broad, making them difficult to define unambiguously. Also, very few NH-side chain cross-peaks are observed, precluding identification of spin systems. However, when the temperature is raised to 15°C, the peaks sharpen substantially, and it is possible to make sequential assignments (Wüthrich 1986) by combined analyses of homonuclear TOCSY, NOESY, and heteronuclear 15N-1H HSQC-TOCSY and 15N-1H HSQC-NOESY spectra. Assignments of all backbone and ∼95% of side chain protons of folded (`f') conformations were facilitated by use of the selectively 15N-labeled BetaCore proteins in heteronuclear experiments. Additional cross-peaks are observed at 15°C beyond those assigned; all of these are found within the random coil envelope and are likely associated with unfolded (`u') conformations, as discussed in detail below.
Folded and unfolded conformations of BetaCore are in slow exchange
15N HSQC spectra of labeled BetaCore molecules were recorded at different temperatures (1, 5, 15, 25, 35, 45, and 55°C, and back to 1°C). As already indicated, the number of peaks at 1°C matches exactly the number of labeled residues, and all can be assigned in the clearly asymmetric cross-linked CM dimer (Fig. 2a ▶). As temperature is increased, a new set of peaks with random coil chemical shifts start to appear, and at 35°C, the number of peaks is exactly double the number of labeled residues (Fig. 2b ▶). At 55°C, the number of peaks again matches the number of labeled residues; all of the peaks observed at low temperature are absent and the only peaks now present are those that grew in with increasing temperature (Fig. 2c ▶). When a sample evaluated at 55°C is returned to 1°C, the original low temperature spectrum (Fig. 2a ▶) is observed.
These data are consistent with fully reversible transitions from folded (`f', at 1°C) to unfolded (`u', at 55°C) conformations. Assignments of backbone and side chains of labeled residues for the unfolded protein were made based on heteronuclear 15N-1H HSQC-TOCSY and 15N-1H HSQC-NOESY. The important conclusion for the 19 residues carrying an 15N-label is that not only are the chemical shifts observed at 55°C all in the random coil region, but also the sequence symmetry of the CM covalent dimer is observed in the thermally denatured form. That is, identical residues at the same sequence positions in CM I and CM II do not have the same chemical shifts in folded BetaCore at 1°C, but the shifts are changed and overlapping when BetaCore is unfolded at 55°C.
When two protein conformations interconvert slowly on the NMR time scale (in the millisecond range), each gives rise to a separate peak corresponding to that conformation (Roberts 1993). The fact that throughout the unfolding transition each resonance is represented by two different peaks, one with a chemical shift well resolved and away from the random coil envelope, and the other with a random coil chemical shift, implies that BetaCore is in slow conformational exchange on the NMR time scale between folded and unfolded forms. Indeed, interconversion between these two conformations is much longer than milliseconds, as supported by the absence of exchange cross-peaks (Falzone et al. 1991) under a variety of experimental conditions (see Materials and Methods). As a consequence, full assignments of unfolded conformations have not been possible.
Sequential NOEs and coupling constants imply β-sheet structure
In the dominant folded `f' conformation of BetaCore, for all residues designed to be in a strand, interresidue NHi–NHi+1 NOE cross-peaks are very weak or entirely absent, whereas CαHi–NHi+1 NOE cross-peaks are intense (Fig. 3 ▶). This pattern of sequential NOE connectivities implies β-sheet structure, but does not impart information about strand alignment.
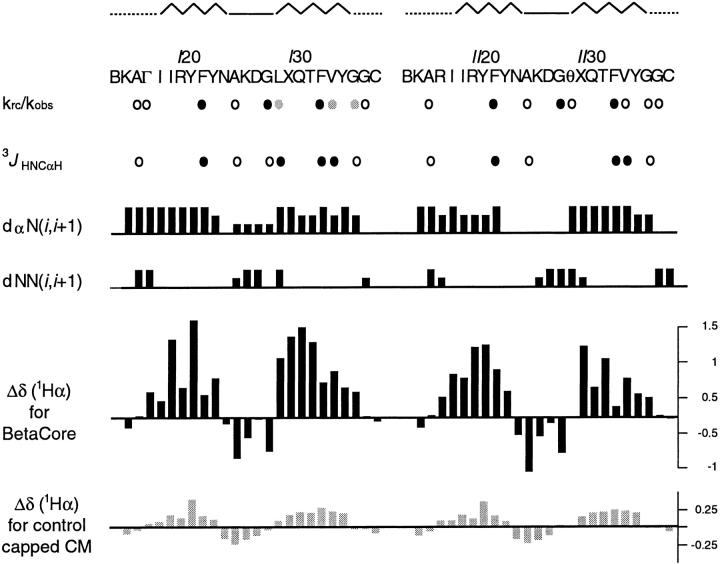
Fig. 3.
NMR-reported secondary structure of folded BetaCore. At the top, the sequence location of β-strands (zig-zag), β-turns (solid), and loops (dotted) are indicated followed by the amino acid sequences in CM I and CM II. Next, krc/kobs observed for 15N-labeled sites of BetaCore at 5°C and pH 3. Filled black circles identify residues highly protected from exchange, filled gray circles identify moderately protected residues, and open circles identify residues that exchange relatively fast. The krc/kobs reported for residues ΓI17 and ϑII29 refer to the Gly that are part of the cross-link. Next, 3JHNCαH observed for 15N-labeled sites of BetaCore at 15°C and pH 3. Filled black circles identify residues with 3JHNCαH > 8 Hz, indicative of residues in extended chain conformation, and open circles identify residues with 3JHNCαH < 6 Hz, indicative of local α-type conformation. Next, summary of NOE sequential connectivities observed for BetaCore at 15°C and pH 3. The height of each line reflects the intensity of the NOE, that is, weak, medium, and strong. Next, plot of chemical shift deviations of CαH protons as a function of CM and residue for BetaCore (black bars), and capped CM I and CM II (gray bars). Capped CMs refer to the building blocks of BetaCore, which instead of being ligated to form BetaCore, have been converted with the appropriate low-molecular weight compound (i.e., acetone with CM I, CH3ONH2 with CM II) to create an oxime (Carulla et al. 2001).
A sample of BetaCore in which ten residues in CM I and nine residues in CM II are labeled with 15N was used to measure vicinal coupling constants 3JHNCαH representative of different regions. All coupling constants for potential strand residues are greater than 8 Hz (the value for FII33 is > 9 Hz), consistent with φ ∼ −120° as is characteristic of β-sheet structure (Fig. 3 ▶). In contrast, values for potential turn and loop residues are smaller than 6 Hz (Fig. 3 ▶). In particular, the 3JHNCαH for AI25 and AII25 are 4.4 and 5.1 Hz, respectively, in agreement with the 3JHNCαH of 4.1 Hz for type I β-turn residue A25 in native BPTI.
Long-range NOEs indicate that BetaCore is a four-stranded β-sheet
Long-range NOEs generally provide the most conclusive evidence for tertiary structure in solution. The NOE cross-peaks characteristic of antiparallel β-sheet are those involving CαHi–CαHj, CαHi–NHj+1, and NHi+1–NHj−1 protons, where i and j are residues that face each other in adjacent β-strands and the CαH protons point towards the interior. The shortest distances, and correspondingly, the most intense NOEs, are between CαHi–CαHj protons; these are the main diagnostic for a β-sheet (Wüthrich 1986). Unambiguously identified CαH–CαH NOEs between residues in the same CM, i.e., YI23–XI30, YI21–TI32, II19–VI34, RI17–GI36, YII23–XII30, YII21–TII32, III19–VII34, and RII17–GII36 (Fig. 4 ▶, thick arrows), are the same as in native BPTI (Wagner et al. 1987), providing compelling evidence that each CM unit in BetaCore samples native-like 4:4 β-hairpin structure. No NOEs in spectra of BetaCore are consistent with nonnative β-sheets. This is in contrast to the `oxidized core module' by itself, which is an equilibrium ensemble of conformations among which a major population is similar to the native-like 4:4 β-hairpin and a minor population approximates 3:5 β-hairpins (Carulla et al. 2000).
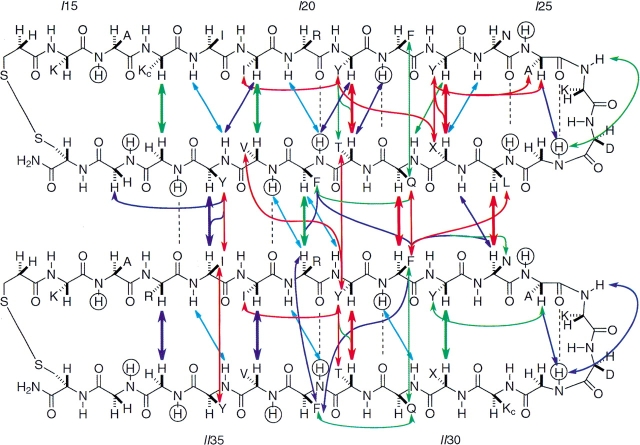
Fig. 4.
Schematic representation of long-range CαH–CαH, NH–CαH, and aromatic-aliphatic NOEs for BetaCore, which is drawn to indicate the proposed four-stranded antiparallel β-sheet. The color of the arrow indicates NOE strength: strong = red; medium = green; weak = purple; very weak = light blue. CαH–CαH NOEs are shown as thicker arrows. Amide protons evaluated in H/D exchange experiments (krc/kobs in Fig. 3 ▶) with 15N-labeled BetaCore are circled; for those showing significant protection, the corresponding deduced hydrogen bonds are indicated by dashed lines. One-letter abbreviations indicate amino acid residue side chains, with Kc at positions 17 of CM I and 29 of CM II being lysines that are elaborated further to create the oxime cross-link (cf. Fig. 1 ▶). In calculated structures of BetaCore (Fig. 5b ▶), the cross-link is primarily but not exclusively beneath the plane shown in the present Figure.
In addition, unambiguously identified CαH–CαH NOEs are observed between residues in different CMs comprising BetaCore. These NOEs: LI29–NII24, QI31–FII22, F133–RII20, and YI35–III18 (Fig. 4 ▶, thick arrows) are all between the second strand (C-terminal half) of CM I and the first strand (N-terminal half) of CM II; importantly, no corresponding NOEs between the second strand of CM II and the first strand of CM I are observed. The data are accommodated by positioning the two CMs in a four-stranded antiparallel β-sheet (Fig. 4 ▶). Although the strands themselves are antiparallel, note how the two CM units are aligned laterally and parallel to each other. The cross-link extends from a position near the N-terminal of the first strand of CM I to a position past the turn on the second strand of CM II.
A significant number of long-range contacts are observed between side chains of residues on adjacent strands within the same CM (e.g., YI23–XI30, YI21–TI32, and YII21–TII32; a total of 21 NOEs) or between different CMs (e.g., QI31–FII22, YI35–III18; a total of 15 NOEs) comprising BetaCore (Fig. 4 ▶); these are all consistent with the proposed four-stranded antiparallel β-sheet. In addition, a number of i, i+2 NOEs are observed between side chains (e.g., YI23–AI25, II19–YI21, YII23–AII25, and III19–YII21); these are characteristic of extended conformations (Fig. 4 ▶).
Chemical shift data corroborate BetaCore four-stranded β-sheet folded conformations
Chemical shift data for α-protons (δHα) provide further evidence that BetaCore adopts four-stranded antiparallel β-sheet conformations in aqueous solution. Secondary structure profoundly affects δHα, with β-sheet protons shifted downfield and turn protons shifted upfield relative to expected random coil values (Ösapay and Case 1994). The values of ΔδHα = δHα observed − δHα random coil (Merutka et al. 1995; Wishart et al. 1995; Andersen et al. 1997) for BetaCore at pH 3 and 15°C, plotted as a function of residue position (Fig. 3 ▶), can be compared to those reported previously (Carulla et al. 2001) for the corresponding single (unlinked) CM units. Overall shapes of these graphs follow expectations from the native-like BPTI β-sheet, that is, downfield shifts for residues 18–24 and 29–35 and upfield shifts for residues 25–28. The absolute values of the deviations from random coil values are much higher for residues from BetaCore (e.g., YI21 has ΔδHα = 1.38) than for corresponding residues in CM units (e.g., YI21 has ΔδHα = 0.38), pointing to considerable stabilization of structure associated with combining two CMs through a covalent cross-link. In comparison, the maximum ΔδHα in the β-sheet core of native BPTI, recorded at 68°C, is ∼1.15 (Wagner and Wüthrich 1982).
Analysis of the fine structure of the chemical shift deviation profile (Fig. 3 ▶) provides additional insights. Note the essential mirror symmetry with respect to the two core modules that form BetaCore: zig-zag shapes are observed for the first strand of CM I and the second strand of CM II, and smoother shapes are observed for the second strand of CM I and the first strand of CM II. Theoretical calculations (Ösapay and Case 1994) provide a precedent for the zig-zag shape matching the hydrogen-bonding pattern in simple β-hairpins; thus, the alternating residues involved in cross-strand contacts (e.g., residues II19, YI21, YI23, XII30, TII32, and VII34 in Fig. 4 ▶) have a more positive δHα proton shift than residues having no interactions with a neighboring strand (e.g., residues II18, RI20, FI22, QII31, FII33, and YII35 in Fig. 4 ▶). Such two-fold i, i+1 periodicity is also found in several native proteins, including tendamistant, plastocyanin, and interleukin 1β. However, when a strand is located in the middle of a β-sheet, each consecutive residue is involved in a contact with one or the other of the two neighboring strands, so the aforementioned i, i+1 periodicity is replaced by a smoother profile (i.e., as actually observed for the second and first strands of CM I and CM II, respectively, in the BetaCore protein; see Figs. 3 and 4 ▶ ▶). All of these data are consistent with the proposed four-stranded antiparallel β-sheet conformation.
The chemical shift deviation profile (Fig. 3 ▶) can even be used to pinpoint the β-turn and distinguish among possible types. Turn residues are calculated and found (Ösapay and Case 1994) to always show a negative ΔδHα, with the sole exception that the chemical shift of position 3 of a type I β-turn is expected to be very close to the random-coil value. Indeed, this is found for both turns of BetaCore (i.e., focus on DI27 and DII27) and agrees with the type I β-turn found in native BPTI.
H/D exchange in BetaCore supports compact folded structure
Hydrogen isotope exchange experiments were conducted at pH 3 and 5°C, recording 2D 15N HSQC spectra as a function of time on a sample of BetaCore in which ten residues in CM I and nine residues in CM II are labeled with 15N. Exchange rate constants (kobs) for all 19 15N-bound amide hydrogens were measured; comparisons to the rates expected in a random coil (krc) calculated based on the pH, temperature, residue, and N-side neighboring residue (Bai et al. 1993) gives protection factors (Fig. 3 ▶). The observed protection factors in BetaCore are at least an order of magnitude higher than those of corresponding residues in the `oxidized core module' (Carulla et al. 2000), and of the same order of magnitude as reported for a partially folded protein such as [14–38]Abu (Barbar et al. 1995).
The exchange pattern in BetaCore is in good agreement with predictions from the four-stranded antiparallel β-sheet model (Fig. 4 ▶). Highly protected residues (i.e., FI22, FII22, FI33, and FII33; krc/kobs = 30–75) engage in hydrogen bonds in the middles of the intramodule strands (two each for CM I and CM II), as well as the signature hydrogen bond of the native-like BPTI type I β-turn (i.e., GI28 and GII28; krc/kobs 100 and 190, respectively). Moderately protected residues (i.e., LI29, VI34, and GI36; krc/kobs = 17–20) are those that engage in intrastrand hydrogen bonds, one within CM I and two between CM I and CM II. Residues that exchange relatively rapidly (i.e., AI16, AII16, GcI17, AI25, AII25, GcII29, VII34, GII36, GI37, and GII37; krc/kobs = 3–10) include representatives from loop and turn regions, the cross-link, and a solvent-exposed side of β-strand, none of which are expected to be involved in hydrogen bonding.
Calculation of the family of BetaCore folded structures
Compatibility of the proposed four-stranded antiparallel β-sheet with all experimentally observed NOE distance constraints, those dihedral angles supported by 3JHNCαH measurements, and the hydrogen bonds deduced from H/D exchange studies, was checked by calculating a three-dimensional model structure (Fig. 5 ▶, Table 1). The most ordered backbone regions are the four strands; there is some flexibility in the turn regions, and the disulfide-bridged loop portions are considerably disordered (Fig. 5a ▶). The most disordered section of BetaCore is the essential long cross-link, as best appreciated by viewing the construct from the side (Fig. 5b ▶). Thus, the cross-link can traverse either face of the four-stranded sheet, with a statistical preference (15 of the best 20 calculated structures) for the face defined by the side chains of YI21, YI23, and YII21. Further structural definition of the cross-link is precluded by the absence of NOEs involving other parts of the molecule. Position ϑII29, which anchors the cross-link onto CM II, has a ΔδHα suggestive of random coil conformation (Fig. 3 ▶), and perturbs neighboring residues comprising the CM II turn so that it is somewhat less defined than the corresponding turn in CM I (Figs. 4 and 5a ▶ ▶). The structural calculations are not sufficiently resolved to determine whether the BPTI native-like `twist' is reproduced in β-hairpins of either CM I or CM II.
Fig. 5.
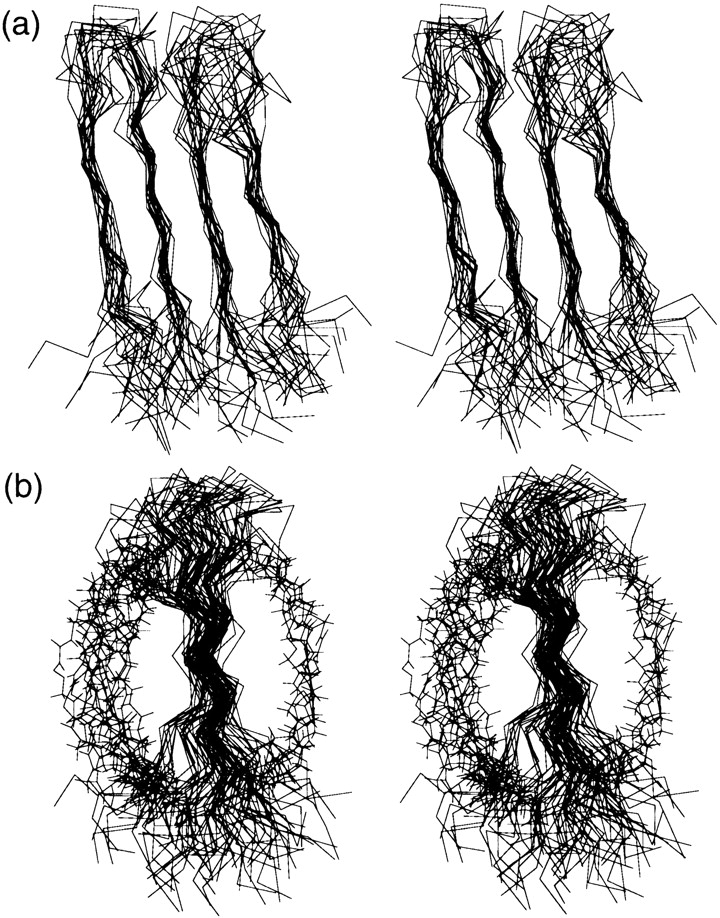
Stereoviews of (a) backbone and (b) backbone plus cross-link superposition of NMR-derived structural ensemble for BetaCore. The 20 best calculated structures are shown. In (a), CM I is on the left and CM II is on the right, the turns are at the top, and the disulfide-spanned loops are at the bottom. In (b), CM I is in front of CM II, the turns are at the top, and the disulfide-spanned loops are at the bottom. The cross-links all start from the loop area of CM I (lower front) and end in the turn region of CM II (upper rear); the majority of them (15 of 20 structures) are on the left, with the remainder (5 of 20 structures) on the right.
Table 1.
Statistics for NMR calculations
| No. of residues | 50 |
| No. of distance constraints | |
| NOEs | |
| Intraresidue | 33 |
| Sequential | 57 |
| i, i + 2 | 16 |
| Cross-strand intermodule | 23 |
| Cross-strand intramodule | 43 |
| Total | 172 |
| No. of dihedral angle constraints | 14 |
| No. of hydrogen bond constraints | 9 |
| Ramachandran analysis | |
| Most favored regions | 49.3% |
| Additional allowed regions | 43.1% |
| Generously allowed regions | 6.2% |
| Disallowed regions | 1.4%a |
| Rmsd deviations (Å) for | |
| All residues (I14-I38, II14-II38) | |
| Backbone atoms | 2.9 |
| Heavy atoms | 3.8 |
| Without termini (I18-I35, II18-II35) | |
| Backbone atoms | 1.9 |
| Heavy atoms | 3.1 |
| β-Strands only (I18-I24, I29-I35, II18-II24, II29-II35) | |
| Backbone atoms | 1.5 |
| Heavy atoms | 2.8 |
| β-Strands only (I18-I23, I30-I35, II18-II23, II30-II35) | |
| Backbone atoms | 1.4 |
| Heavy atoms | 2.3 |
a No NOE constraints were found for residues in these regions.
Beyond the key, highly specific intra- and intermodule backbone interactions that define the four-stranded antiparallel β-sheet conformation, and regardless of which face is traversed by the long cross-link, the most organized regions of the BetaCore structure are stabilized by packing of predominantly hydrophobic side chains on both faces. The most important interactions on one face involve the side chains of YI21, YI23, XI30, TI32, YII21, and TII32, while the other face features interactions of FI22, QI31, FI33, YI35, FII22, QII31, FII33, and YII35. These patterns reflect the parallel arrangement of the two CMs: two intramodule cross-strand A–B interactions (in CM I and CM II) are always in register with a corresponding intermodule cross-strand B–A interaction (between CM I and CM II). Moreover, when intramodule A–B are a backbone-hydrogen bonded pair, the intermodule B–A are a nonhydrogen-bonded pair, and vice versa (Wouters and Curmi 1995).
Unfolding transitions of BetaCore
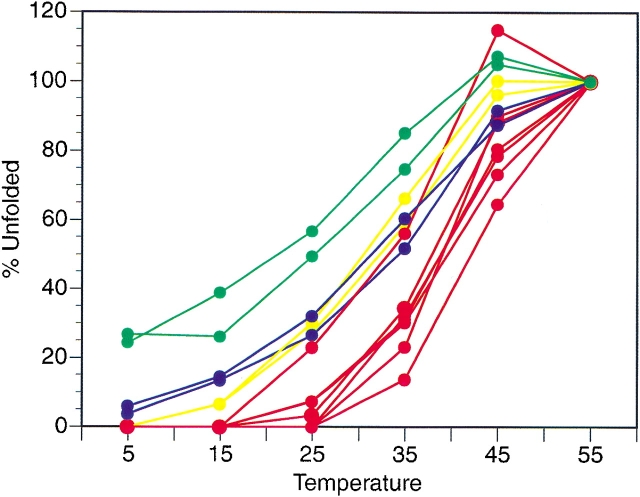
15N HSQC spectra of BetaCore provide evidence for folded (`f') and unfolded (`u') conformations (Fig. 2 ▶). Reversible thermal unfolding is monitored by plotting, as a function of temperature, the volume of each unfolded cross-peak, referenced to the volume of the peak for the same residue at 55°C where the protein is completely unfolded (Fig. 6 ▶). Although for each probe examined the curve is roughly sigmoidal, indicating some cooperativity, the transition midpoints (Tm) vary considerably and hence unfolding is non-two-state. Residues in the strands (i.e., FI22, LI29, FI33, VI34, and FII33) have a Tm ≈ 39°C, except for FII22 which has Tm ≈ 42°C and VII34 which has Tm ≈ 33°C (VII34 is not involved in interstrand hydrogen bonding). Residues in the loops (i.e., AI16 and AII16) and residues in the turn (i.e., AI25 and AII25) have a Tm ≈ 33°C. Glycines in the cross-link have a Tm ≈ 25°C; these are the only residues with significant levels of `u' at 5°C. For all of these residues, corresponding curves showing decreasing `f', rather than increasing `u', give qualitatively similar results, but Tm estimations are not possible due to ambiguity in the folded baselines.
Fig. 6.
Appearance of unfolded (`u') conformations of BetaCore reported in 15N HSQC experiments as a function of temperature. Residues located in the proposed β-sheet strands are shown in red, those in the β-turn are yellow, those in the loops are blue, and those in the cross-link are green; residue identification and apparent Tm are covered in text. These plots are not made for GI36, GI37, GI28, GII36, GII37, and GII28 because assignments of their unfolded (`u') conformations are ambiguous.
Other aspects of the thermal unfolding also indicate non-two-state behavior. First, for any given residue, the total 15N HSQC volume of `f' plus `u' cross-peaks at 5°C is smaller than the volume of `u' at 55°C. This ratio of volumes ranges from ∼0.8 for the Gly residues that are part of the cross-link, to ∼0.5 for loop residues, to ∼0.25–0.5 for residues in the strands and turns. Second, the absolute values of cross-peak volumes of `u' peaks at 55°C vary over a twofold range, not correlated in any obvious way to the nature or environment of the residue. Third, a subset of the residues examined (i.e., FI33, FII22, FI22, LI29, GI36, and AI25) show a 10%–30% increase in the 15N HSQC volume of `f' as the temperature is raised from 5° to 15°C. Fourth, spectra of BetaCore show line width broadening for some but not all peaks. The line widths of the `f' conformation at 5°C range from ∼15 Hz for cross-link Gly residues, to 16–22 Hz for loop residues, to ∼23–34 Hz for strand or turn residues; all become sharper as temperature is increased. The line widths of the `u' conformation at 35°C range from ∼16 Hz for cross-link Gly residues, to 12–20 Hz for loop and turn residues, to ∼24–34 Hz for residues in the strands; again, all sharpen at 55°C. Line broadening and volume reduction reflect the presence of multiple conformations that have different chemical shifts for the same nucleus and interconvert on an intermediate NMR time scale (millisecond to microsecond range) (Roberts 1993). Applying these ideas to BetaCore, we conclude that the ensemble structure at low temperature consists of a dominant, folded conformation in equilibrium with minor conformations that may be exchange broadened and/or sparsely populated. Thermal unfolding of the dominant family of conformations is global, and non-two-state. Different parts of four-stranded BetaCore become random coil-like at different temperatures, and/or the `u' ensemble varies with temperature.
Role of cross-link in structure and stability of BetaCore
The components of BetaCore are two units of a CM that, by itself, is monomeric and favors native-like β-sheet structure (Carulla et al. 2000). When combined covalently by a reasonably optimized long oxime cross-link, each constituent CM is individually stabilized toward native-like conformation (while minor nonnative conformations are eliminated), and new highly specific intermodule interactions emerge leading to a monomeric collapsed structure with substantially enhanced global stability. The primary role of the cross-link is to make the system unimolecular, thereby increasing the probability of packing interactions between two CMs. The cross-link itself is very flexible, as evidenced by the absence of NOEs, as well as the rapid H/D exchange and low Tm values of reporter residues; it is not in the vicinity of stable, organized structure. Furthermore, the cross-link used here has polar moieties which are likely to facilitate the overall water solubility of BetaCore, in contrast to conceivable more lipophilic cross-linkers that might nucleate aggregation.
The length and flexibility of the cross-link has additional advantages in this system: it allows the two CMs to sample various relative orientations and does not interfere with essential packing. The nonsymmetrical CM–CM association actually accessed in BetaCore features a four-stranded antiparallel β-sheet, where the cross-link connects points on outside strands (i.e., the N-terminal strand of CM I and the C-terminal strand of CM II). Several CM covalent dimers with shorter and/or differently positioned cross-links that were synthesized and evaluated in pilot work gave no or less indication for compact structure (Carulla et al. 2001), findings that in view of the present knowledge of BetaCore structure become plausible. Relatedly, the observed preferred structure, featuring its antiparallel and asymmetric arrangement of chains corresponding to parallel alignment of two CMs, differs from symmetric, antiparallel CM dimer interfaces anticipated in earlier stages of the design process.
Further insights can be gleaned from simulated annealing calculations showing that the cross-link can be accommodated on either face of the β-sheet structure of BetaCore. This is consistent with the idea that the cross-link used here has no direct role in organizing structure beyond its effect to combine two CMs into a single molecule. It raises new questions about what other cross-link motifs (in terms of structure, length, and positioning) could lead to similar or even enhanced formation of and stabilization of collapsed structure. For example, the C-terminal strand of CM I and the N-terminal strand of CM II could be connected by a rather short cross-link, compatible with the observed four-stranded antiparallel β-sheet geometry (proximal effect). Alternatively, it may be possible to develop a long transverse cross-linker (distal effect) that packs in a more specific way to a given face of β-sheet structure, hence mimicking themes found in naturally occurring proteins such as ubiquitin and the immunoglobulin binding domains of proteins G and L.
Significance of BetaCore
Cross-linking core modules can indeed lead to their mutual stabilization to native-like conformations that are significantly more stable than other accessible conformations. BetaCore achieves a four-stranded antiparallel β-sheet conformation, as supported by well-dispersed chemical shifts, i, i+1 periodicity, numerous long-range NOEs, and slowed amide hydrogen isotope exchange patterns. This conformation reinforces features expected from the structure of the BPTI source on which the CMs are based, but also shows significant interactions not found in BPTI. To the best of our knowledge, this is the first report that a designed protein adopts a four-stranded antiparallel β-sheet conformation in water.
Folded BetaCore undergoes reversible, global, moderately cooperative, non-two-state thermal transitions to an equilibrium ensemble of unfolded `u' conformations. Folded and unfolded interconvert slowly on the NMR time scale, indicating a significant energy barrier between them. Thus, BetaCore has properties that compare favorably to those of some of the more successful designed proteins in the literature, and approach those of native proteins.
The BetaCore system, as introduced in this paper, provides ample possibilities for further optimization, including the length/positioning of the cross-link and packing of side chains. The roles of individual residues and residue pairs from the strands and turns in affecting the formation and stabilities of β-sheets, and in controlling the balance between intramolecular interactions leading to collapsed monomeric structure and corresponding intermolecular interactions leading to aggregation, can be evaluated systematically. With respect to the latter, it is possible that the current long transverse cross-link helps provide steric barriers to intermolecular aggregation.
Another research direction suggested by the present work involves identification and synthesis of further core modules with known conformational preferences (α-helix as well as β-hairpins and sheets)—derived from proteins for which structural and H/D exchange data are available—and then combining these covalently through use of appropriate cross-links to test the hypothesis that more stable, compact, folded protein structures will result.
Materials and methods
Chemical synthesis
Unlabeled and 15N-selectively labeled samples of BetaCore were prepared by automated Fmoc solid-phase peptide synthesis according to methods described previously (Carulla et al. 2001). Three 15N-selectively labeled variants of BetaCore were prepared: 15NI–II has ten labels in CM I, I–15NII has nine labels in CM II, and 15NI–15NII has all 19 labels, ten in CM I and nine in CM II. 15N is incorporated in the prospective loops (AI16, AII16, GI36, GII36, GI37, and GII37), turns (AI25, AII25, GI28, and GII28), cross-link (GcI17 and GcII29), and antiparallel β-sheets (FI22, FII22, LI29, FI33, FII33, VI34, and VII34). Purified proteins were evaluated by analytical C-4 reversed-phase HPLC (>95%), and amino acid analyses (agreement with theoretical values) and ESMS: for I–II, calcd average mass: 5919.92, found: 5920.31 ± 0.48; for 15NI–II, calcd average mass: 5929.99, found: 5930.10 ± 0.37; for I–15NII, calcd average mass: 5928.99, found: 5928.21 ± 1.02; and for 15NI–15NII, calcd average mass: 5939.07, found: 5937.31 ± 1.65.
Sedimentation equilibrium analysis
Sedimentation equilibrium experiments were performed on a Beckman Optima XL-A analytical ultracentrifuge. Experiments were conducted at 5°C, with protein concentrations ranging from 2.5 μM to 0.05 mM in water at pH 3 or 20 mM NaCl at pH 3, and two rotor speeds (30,000 and 44,000 rpm). Data were analyzed by nonlinear least square techniques using the program Kdalton (Philo 2000). This program fits data to the following general equation, giving the total concentration at radial position r for a nonideal, reversible association of monomer to N-mer:
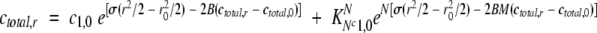 |
where r0 is a reference position, c1,0 is the monomer concentration at that reference position, B is the second virial coefficient, and KN is the association constant for formation of N-mer from monomer (Johnson et al. 1981). In this equation, σ is the reduced molecular mass given by [M(1 − νρ)ω2]/(RT), where M is the monomer mass, v is the peptide partial specific volume, ρ is the solvent density, ω is the rotor angular velocity, and T is temperature. As special cases, B is 0 for ideal solutions, and KN is 0 when the fit is to a single species. The partial specific volume of 0.7284 at 5°C was calculated with the program Sednterp (Laue et al. 1992) using the following sequence of natural amino acids as a model for BetaCore: C K A K G G I I R Y F Y N A K D G L V Q T F V Y G G C C K A R I I R Y F Y N A K D G K G G V Q T F V Y G G C. The same program calculates ρ = 0.99999 and 1.00082 g/mL respectively for samples in water and 20 mM NaCl.
NMR spectroscopy
NMR samples for Betacore were 0.4 mM at pH 3. Samples in D2O were dissolved while working in a glove bag under argon. Spectra of BetaCore were obtained in 90:10 H2O/D2O and 99.9% D2O at 5, 10, 15, 25, 35, 45, and 55°C on a Varian 600 MHz or 800 MHz Inova instrument. Spectra were acquired with the following number of complex points and spectral widths: TOCSY (τm = 65 msec) (Griesinger et al. 1988) and NOESY (τm = 200 msec) (Kumar et al. 1980), F1 (1H) 256, 9000 Hz; F2 (1H) 2048, 9000 Hz; 64 transients; 15N HSQC (Kay et al. 1992), F1 (15N) 128, 2200 Hz; F2 (1H) 1024, 9000 Hz; 16 transients; 15N-1H HSQC-TOCSY (τm = 65 msec) (Zhang et al. 1994) and 15N-1H HSQC-NOESY (Zhang et al. 1994) (τm = 200 msec), F1 (1H) 1024, 9000 Hz; F2 (1H) 96, 9000 Hz; F3 (15N) 32, 2200 Hz; 8 transients; HNHA experiments (Kuboniwa et al. 1994) F1 (1H) 796, 7000 Hz; F2 (1H) 64, 7000 Hz; F3 (15N) 32, 2200 Hz; 32 transients. Suppression of intense solvent resonances was achieved by presaturation or use of the WATERGATE sequence (Piotto et al. 1992). Data were processed and analyzed using the programs NMRPipe (Delaglio et al. 1995) and NMRView (Johnson and Blevins 1994). Data points were weighted using either a 54° or 72° shifted square sine bell in each dimension. Two-dimensional datasets were zero-filled to form 4K × 2K real matrices. Baseline corrections were applied in both dimensions. Three-dimensional datasets were zero filled to form 2K × 0.5K × 0.1K real matrices. Hydrogen isotope exchange rates were obtained by measuring peak volumes versus time in a series of two-dimensional 15N HSQC spectra at pH 3 and 5°C. Pseudo-first-order rate constants were determined from nonlinear least-squares fit of an exponential rate equation to experimental data. Thermal unfolding curves were obtained by measuring peak volumes versus temperature in a series of two-dimensional 15N HSQC spectra. A relaxation delay time of 3 sec was used to permit quantitative volume integration. Reversibility of thermal denaturation was verified by comparison of low-temperature spectra acquired before and after unfolding. Exchange cross-peaks between folded and unfolded conformations are absent under a variety of experimental conditions examined, including: TOCSY (τm = 30 msec, 50 msec, 70 msec), NOESY (τm = 150 msec, 200 msec, 400 msec), 15N-1H HMQC-TOCSY (τm = 30 msec, 50 msec, 70 msec), and 15N-1H HMQC-NOESY (τm = 150 msec, 200 msec, 400 msec) at 15, 28 and 35°C.
Structure calculation
NOE cross-peaks obtained from NOESY spectra taken at pH 3 and 15°C with 200 msec mixing time in H2O:D2O (9:1 by vol) and D2O were integrated and converted to distance constraints (strong, medium, and weak, corresponding to upper limits of 2.8, 3.4, and 5 Å. Pseudoatoms were defined for distances involving nonstereospecifically assigned protons, and upper limits were corrected appropriately. Dihedral angle constraints were deduced from 3JHNCαH coupling constants obtained from HNHA experiments. The φ angles were constrained to −120 ± 30° for residues with 3JHNCαH > 8 Hz, to −60 ± 30° for turn residues with 3JHNCαH < 5 Hz, to −60 ± 50° for loop residues with 3JHNCαH < 6 Hz, and to 65 ± 30° for GI28 in the turn (since this position in a type I β-turn favors 65 ±15° φ angle and the observed 3JHNCαH = 5.2 and 4.7 were consistent with this value). Hydrogen bonds implied by H/D exchange experiments were also input as constraints.
Structures (total = 200) of BetaCore were calculated by the program X-Plor 3.851 (Brunger 1993) on Silicon Graphics Octane workstations. The files topallhdg.pro and parallhdg.pro were modified to account for the geometries of the cross-link and the unusual amino acids B and X (Fig. 1 ▶), as described in the electronic supplemental material. Parameters for the standard simulated annealing protocol, sa.inp, were 16,000 steps of 1.5 fsec at 2000°K, followed by cooling to 300°K in 10,000 cooling steps of 1.5 fsec. The resulting structures were further refined using the protocol refine. inp a total of three times; this involved each time starting at 2000°K, and cooling to 300°K in 10,000 cooling steps of 1.5 fsec. Converged structures (total = 20) were selected on the basis of no constraint violations greater than 0.5 Å for NOEs and 5° for dihedrals, lowest total energy, and compatibility of Ramachandran plots. Structures were visualized using the program InsightII (Biosym) and analyzed with the program Procheck (Laskowski et al. 1993).
Accession numbers
The coordinates and constraint files for the ensemble of 20 structures (Fig. 5 ▶) have been deposited in the Protein Data Bank (accession code: 1K09). Chemical shifts and coupling constants have been deposited in the BioMagResBank (accession code: 5183)
Electronic supplemental material
Tables of 15N and 1H chemical shifts of BetaCore `f' conformation and of 15N-labeled residues in BetaCore `u' conformation at 15°C and pH 3. Tables of coupling constants, hydrogen exchange rate constants, and protection factors for 15N-labeled residues in BetaCore at 5°C and pH 3. Topology and parameter files for X-Plor that take into account the cross-link (lys1 and lys2), abu, and mpa. BetaCore NH–CαH region of TOCSY spectra taken at 5°C and 15°C and pH 3 and BetaCore CαH–CαH region of NOESY spectrum taken at 15°C and pH 3.
Acknowledgments
We thank Dr. Gianluigi Veglia for help in structure calculations as well as for helpful discussions, Dr. David Live for help with NMR experiments, Dr. Elisar Barbar for critical reading of the manuscript, Dr. John Philo for running the sedimentation equilibrium measurements, and Dr. Cristobal Alhambra for help with the cross-link parameterization. NMR experiments were performed at the University of Minnesota High Field NMR Laboratory. Workstation use was supported in part by the University of Minnesota Supercomputing Institute. The analytical ultracentrifuge was at Alliance Protein Laboratories, Thousand Oaks, CA. N.C. was the recipient of a Doctoral Dissertation Fellowship from the Graduate School of the University of Minnesota. Supported by National Institutes of Health GM 51628 (G.B. and C.W.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
1D 1H, one-dimensional proton
Abu or X, α-amino-n-butyric acid
BPTI, bovine pancreatic trypsin inhibitor
CM, oxidized core module
δHα, chemical shift for α-protons
ESMS, electrospray mass spectrometry
Fmoc, 9-fluorenylmethoxycarbonyl
HPLC, high-performance liquid chromatography
HSQC, 15N-1H heteronuclear single quantum coherence
3JHNCαH, vicinal coupling constant
kobs, exchange rate constant
krc exchange rate constant expected in a random coil
Mpa or B, β-mercaptopropionic acid
NMR, nuclear magnetic resonance
NOESY, nuclear Overhauser effect spectroscopy
SEC, size exclusion chromatography
Tm, transition midpoints
TOCSY, total correlation spectroscopy
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.4440102.
References
- Andersen, N.H., Neidigh, J.W., Harris, S.M., Lee, G.M., Liu, Z., and Tong, H. 1997. Extracting information from the temperature gradients of polypeptide NH chemical shifts. 1. The importance of conformational averaging. J. Am. Chem. Soc. 119 8547–8561. [Google Scholar]
- Bai, Y., Milne, J.S., Mayne, L., and Englander, S.W. 1993. Primary structure effects on peptide group hydrogen exchange. Proteins: Structure, Function, and Genetics 17 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar, E., Barany, G., and Woodward, C. 1995. Dynamic structure of a highly ordered β-sheet molten globule: Multiple conformations with a stable core. Biochemistry 34 11423–11434. [DOI] [PubMed] [Google Scholar]
- Beasley, J.R. and Hecht, M.H. 1997. Protein design: the choice of de novo sequences. J. Biol. Chem. 272 2031–2034. [DOI] [PubMed] [Google Scholar]
- Blanco, F., Ramirez-Alvarado, M., and Serrano, L. 1998. Formation and stability of β-hairpin structures in polypeptides. Curr. Opin. Struct. Biol. 8 107–111. [DOI] [PubMed] [Google Scholar]
- Brunger, A. 1993. X-Plor version 3.1: A system for X-ray crystallography and NMR, Yale University Press, New Haven, CT.
- Carulla, N., Woodward, C., and Barany, G. 2000. Synthesis and characterization of a β-hairpin peptide that represents a `core module' of Bovine Pancreatic Trypsin Inhibitor (BPTI). Biochemistry 39 7927–7937. [DOI] [PubMed] [Google Scholar]
- Carulla, N., Woodward, C., and Barany, G. 2001. Towards new designed proteins derived from Bovine Pancreatic Trypsin Inhibitor (BPTI): Covalent cross-linking of two `core modules' by oxime-forming ligation. Bioconjug. Chem. 12 726–741. [DOI] [PubMed] [Google Scholar]
- Dahidat, B.I. and Mayo, S.L. 1997. De novo protein design: Fully automated sequence selection. Science 278 82–87. [DOI] [PubMed] [Google Scholar]
- Das, C., Nayak, V., Raghothama, S., and Balaram, P. 2000. Synthetic protein design: Construction of a four-stranded β-sheet structure and evaluation of its integrity in methanol-water systems. J. Peptide Res. 56 307–317. [DOI] [PubMed] [Google Scholar]
- Das, C., Raghothama, S., and Balaram, P. 1998. A designed three-stranded β-sheet peptide as a multiple β-hairpin model. J. Am. Chem. Soc. 120 5812–5813. [Google Scholar]
- De Alba, E.D., Santoro, J., Rico, M., and Jimenez, M.A. 1999. De novo design of a monomeric three-stranded antiparallel β-sheet. Protein Sci. 8 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrado, W.F., Summa, C.M., Pavone, V., Nastri, F., and Lombardi, A. 1999. De novo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem. 68 779–819. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Falzone, C.J., Wright, P.E., and Benkovic, S.J. 1991. Evidence for two interconverting protein isomers in the methotrexate complex of dihydrofolate reductase from Escherichia coli. Biochemistry 30 2184–2191. [DOI] [PubMed] [Google Scholar]
- Gallagher, W.H. and Woodward, C.K. 1989. The concentration dependence of the diffusion coefficient for Bovine Pancreatic Trypsin Inhibitor: A dynamic light scattering study of a small protein. Biopolymers 28 2001–2024. [DOI] [PubMed] [Google Scholar]
- Gellman, S.H. 1998. Minimal model systems for β -sheet structure in proteins. Curr. Opin. Chem. Biol. 2 717–725. [DOI] [PubMed] [Google Scholar]
- Griesinger, C., Otting, G., Wüthrich, K., and Ernst, R.R. 1988. Clean TOCSY for 1H spin system identification in macromolecules. J. Am. Chem. Soc. 110 7870–7872. [Google Scholar]
- Hill, R.B., Raleigh, D.P., Lombardi, A., and DeGrado, W.F. 2000. De novo design of helical bundles as models for understanding protein folding and function. Acc. Chem. Res. 33 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, R.S. 1996. De novo design of α-helical proteins: Basic research to medical applications. Biochem. Cell Biol. 74 133–154. [DOI] [PubMed] [Google Scholar]
- Ilyina, E., Roongta, V., Pan, H., Woodward, C., and Mayo, K.H. 1997. A pulsed-field gradient NMR study of Bovine Pancreatic Trypsin Inhibitor self-association. Biochemistry 36 3383–3388. [DOI] [PubMed] [Google Scholar]
- Imperiali, B. and Ottesen, J.J. 1999. Uniquely folded mini-protein motifs. J. Peptide Res. 54 177–184. [DOI] [PubMed] [Google Scholar]
- Jiang, X., Kowalski, J., and Kelly, J.W. 2001. Increasing protein stability using a rational approach combining sequence homology and structural alignment: Stabilizing the WW domain. Protein Sci. 10 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B.A., and Blevins, R.A. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Johnson, M.L., Correia, J.J., Yphantis, D.A., and Halvorson, R.R. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least squares techniques. Biophys. J. 36 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, L.E., Keifer, P., and Saarinen, T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc 114 10663–10665. [Google Scholar]
- Koepf, E.K., Petrassi, H.M., Sudol, M., and Kelly, J.W. 1999. WW: An isolated three-stranded antiparallel β-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 8 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme, T., Ramirez-Alvarado, M., and Serrano, L. 1998. Design of a 20-amino acid, three-stranded β-sheet protein. Science 281 253–256. [DOI] [PubMed] [Google Scholar]
- Kuboniwa, H., Grzesiek, S., Delaglio, F., and Bax, A. 1994. Measurement of HN-Hα J couplings in calcium-free Calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR 4 871–878. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Ernst, R.R., and Wüthrich, K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Comm. 95 1–6. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26 283–291. [Google Scholar]
- Laue, T.M., Shah, B.D., Ridgeway, B.D., and Pelletier, S.L. 1992. Analytical ultracentrifugation in biochemistry and polymer science, Royal Society of Chemistry, Cambridge.
- Li, R. and Woodward, C. 1999. The hydrogen exchange core and protein folding. Protein Sci. 8 1571–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra, S. and Nowick, J. S. 2000. β-Sheet interactions between proteins. In The amide linkage: Structural significance in chemistry, biochemistry, and materials science, (eds. A. Greenberg, C. M. Breneman, and J. F. Liebman), pp. 495–518. John Wiley & Sons, New York.
- Merutka, G., Dyson, J. H., and Wright, P. E. 1995. `Random coil' 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J. Biomol. NMR 5 14–24. [DOI] [PubMed] [Google Scholar]
- Ösapay, K. and Case, D.A. 1994. Analysis of proton chemical shifts in regular secondary structure of proteins. J. Biomol. NMR 4 215–230. [DOI] [PubMed] [Google Scholar]
- Ottesen, J.J. and Imperiali, B. 2001. Design of a discretely folded mini-protein motif with predominantly β-structure. Nature Struct. Biol. 8 535–539. [DOI] [PubMed] [Google Scholar]
- Philo, J.S. 2000. Sedimentation equilibrium analysis of mixed associations using numerical constraints to impose mass or signal conservation. Meth. Enzymol. 321 100–120. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solution. J. Biomol. NMR 2 661–665. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S., Richardson, D.C., Tweedy, N.B., Gernert, K.M., Quinn, T.P., Hecht, M.H., Erickson, B.W., Yan, Y., McClain, R.D., and Surles, M.C. 1992. Looking at proteins: Representations, folding, packing, and design. Biophys. J. 63 1186–1209. [PMC free article] [PubMed] [Google Scholar]
- Roberts, G.C.K. 1993. Effects of chemical exchange on NMR spectra. In NMR of macromolecules: A practical approach, (ed. G.C.K. Roberts), Oxford University Press, New York, pp. 153–182.
- Schenck, H.L. and Gellman, S.H. 1998. Use of a designed triple-stranded antiparallel β-sheet to probe β -sheet cooperativity in aqueous solution. J. Am. Chem. Soc. 120 4869–4870. [Google Scholar]
- Smith, C. K. and Regan, L. 1997. Construction and design of β-sheets. Acc. Chem. Res. 30 153–161. [Google Scholar]
- Tuchscherer, G., Scheibler, L., Dummy, P., and Mutter, M. 1998. Protein design: On the threshold of functional properties. Biopolymers 47 63–73. [DOI] [PubMed] [Google Scholar]
- Vita, C., Vizzavona, J., Drakopoulou, E., Zinn-Justin, S., Gilquin, B., and Méndez, A. 1998. Novel miniproteins engineered by the transfer of active sites to small natural scaffolds. Biopolymers 47 93–100. [DOI] [PubMed] [Google Scholar]
- Wagner, G., Braun, W., Havel, T.F., Schaumann, T., Go, N., and Wüthrich, K. 1987. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the Basic Pancreatic Trypsin Inhibitor determined using two different algorithms, DISGEO and DISMAN. J. Mol. Biol. 196 611–639. [DOI] [PubMed] [Google Scholar]
- Wagner, G. and Wüthrich, K. 1982. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra basic pancreatic trypsin inhibitor. J. Mol. Biol. 155 347–366. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Bigam, C.G., Holm, A., Hodges, R.S., and Sykes, B.D. 1995. 1H, 13C, 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5 67–81. [DOI] [PubMed] [Google Scholar]
- Woodward, C., Barbar, E., Carulla, N., Battiste, J., and Barany, G. 2001. Experimental approaches to protein folding based on the concept of a slow hydrogen exchange core. J. Mol. Graphics Modell. 19 94–101. [DOI] [PubMed] [Google Scholar]
- Wouters, M.A. and Curmi, P.M.G. 1995. An analysis of side chain interactions and pair correlations within antiparallel β-sheets: The differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins: Structure, Function, and Genetics 22 119–131. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids, J. Wiley & Sons, New York.
- Zhang, O., Kay, L.E., Olivier, J.P., and Forman-Kay, J.D. 1994. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4 845–858. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz, P., Georgalis, Y., and Saenger, W. 1991. Self-association of Bovine Pancreatic Trypsin Inhibitor: Specific or nonspecific? Biopolymers 31 1347–1349. [DOI] [PubMed] [Google Scholar]