Abstract
To examine how a short secondary structural element derived from a native protein folds when in a different protein environment, we inserted an 11-residue β-sheet segment (cassette) from human immunoglobulin fold, Fab new, into an α-helical coiled-coil host protein (cassette holder). This de novo design protein model, the structural cassette mutagenesis (SCM) model, allows us to study protein folding principles involving both short- and long-range interactions that affect secondary structure stability and conformation. In this study, we address whether the insertion of this β-sheet cassette into the α-helical coiled-coil protein would result in conformational change nucleated by the long-range tertiary stabilization of the coiled-coil, therefore overriding the local propensity of the cassette to form β-sheet, observed in its native immunoglobulin fold. The results showed that not only did the nucleating helices of the coiled-coil on either end of the cassette fail to nucleate the β-sheet cassette to fold with an α-helical conformation, but also the entire chimeric protein became a random coil. We identified two determinants in this cassette that prevented coiled-coil formation: (1) a tandem dipeptide NN motif at the N-terminal of the β-sheet cassette, and (2) the hydrophilic Ser residue, which would be buried in the hydrophobic core if the coiled-coil structure were to fold. By amino acid substitution of these helix disruptive residues, that is, either the replacement of the NN motif with high helical propensity Ala residues or the substitution of Ser with Leu to enhance hydrophobicity, we were able to convert the random coil chimeric protein into a fully folded α-helical coiled-coil. We hypothesized that this NN motif is a "secondary structural specificity determinant" which is very selective for one type of secondary structure and may prevent neighboring residues from adopting an alternate protein fold. These sequences with secondary structural specificity determinants have very strong local propensity to fold into a specific secondary structure and may affect overall protein folding by acting as a folding initiation site.
Keywords: β-sheet to α-helix transition, structural cassette mutagenesis, hydrophobic effect, NN motif, coiled-coil, protein folding
Although the three-dimensional structure of a protein is determined by its amino acid sequence, the difficulty in understanding protein folding is unravelling the role each amino acid plays in determining the final fold and in stabilizing that fold. Any given residue in a protein may participate in a multitude of interactions, including those localized within a secondary structural element as well as longer range contacts with other regions of the protein a considerable distance away in the amino acid sequence. A further complication lies in the inherent degeneracy in the coding of protein folding, because a specific protein fold can be encoded by different amino acid sequences. For example, in the T4 lysozyme, the buried hydrophobic residues are largely interchangeable with other nonpolar amino acids, causing little structural perturbation (Xu et al. 1998; Ohmura et al. 2001). Thus, an emerging hypothesis is that not all residues are equally important for protein folding; that is, some amino acid interactions are the determinants of a particular folding pathway, while others merely serve to stabilize the specified fold (Lattman and Rose 1993; Dill 1999). Another example of degeneracy in protein folding can be found when comparing thermophilic homologs of mesophilic enzymes, where multiple surface-exposed amino acid mutations enhance protein stability with little or no change in overall enzyme architecture (Lehmann et al. 2000). Therefore, we need to identify the "structurally important residues;" that is, residues that dictate the protein fold. Distinguishing the determinants of protein folding is difficult because the folding process is context-dependent. For example, the local propensity of most short peptide sequences to adopt a defined structure is very weak, because they have little structure in aqueous solution isolated from their native protein scaffold.
The structural ambivalence of short peptide sequences is illustrated by the identification of chameleon sequences, that is, a segment of amino acids that can be stabilized as alternate secondary structures depending on protein environment (Minor and Kim 1996; Meizi 1998). Minor and Kim (1996) have designed an 11-residue peptide which is unstructured in a benign aqueous environment but, when inserted into different regions of the B1 domain of protein G (GB1), can be stabilized either as an α-helix or a β-sheet depending on the nonlocal tertiary contacts. In an alternate approach, using a de novo designed "structural cassette mutagenesis" model protein, our laboratory inserted an 11-residue β-sheet secondary structural element (cassette) from human immunoglobulin Fab into an α-helical coiled-coil host protein (or cassette holder) (Kwok et al. 1998a). This β-sheet cassette, when inserted with optimal hydrophobic alignment with the coiled-coil hydrophobic 3–4 repeat of the host protein, folded into a fully helical conformation, illustrating the stabilization of the native β-strand sequence in a nonnative α-helical secondary structure. A statistical survey of the Brookhaven Protein Databank (Argos 1987) showed that sequence-similar pentapeptides can exist in unrelated tertiary structures in different proteins, highlighting the importance of secondary structure adaptation to its protein context. Interestingly, the conformational switching phenomenon is well established in protein misfolding diseases. For instance, the normal cellular prion protein (PrPC) is a normal isoform with high α-helical content, but the abnormal disease-causing β-sheet-rich (PrPSc) isoform is associated with formation of insoluble protein aggregates, leading to senile plaques (for review, see Cohen 1999; Prusiner 1997). Conformational transformation is also observed with Alzheimer's β-amyloid peptide (Aβ), where it generally exhibits α-helical content in organic solvents, but exhibits predominantly β-sheet structure in water.
Cregut et al. (1999) approached the problem of studying short- versus long-range interactions by engineering mutant GB1 proteins, where the region corresponding to the native α-helix was replaced by different stable β-hairpin secondary structural elements. The resulting mutant proteins had decreased stability compared to wild-type protein (up to −5 kcal/mol), but all of the 13-residue β-hairpin insertions were converted to an α-helical conformation (i.e., they exhibited chameleon-like behaviors) by the tertiary hydrophobic interactions that favor the stabilization of helical secondary structure, despite the observation that these sequences form as β-hairpins in their native environment. Those authors subsequently concluded that, although local interactions play an important role in protein stability, they do not confer the specificity of folding, which was concluded to be largely determined by nonlocal tertiary interactions within an existing protein environment. From these observations, a natural question arises: are there sequences that are specific for one type of secondary structure and cannot fold into a different secondary structure (i.e., nonchameleon sequences), thus serving as important determinants of protein folding? Kammerer et al. (1998) suggested that an important determinant of folding of many coiled-coils is a "trigger sequence", or nucleating sequence where the coiled-coils would not fold without this sequence. However, Lee et al. (2001) subsequently demonstrated that a specific sequence is not essential for coiled-coil folding but rather a coiled-coil will fold when its overall stability exceeds a critical threshold. This does not rule out the possibility that very stable sequences can nucleate or trigger folding. These results lend support to our hypothesis that polypeptide sequences which are highly specific for only one type of secondary structure do exist, containing features or "secondary structural specificity determinants" (SSS determinants) that prevent the sequence from folding into alternate secondary structures. To test such a hypothesis, we believe it is necessary to use a well defined model protein which can test the contribution to protein folding of both short- and long-range interactions while minimizing or eliminating any ambiguity in interpretation of results which may arise from using more complex native proteins. Thus, our favored approach to solving the protein folding problem is to insert a small, defined secondary structural element into a larger host protein with different secondary structural propensity, followed by identification of which residues profoundly affect the structure and stability of the host protein. As described previously (Kwok et al. 1998a), our minimalist structural cassette approach is based on a de novo designed α-helical coiled-coil motif, recognized as one of nature's favorite ways of creating a dimerization motif (Hodges 1996; Micklatcher and Chmielewski 1999). A major advantage of such a model is that there is only one type of secondary structure present in the host protein (i.e., the α-helix) and the effects of single residue substitutions on protein folding and stability are more straightforward to quantify than mutations in native globular proteins. Extensive work in our laboratory and others has led to a good understanding of interactions which stabilize coiled-coils, including: stability contributions of the hydrophobic core positions a (Wagschal et al. 1999) and d (Tripet et al. 2000) in the hydrophobic gabcdef heptad repeat characteristic of this motif (Hodges et al. 1981; Hodges 1996); polypeptide chain length effects on coiled-coil stability (Su et al. 1994; Fairman et al. 1995); intrinsic amino acid side-chain propensities for α-helix or β-sheet structure (Lyu et al. 1990; O'Neil and Degrado 1990; Chakrabartty et al. 1994; Zhou et al. 1994; Monera et al. 1995; Minor and Kim 1994); helix capping and termination signals (Aurora and Rose 1998; Lu et al. 1999); hydrogen bonding (Borg et al. 2001); lactam-bridge stabilization (Houston et al. 1996; Kwok et al. 2001); and side-chain rotamer entropic effects (Yu et al. 1999; Penel and Doig 2001). In addition, the three-dimensional structures of many natural helical protein motifs have been characterized by both crystallography and NMR experiments (for review, see Kohn et al. 1997), thus providing a wealth of structural data for comparison with experimental observation. The insertion of an amino acid sequence ("cassette") which exhibited β-strand structure in its native protein into our host coiled-coil protein results in an α-helix-β-sheet-α-helix (host-guest-host) chimeric protein arrangement and may result in three possible folding scenarios: (1) the α-helices in the host protein induce or nucleate the β-sheet cassette to fold into an α-helical conformation; (2) the sequence of the β-sheet cassette prevents itself from being induced by the host protein into α-helical structure, but the nucleating α-helices of the host protein still fold; and (3) the sequence of the β-sheet not only does not fold into an α-helical conformation, but is able to prevent the host protein from folding into α-helical structure. The present study extends our investigation of the potential of our SCM model to examine short-range and long-range interactions in the stabilization of secondary structural folds. Specifically, we describe its efficacy in identifying and characterizing novel secondary structure specificity determinants as well as quantifying their contributions to protein stability.
Results
Design of the α-helical coiled-coil host protein as a cassette holder
A detailed description of the de novo design and application of the SCM model has been reported (Kwok et al. 1998a). To summarize, we designed a very stable α-helical coiled-coil host protein with a repeating heptad sequence (gabcdef) of E-V-E-A-L-K-K, where the hydrophobic core is occupied by Val and Leu residues in positions a and d, respectively, forming a 3–4 (or 4–3) hydrophobic repeat (Fig. 1 ▶). The N-terminal α-helical segment of the host protein contains an interchain disulfide bridge, due to a Cys-Gly-Gly linker sequence at the N-terminal of each polypeptide chain, this linker facilitating coiled-coil folding and increasing stability. In addition, this flexible linker eliminates the concentration-dependent monomer-dimer equilibrium in the formation of a two-stranded coiled-coil, thus making chemical denaturation studies independent of protein concentration (Zhou et al. 1992). Furthermore, the disulfide bridge also ensures that the polypeptide chains are in-register and parallel. As seen in Figure 1 ▶, the N-terminal α-helical segment of the polypeptide chain contains three hydrophobes in the hydrophobic core, and the C-terminal α-helical segment contains four hydrophobes, thus maintaining the continuity of the 3–4 hydrophobic repeat characteristic of coiled-coils throughout the cassette holder. Besides hydrophobic stabilization by large hydrophobes (Val and Leu in a and d positions, respectively), inter- and intrachain ionic interactions were introduced to stabilize the coiled-coil. Interchain salt bridges were engineered by placing Lys and Glu residues at positions e and g of the α-helical segments, resulting in ionic stabilization due to electrostatic attractions (i → i` + 5 or g to e`) between negatively charged Glu side chains and positively charged Lys side chains. In addition to such interchain electrostatics for stabilization of the coiled-coil, the potential to form intrachain ionic interactions between Lys residues at either positions e or f to Glu at position b (i to i + 3 or i to i + 4) enhances the overall helix nucleation potential. Ala residues were chosen at positions c and f because Ala has the highest intrinsic helical propensity (Zhou et al.1994), and a relatively small, nonbulky side chain (for example Ala 11 and Ala 25) that minimizes the introduction of extraneous context-dependent interactions with the cassette. These helix stabilizing interactions were engineered to create a very stable coiled-coil host protein, where the α-helical segments have the potential to nucleate the induction of α-helical structure in the cassette region. Indeed, insertion of an 11-residue control cassette with a sequence identical to that of the cassette holder (E-A-L-K-K-E-V-E-A-L-K) into the center of the host protein resulted in a very stable (GdnHCl denaturation midpoint of 5.6 M) and fully folded α-helical coiled-coil (the 11-residue cassette length was chosen as representative of the average length of α-helical and β-strand segments; Chou and Fasman 1974, 1978). This central region of the coiled-coil host protein was designed to be the region of cassette insertion, because perturbing the central region of a coiled-coil has been shown to have the greatest destabilizing effect on overall protein conformation (Zhou et al. 1992; Harbury et al. 1993). In addition, insertion of a cassette into the central location permits optimal helix nucleation as the flanking coiled-coil regions have the potential to direct folding of the cassette sequence (in our case a β-sheet sequence) into helical structure from both ends.
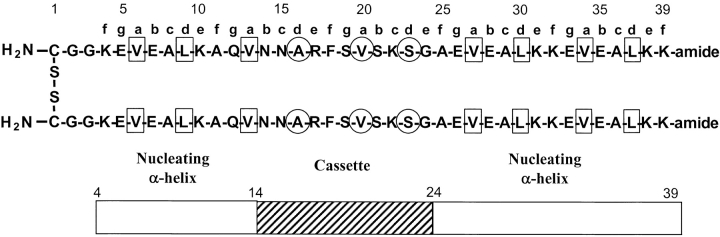
Fig. 1.
Model of the 78-residue disulfide-bridged two-stranded coiled-coil host protein. This model peptide consists of two nucleating α-helices (residues 4-13, 25-39; open rectangles) at the N- and C-termini of each 39-residue polypeptide chain. The hydrophobic residues at positions a and d of the coiled-coil heptad repeat abcdefg (denoted above the polypeptide chain), forming the hydrophobic core of the two nucleating α-helices, are boxed. The 11-residue cassette (residues 14-24; hatched rectangle) is inserted into the center of the cassette holder, and the residues in this immunoglobulin β-strand cassette (Fig. 2 ▶) that would be buried in the hydrophobic core if the entire cassette holder fold into a coiled-coil are circled.
The effectiveness of our model to induce α-helical structure in a peptide sequence originally exhibiting a β-sheet conformation in its native protein was described by Kwok et al. (1998a) for a β-strand sequence from immunoglobulin Fab, a protein consisting predominantly of β-sheet secondary structure. When this 11-residue β-strand sequence (7Fab:64–74) was inserted as a cassette into our model coiled-coil cassette holder, the entire chimeric protein folded into an α-helical coiled-coil conformation. Even the subsequent substitution of five Thr residues (Thr having a low α-helical propensity and the highest β-sheet propensity of the amino acids; Minor and Kim 1994; Zhou et al. 1994) into the cassette, in addition to the three Thr residues already present, failed to overcome the strong α-helix nucleating effect of the cassette holder (Kwok et al. 1998b). Thus, even this modified cassette sequence with significantly enhanced β-sheet potential was fully induced into α-helical structure by the nucleating α-helices. Although such a result might suggest that our model host protein has such strong α-helical potential that all β-sheet segments from native proteins may potentially be induced into α-helical structure, we hypothesized that specific sequences exist which cannot be induced in this way. Indeed, the identification of a peptide sequence which exhibited β-sheet structure in its native protein and which was shown either not to fold into an α-helix when inserted into our model cassette holder, or even to disrupt this very robust model which has been shown to be extremely effective in inducing α-helical structure, would be a sequence worthy of further study.
Selection of the cassette
As noted above, the first objective of the present study was to determine whether a native β-sheet sequence could be identified which can override the strong nucleating potential of the flanking α-helical coiled-coil regions of the cassette holder, thus illustrating an example of local propensity overriding the longer-range tertiary stabilization of the coiled-coil 3–4 hydrophobic repeat. The second objective was to identify the sequence-dependent features responsible for prevention of α-helix formation. Upon closer examination of the amino acid sequences in immunoglobulin Fab, an amphipathic β-strand (7Fab:53–63; Fig. 2 ▶) was identified which immediately precedes the "chameleon" β-sheet strand previously used as the cassette (Kwok et al. 1998a). Interestingly, this sequence included an adjacent pair of Asn residues, Asn being a very common helix capping residue (Aurora and Rose 1998). Thus, with this feature in mind, this β-strand sequence was chosen as our initial candidate cassette for the present study.
Fig. 2.

Molscript drawing of residues 1-100 of immunoglobulin λ light chain fragment Fab New (PDB designation: 7FAB). The top panel shows the immunoglobulin fold with the β-strand (white) that was used as a cassette for insertion into the two-stranded α-helical coiled-coil cassette holder. The bottom panel is a side view of this strand showing the buried side chains (white), Asn 54, Arg 56, Phe 57, Val 59, and Lys 61 and the solvent exposed side chains (green), Asn 53, Ala 55, Ser 58, Ser 60, and Ser 62. The sequence of this 11-residue β-strand cassette (7FAB: 53-63) is shown in the bottom panel with the residues that would form a continuous 3-4 hydrophobic repeat with the cassette holder (yellow). The orange arrows denote the solvent-exposed residues. The host protein containing this sequence is referred to as the Parent peptide in Fig. 3 ▶ (residues 14-24).
As shown in Figure 2 ▶, in its native conformation in the Fab molecule, this β-strand buries two large hydrophobic residues in the hydrophobic core of the immunoglobulin fold (Phe 57 and Val 59), obviously to help stabilize the sequence in a β-sheet conformation. In a similar fashion, we optimized the alignment of the cassette to create the greatest possible burial of hydrophobic residues in the hydrophobic core of the coiled-coil if the sequence were to adopt an α-helical structure. Thus, the 3–4 hydrophobic repeat through the cassette would be as hydrophobic as possible. Significantly, if this sequence was induced into an α-helix by the nucleating α-helical segments of the cassette holder, a small hydrophobe (Ala 55) and a larger hydrophobe (Val 59) would occupy positions 16d and 20a, respectively, thus maintaining the 3–4 hydrophobic repeat of the host protein; that is, these two hydrophobes would be buried in the hydrophobic core of the potentially fully folded α-helical coiled-coil protein model. Thus, despite the presence of the polar Ser 62 from the Fab protein at hydrophobic position 23d, this cassette sequence alignment represents the highest potential for the strongly nucleating α-helices to induce this β-strand sequence (denoted Parent cassette; Fig. 3 ▶) into α-helical structure as well as maximize potential coiled-coil structure and stability.
Fig. 3.
Amino acid sequences of the cassettes that were inserted into the center of the two-stranded α-helical coiled-coil host protein. The Parent peptide contains the β-sheet cassette from the immunoglobulin fold (Fig. 2 ▶). Peptide nomenclature is based on the position of substitutions. For example, S23L denotes a leucine replacement of serine at position 23 of the cassette. These substitutions are boxed. The heptad repeat is denoted as abcdefg, where positions a and d are the positions in the hydrophobic core of the coiled-coil. The dotted lines denote the nucleating α-helices of the host protein (Fig. 1 ▶).
Characterization of the host protein following insertion of Parent cassette
To determine the effect of insertion of the Parent cassette into the cassette holder, we characterized the structure of our model protein using circular dichroism (CD). As seen in Figure 4 ▶, not only did the Parent cassette not fold into an α-helix under benign conditions when inserted into the cassette holder, but the nucleating helices of the host protein were also highly disrupted (only 23% overall helicity for the entire model protein; Table 1). Significantly, even in the presence of helix-inducing solvent (50% TFE), only 69% helicity was achieved, strongly suggesting the presence of features specific to the Parent cassette which are able not only to override the strong helix-inducing effects of the nucleating α-helices but the helix-inducing properties of an excellent helix-inducing solvent. Indeed, with the 11-residue Parent cassette representing about a third of overall polypeptide length, this helicity value indicates that two-thirds of the polypeptide sequence (the nucleating α-helices) are helical in the presence of 50% TFE, whereas the cassette retains random structure under these conditions.
Fig. 4.
Circular dichroism spectra of the cassette holder (Fig.1) containing different cassette sequences at 25°C (Fig. 3 ▶). (A) CD experiments were carried out in a 50mM PO4 (K2HPO4/KH2PO4), 100mM KCl, pH 7.0 buffer with the following peptides: Parent (open circles), S23L (open squares), and N14A,N15A (open triangles). (B) CD scans of Parent and N14A,N15A peptides were carried out in a 50mM PO4, 100mM KCl, pH 7.0 buffer in the absence (benign) or presence of 50% (v/v) trifluoroethanol (+TFE); Parent benign (open circles), Parent + TFE (closed circles), N14A,N15A benign (open triangles) and N14A,N15A + TFE (closed triangles). Concentration of peptides ranged from 87 to 104μM.
Table 1.
Circular dichroism results of the oxidized two-stranded peptides
| [θ]222b | % Helixd | ||||||||
| Peptide number | Peptide namea | Benign (25° C) | 50% TFE | [θ]222/208c | Benign (25° C) | 50% TFE (25° C) | [Gdn•HCl]1/2e (M) | mf (kcal•mol•M−1) | ΔΔGug (kcal•mol −1) |
| 1 | Parent | −8,100 | −24,100 | 0.64 | 23 | 69 | –h | –h | –h |
| 2 | S23L | −32,800 | −29,200 | 1.01 | 95 | 89 | 1.50 | 1.70 | −1.8 |
| 3 | N14A, N15A | −33,400 | −30,600 | 1.02 | 96 | 88 | 1.00 | 1.67 | −2.6 |
| 4 | N14A, N15A, S23A | −33,100 | −31,400 | 1.03 | 95 | 90 | 1.30 | 1.73 | −2.2 |
| 5 | N14A, N15A, S23L | −33,900 | −34,100 | 1.03 | 97 | 99 | 2.55 | 1.73 | 0.0 |
| 6 | S19A, S21A, S23L | −31,200 | −32,000 | 1.01 | 90 | 92 | 1.80 | 1.68 | −1.3 |
| 7 | N14A, S23L | −32,600 | −32,500 | 1.02 | 94 | 93 | 2.55 | 1.59 | 0.0 |
| 8 | N15A, S23L | −33,200 | −32,800 | 1.03 | 95 | 94 | 2.70 | 1.64 | +0.2 |
a Peptide cassette sequences and nomenclature are shown in Figure 3 ▶.
b [θ]222 is the mean residue molar ellipticity (degrees • cm2 • dmol−1) measured at 222 nm in a 100 mM KCl, 50 mM PO4 (K2HPO4/KH2PO4) buffer, pH 7.0 in the absence (Benign) or presence of 50% trifluoroethanol (50% TFE) (v/v). Concentration of peptides ranges from 87 to 104 μM. The uncertainty in the molar ellipticity values is +/− 300.
c The helical ratio [θ]222/208 was calculated by dividing the observed molar ellipticity value at 222 nm ([θ]222) by the observed molar ellipticity value at 208 nm ([θ]206) in benign buffer.
d % Helix was calculated from [θ]222 based on an ellipticity value of −36,000 for 100% α-helical content derived from the equation XHn = XH∞ (1−k/n), where XH∞ is −37,400, the wavelength dependent constant, k, is 2.5, and n is 36 for the number of helical residues (Chen et al. 1974).
e [Gdn•HCl]1/2 is the denaturation midpoint of the two-state unfolding of an α-helical coiled-coil to a random coil. Guanidinium denaturations were carried out at 25 °C after overnight equilibration at room temperature. [GdnHCl]1/2 is reproducible within +/− 0.05 M when the same stock solution of guanidinium hydrochloride was used. Values were rounded off to the nearest 0.05 M. Concentration of peptides ranges from 87 to 104 μM.
fm is the slope term defined by the linear extrapolation equation ΔGu = ΔGu(H2O) − m [denaturant], and ΔGu(H2O) is the free energy of denaturation in the absence of denaturant, whereas ΔGu is the free energy of unfolding at a given denaturant concentration. (See Materials and Methods).
g ΔΔGu, the change of the free energy of unfolding relative to peptide N14A,N15A,S23L, derived from the equation: ΔΔGu=([denaturant])1/2.a − [denaturant]1/2.b)(ma+mb)/2, (Serrano et al. 1990; see Materials and Methods). The ΔGu(H2O) for this analog is 2.8 kcal•mol−1.
h Not determined because Parent peptide is mostly unfolded in benign conditions.
Two regions of the β-strand sequence were deemed likely to be responsible for the prevention of helical structure in the cassette: (1) the NN motif was suspect due to the low intrinsic helical propensity of Asn, in conjunction with the helix capping effects of Asn residues; and (2) the unfavorable burial of a polar residue, Ser 23, in the hydrophobic core of the coiled-coil if the cassette was to fold into an α-helical structure. These two features were therefore studied further.
Identification of potential determinants that prevent α-helix formation in the cassette
Two analogs of the Parent sequence were then synthesized to delineate the potential contribution of the two regions noted above to the prevention of α-helix induction: (1) S23L, where the polar Ser residue in position 23 of the model protein is replaced by a hydrophobic Leu residue, thus maintaining a continuous 3–4 hydrophobic repeat throughout the entire length of the protein sequence; and (2) N14A, N15A, where there is a tandem replacement of two residues with low helical propensity (Asn) with two residues of high α-helical propensity (Ala). The sequences of these analogs are shown in Figure 3 ▶ (peptides 2 and 3).
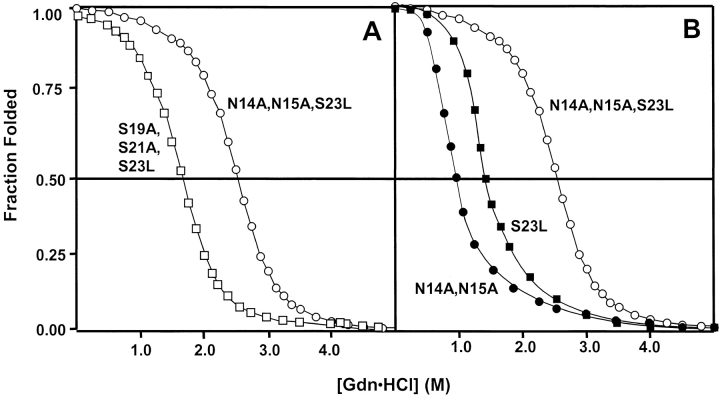
As noted in Table 1, the S23L analog (peptide 2) forms a fully folded α-helical coiled-coil under benign conditions; that is, the cassette holder has induced an α-helical conformation in the S23L cassette sequence, the entire protein now exhibiting a [θ]222/208 ratio > 1.0 indicative of coiled-coil formation (Lau et al. 1984; Yu et al. 1996), 95% helicity, and a molar ellipticity of −32,800° (Fig. 4A ▶; Table 1) which is not enhanced further by the addition of 50% TFE. Thus, the replacement of a single polar residue (Ser) with a hydrophobic residue (Leu) at position 23d (Fig. 3 ▶) had a profound impact on overall folding of the model protein, converting an essentially unstructured molecule (Fig. 4A ▶; Table 1) into an α-helical coiled-coil with a chemical denaturation midpoint, [GdnHCl]1/2, of 1.50 M (Fig. 5B ▶; Table 1).
Fig. 5.
GdnHCl denaturation profiles of selected peptide analogs. (A, B) Chemical denaturation experiments were carried out at 25°C in a 50mM PO4 (K2HPO4/KH2PO4), 100mM KCl, pH 7.0 buffer with increasing concentrations of GdnHCl as denaturant. The denaturation dilution samples were vortexed and left to equilibrate overnight at RT. The fraction folded of each peptide was calculated as described in Materials and Methods. S23L (closed squares), S19A,S21A,S23L (open squares), N14A,N15A (closed circles), N14A,N15A, S23L (open circles). Concentration of peptides ranged from 87 to 104 μM.
In a manner similar to that of the S23L analog, N14A, N15A (peptide 3, Table 1) also exhibited a fully folded α-helical coiled-coil structure under benign conditions ([θ]222/208 = 1.02; Table 1), with 96% α-helicity and a molar ellipticity of −33,400° (Fig. 4A ▶; Table 1) and a [GdnHCl]1/2 value of 1.00 M (Fig. 5B ▶, Table 1). Again, in a manner similar to that of S23L, no further induction of α-helicity is apparent with the addition of 50% TFE, and a changed [θ]222/208 ratio < 1.0 (0.84, Fig. 4B ▶) for N14A, N15A is quite clear compared to that under benign conditions, indicating dissociation of the coiled-coil structure in the presence of TFE, a solvent known to induce α-helical secondary structure in potentially helical peptide sequences but to denature tertiary and quaternary structure (Cooper and Woody 1990; Sönnichsen et al. 1992). These results suggest that, when acting in concert, both the NN motif and Ser 23 do indeed have a profound preventive effect on helix induction; that is, they appear to be acting as secondary structure specificity determinants (SSS) by overcoming the strong helix-inducing effect of the flanking nucleating α-helices. We next wanted to determine the relative contribution of the two regions to resistance to helix induction, as well as the requirement or otherwise of the NN motif, where Asn residues have their effect in tandem, as opposed to the Asn residues having their effect independently as single residues. Concomitant with these issues is what specific features of the two regions under scrutiny contribute to coiled-coil stability or instability when one or the other region has been removed. Although both the S23L and N14A, N15A analogs form fully folded α-helical coiled-coils under benign conditions (Fig. 4 ▶; Table 1), these molecules exhibit only moderate stability ([GdnHCl]1/2 values of 1.50 M and 1.00 M, respectively; Table 1). Thus, we designed and synthesized more analogs to examine the quantitative effect of mutations in the NN motif and the hydrophobic core of the coiled-coil (residue 23d) on stabilization or destabilization of the coiled-coil.
Verification of the NN motif as a helix-destabilizing secondary structure specificity determinant
We designed two peptide analogs, N14A, S23L and N15A, S23L (Fig. 3 ▶) to elucidate the role of the individual residues in the NN motif with regard to the helix disruptive properties of the Parent cassette. We used the S23L sequence to ensure folding of the cassette and increase stability of the coiled-coil to be able to measure substitution effects in the NN motif accurately. As shown in Table 1, both analogs were fully folded α-helical coiled-coils, with similar [GdnHCl]1/2 values of 2.55 M and 2.70 M for N14A, S23L and N15A, S23L, respectively; that is, removal of either Asn residue significantly increased coiled-coil stability relative to S23L ([GdnHCl]1/2 = 1.50 M; Table 1), where both Asn residues are present. These results support a synergistic contribution of both Asn residues to destabilizing effects on α-helical structure, as well as further emphasizing the role of the NN motif as a helix-destabilizing SSS determinant.
Contribution of SSS regions to coiled-coil stability
Two analogs, N14A, N15A, S23L and N14A, N15A, S23A (Fig. 3 ▶), were synthesized to examine further the contribution to coiled-coil stability of residue hydrophobicity when substituted into the hydrophobic core at position 23d. Both analogs were fully folded α-helical coiled-coils under benign conditions, with [GdnHCl]1/2 values of 2.55 M (N14A, N15A, S23L; Table 1, Fig. 3 ▶) and 1.30 M (N14A, N15A, S23A). Thus, the stability of the analogs increased with increasing hydrophobicity of the residue substituted into the hydrophobic core of the coiled-coil at position 23d: from a [GdnHCl]1/2 value of 1.00 M for the polar Ser at this position (N14A, N15A; Fig. 5 ▶, Table 1) to 1.30 M for the minimally hydrophobic Ala (N14A, N15A, S23A; Table 1) to 2.55 M for the highly hydrophobic Leu (N14A, N15A, S23L). Aside from the expected coiled-coil stabilizing effect of increasing residue hydrophobicity at the nonpolar 23d position (Tripet et al. 2000), the destabilizing effect of a polar residue (Ser in this case) is also quite clear. Interestingly, the stability of N14A, N15A, S23L appeared to correlate closely with the additive (and in concert) destabilizing effects of the unfavorable Ser 23 burial in the coiled-coil core and the NN motif, with the S23L and N14A, N15A analogs exhibiting [GdnHCl]1/2 values of just 1.50 M and 1.00 M, respectively (Table 1).
Finally, although it has been verified that the NN motif is indeed an SSS determinant that resists induction of the Parent cassette into helical structure, the tandem nature of the NN dipeptide makes it more difficult to assess its destabilizing contribution to overall coiled-coil stability, because the replacement of two Asn residues with two Ala residues (e.g., N14A, N15A, S23L) changes two factors simultaneously: (1) the NN motif is removed, and (2) there is an overall increase in helical propensity, because Ala has a significantly higher α-helical propensity compared to Asn and it was shown previously that amino acid α-helical propensity is an important factor governing protein stability (Monera et al. 1995). Thus, to differentiate the destabilizing contribution attributed to amino acid propensity changes (as a result of the two Asn to two Ala replacements) from the intrinsic helix-destabilizing nature of the NN motif, we prepared analog S19A, S21A, S23L (Fig. 3 ▶). Since Ser and Asn have the same intrinsic α-helical propensity (Zhou et al. 1994), this analog should contain the same overall α-helical propensity as that of N14A, N15A, S23L (note that, similar to the two Asn residues, neither of the Ser residues substituted at positions 19 and 21 of S19A, S21A, S23L are situated in the hydrophobic core of the coiled-coil). The S19A, S21A, S23L analog forms a fully folded α-helical coiled-coil under benign conditions, with a [GdnHCl]1/2 value of 1.80 M (Table 1; Fig. 5 ▶). As is clear from Figure 5 ▶, this stability value for S19A, S21A, S23L is considerably less than that of N14A, N15A, S23L, ([GdnHCl]1/2 = 2.55 M; Table 1) where the highly destabilizing NN motif has been removed although overall helical propensity is identical; that is, the difference in stability between the two analogs (N14A, N15A, S23L − S19A, S21A, S23L = 2.55 M − 1.80 M = 0.75 M change in the [GdnHCl]1/2 values) may be attributed to the intrinsic destabilizing contribution of the NN motif separate from any helix propensity concerns.
Discussion
The folding of secondary structure generally correlates with overall protein hydrophobic stabilization and local folding preferences. However, our Parent peptide (Fig. 1 ▶) did not fold because sequence characteristics of the inserted β-sheet cassette prevented helix induction by the flanking α-helices in the host protein, and the strong secondary structure specificity (SSS) determinants in the β-sheet cassette prevented the α-helical host from folding into a coiled-coil conformation. Even in a helix-promoting environment (50% TFE), only 69% helicity (i.e., two-thirds) was achieved by the Parent peptide, underlying the strong helix-disruptive elements of the remaining third of the sequence; that is, the inserted β-strand cassette. It is also worth noting that the β-sheet cassette could not nucleate β-sheet structure in the flanking peptide sequences of the cassette holder because there is no β-sheet preference in these sequences, hence the observed general unfolding of the whole protein. By systematic removal of the helix-disruptive motifs of the Parent cassette, we achieved a β-sheet to α-helix transition of the cassette, allowing the host protein to fold into a coiled-coil. This systematic removal of sequence features with potential helix-disruptive (or helix-preventive) characteristics demonstrated how two adjacent Asn residues ("NN" motif) and an unfavorable burial of a polar Ser residue at the hydrophobic core of a putative coiled-coil acted in concert to prevent helix induction by the nucleating α-helices at either end of the Parent cassette.
Table 2 offers a summary of the effects of systematic mutation of the Parent cassette on subsequent coiled-coil formation and stability. Thus, specific pairs of cassette analogs are compared to highlight the contribution of specific sequence substitutions to coiled-coil stability, reported either as the change in free energy (ΔΔGu) per coiled-coil or ΔΔGu per substitution or interaction (data adapted from ΔΔGu values reported in Table 1). In each comparison of a pair of analogs, the ΔΔGu value per coiled-coil (i.e., the relative change of free energy of unfolding in the coiled-coil due to a particular substitution(s); Table 2) was calculated by subtracting the ΔΔGu value (i.e., the change in free energy of unfolding relative to N14A, N15A, S23L; Table 1) of the less stable analog from the more stable analog. For example, when gauging the contribution of Ala in place of Ser in the hydrophobic core (Table 2), ΔΔGu for N14A, N15A, S23A minus ΔΔGu for N14A, N15A = −2.2 – (−2.6) = 0.4 kcal.mol−1 per coiled-coil or 0.4/2 = 0.2 kcal.mol−1 per substitution; that is, the substitution of Ala for Ser has contributed 0.2 kcal.mol−1 to enhancement of coiled-coil stability for each of the two Ser → Ala substitutions.
Table 2.
The change in free energy for different noncovalent interactions
| Analog comparison | |||||
| More stable | Less stable | Description of contribution in each comparisona | Number of substitutions or interactions per coiled-coilb | ΔΔGu per coiled-coilc (kcal•mol−1) | ΔΔGu per substitution or interactiond (kcal•mol−1) |
| N14A,N15A,S23A | N14A,N15A | Contribution of Ala in place of Ser in the hydrophobic core | 2 | 0.4 | 0.2 |
| N14A,N15A,S23L | N14A,N15A | Contribution of Leu in place of Ser in the hydrophobic core | 2 | 2.6 | 1.3 |
| N14A,N15A,S23L | N14A,N15A,S23A | Contribution of Leu in place of Ala in the hydrophobic core | 2 | 2.2 | 1.1 |
| S19A,S21A,S23L | S23L | Contribution of Ala in place of Ser (increase α-helical propensity) | 4 | 0.5 | 0.1 |
| N14A,N15A,S23L | S23L | Destabilizing effect of the NN motif (includes N → A propensity effect) | 2 | 1.8 | 0.9 |
| N14A,N15A,S23L | S19A,S21A,S23L | Net destabilizing effect of the NN motif | 2 | 1.3 | 0.6 |
| N14A,N15A,S23L | N14A,S23L | Contribution of single Asn(N15) → Ala substitution | 2 | 0.0 | 0.0 |
| N14A,N15A,S23L | N15A,S23L | Contribution of single Asn(N14) → Ala substitution | 2 | −0.2 | −0.1 |
| N14A,S23L | S23L | Destabilizing effect of the NN motif (includes N → A propensity effect) | 2 | 1.8 | 0.9 |
| N15A,S23L | S23L | Destabilizing effect of the NN motif (includes N → A propensity effect) | 2 | 2.0 | 1.0 |
a Physical description of the effects of substitution(s) on coiled-coil stability.
b Number of interactions in a two-stranded α-helical coiled-coil; there are two identical polypeptide chains that are joined by a disulfide bridge in our cassette model.
c The relative change of the free energy of unfolding in the coiled-coil due to the substitution(s).
d The relative change of the free energy of unfolding per substitution or interaction. This is obtained by dividing the ΔΔGu/coiled-coil by the number of substitution(s) or interaction(s) in the coiled-coil, and rounded off to the nearest 0.1 kcal/mol.
Protein core hydrophobic interactions represent a major contributor to protein stability, and this contribution is especially apparent during the nucleation of β-sheet to α-helix structural transition of our Parent cassette following removal of SSS determinants. Thus, the strong helix nucleation potential of our coiled-coil model is attributed to the host protein (cassette holder), possessing a 3–4 hydrophobic repeat, characteristic of α-helical coiled-coils, comprised of large hydrophobic Val and Leu residues. Subsequent insertion of a cassette with proper alignment of hydrophobic residues to allow a continuous 3–4 hydrophobic repeat throughout the whole protein sequence then allowed an accurate assessment of the effect of mutations in the inserted cassette sequence which stabilized or destabilized the hydrophobic core. Thus, the experimentally observed stabilizing effect of a hydrophobic Leu replacement for a polar Ser at position 23d was 2.6 kcal.mol−1 or 1.3 kcal.mol−1 for each Leu → Ser substitution (Table 2). This trend of stability enhancement by removal of Ser from the hydrophobic core was also observed by Tripet et al. (2000), who reported an increase in stability of their coiled-coil model of 2.8 kcal.mol−1 when Ser was replaced by Leu at a d position; in addition, an Ala to Leu replacement increased coiled-coil stability by 1.9 kcal.mol−1. Thus, our experimental values of 2.6 kcal.mol−1 and 2.2 kcal.mol−1 for Ser → Leu and Ala → Leu substitutions, respectively, correlated well with these earlier reported values. Significantly, in T4 lysozyme (i.e., in a globular protein context), the reported loss in stability associated with a Leu to Ala substitution was 2.1 kcal.mol−1, with this value considered to be a measure of the difference in the free energy of transfer of Leu and Ala from the solvent to the protein core (Xu et al. 1998). However, the contribution of large-to-small hydrophobic residue substitution is a more complex issue in a globular protein than in a coiled-coil, because the resulting cavity formation from the loss of van der Waal's interactions can result in dramatic loss of globular protein stability. In the coiled-coil setting, the replacement of Leu with Ala would represent a net loss of 78 Å2 of hydrophobic surface area. By multiplying this hydrophobic area by two because there are two polypeptide chains per coiled-coil, and then by an associated hydrophobic factor of 25 cal/Å2 (Karplus 1997), we estimated that the decrease of stability is 2.0 kcal.mol−1, a value that is in excellent agreement with our observed value of 2.2 kcal.mol−1 (Table 2).
Although the helix-destabilizing property of the NN motif was not reported previously, this helix-disruptive nature correlates with known physical properties of Asn. For instance, Asn is a very common C-terminal helix capping residue (Aurora and Rose 1998), has a strong tendency to form solvent-exposed turns in connecting transmembrane helices (Monne et al. 1999), and has been commonly observed in crystal structure to participate in hydrogen bonding interactions with residues i to i + 2 ahead in the sequence (Wan and Milner-White 1999). However, the present study clearly demonstrated that the helix-destabilizing effect of the NN motif (1.8 kcal.mol−1 per coiled-coil, including helix propensity effect; Table 2) is significantly greater than the contributions of the individual Asn residues. Indeed, when the individual Asn residues in the NN motif were substituted by Ala, the resulting contributions to coiled-coil stability were negligible (0.0 and −0.2 kcal.mol−1 per coiled-coil for Asn 15 and Asn 14, respectively; Table 2). Thus, the adjacent Asn residues of the NN motif are working synergistically to contribute significant helix destabilization.
The presence of the polar Ser residue in the hydrophobic core of the coiled-coil contributes more to coiled-coil destabilization than does the NN motif (compare 2.6 kcal.mol−1 increase in coiled-coil stability for a Leu to Ser substitution with just 1.3 kcal.mol−1 for the net destabilizing effect of the NN motif; Table 2). However, the contributions of both the NN motif and Ser 23 are each of major significance since, as noted above, only when working in concert is the β-strand cassette able to override the helix-nucleating influence of the α-helical host protein. Furthermore, the differences in free energy between the most stable peptide (N14A, N15A, S23L) and the two analogs containing single Asn to Ala substitutions (N14A, S23L and N15A, S23L) were, as noted previously, negligible (−0.2 and 0.0 kcal.mol−1, respectively; Table 2). Thus, if either of these Asn residues was removed from the NN motif, its destabilizing influence is essentially abolished. These observed destabilizing tendencies of the NN motif and Ser burial in the hydrophobic core of the coiled-coil may reflect a more global destabilizing approach found in natural proteins. Table 3 reports the results of a protein databank search for the occurrence of Ser burial in the hydrophobic core of naturally occurring coiled-coils and NN motifs in α-helices. As shown in Table 3, Ser occurs with a frequency of only 11% in the hydrophobic core of natural coiled-coils; this compares with a 100% frequency for Leu. This observed infrequency of Ser in the hydrophobic core of native coiled-coils is indicative of its destabilizing effects on this secondary structure, clearly underlined by the present study. Interestingly, the majority of NN motifs were found to occur in loops, turns, or undefined structure (86%), and much less frequently in β-sheet segments (5.9%) and α-helices (8%) (Table 3); that is, nature favors the NN motif to occur outside of protein secondary structure, and its infrequency in helices may correlate with its intrinsic helix-destabilizing nature.
Table 3.
Numerical analysis of secondary structural specificity determinants
| Non-redundant occurrence of NN motif in the PDBa | 5041 |
| a) found in α-helical structureb | 8.1% |
| b) found in β-sheet structureb | 5.9% |
| c) found in loops, turns or undefined conformationsb | 86.0% |
| Frequency of serine found in hydrophobic core of coiled-coilsc | 11% |
a Performed on a nonredundant version of the Brookhaven Protein Database with 1021 protein sequences.
b Secondary structures were determined by SEQSEE and VADAR.
c Normalized occurrence relative to leucine (=100) (Tripet et al. 2000).
As a final comment, it is interesting to note that, in the native Fab protein, the β-strand chosen as the cassette in the Parent peptide for the present study (Fab:53–63) immediately preceded the "chameleon" sequence (Fab:64–74) chosen as the cassette in our previous study introducing our cassette mutagenesis model (Kwok et al. 1998a). As reported, while exhibiting β-sheet structure in the native protein, this sequence was fully induced into α-helical structure when inserted into the cassette holder, in distinct contrast to the cassette sequence used in the Parent peptide of the pres-ent study. Thus, it is tempting to speculate that the strong resistance to helix induction of the Fab:53–63 sequence may influence to some extent the preference for β-sheet structure of the chameleon sequence Fab:64–74 in the native protein. Certainly, such a possibility is worthy of further investigation.
Secondary structural specificity (SSS) determinants are important characteristics of polypeptide sequence influencing protein folding because they are very selective for one type of sequence and prevent the folding of alternative protein structure. These determinants are also important because they can exert long-range effects on neighboring sequences, in contrast to the current view that short sequences are generally structurally ambivalent (chameleon-like) and play only a minor role in dictating protein conformation. We have shown that a short 11-residue β-sheet sequence can contribute significantly to helix destabilization in the host protein, and can also have a long-range influence on overall protein structure. Future work to characterize other SSS determinants would increase our understanding of the intricate interplay of short- and long-range interactions in determining protein folding.
Materials and methods
Peptide synthesis
Peptides in this study were synthesized by solid-phase methodology, as described by Hodges et al. (1988) and Sereda et al. (1993) by the standard N-t-butyloxycarbonyl (t-Boc) approach (for review, see Merrifield 1997). Peptides were made on copoly(styrene, 1% divinylbenzene)-4-methylbenzhydrylamine-HCl (MBHA) resin with a 100–200 mesh and a substitution of 0.77 mmol amino groups per gram (Novabiochem, Switzerland). The following side chain protecting groups were used: benzyl (Thr, Ser), cyclohexyl (Asp), 4-methylbenzyl (Cys), trityl (Asn), and tosyl (Arg). A peptide-resin core (1.5 gram of MBHA resin, 1.1 mmol) was swelled and washed repeatedly with dichloromethane (DCM) and N,N-dimethylforamide (DMF) in a 25 mL polypropylene solid-phase extraction reservoir. Activation reagent O-benzotriazol-1-yl-1,1,3,3 tetramethyluronium hexafluorophosphate (HBTU, 0.45 M) was dissolved in DMF/DCM/ dimethylsulfoxide (DMSO) (85:10:5 v/v/v) and reacted with excess t-Boc-protected amino acid (1.1 equivalent to resin substitution) and excess diisopropylethylamine (DIEA, 1.5 equivalent to resin substitution) for 5 min. The activated amino acid ester (4.0 equivalent excess compared to resin substitution) was then coupled onto the solid-phase support by agitation for one h. Excess unreacted amino acids were removed by three alternating washes of DCM and DMF. The extent of amino acid coupling was monitored by the Kaiser ninhydrin test (Fontenot et al. 1991), and if the reaction was not complete, the coupling reaction was repeated. Peptide synthesis was continued for 15 residues, at which time the peptide-core was divided into eight equal aliquots for the synthesis of different analogs. Cleavage of the t-Boc and side chain protecting groups and the subsequent release of completed peptides from the MBHA resin support were achieved by hydrogen fluoride (HF) cleavage in the presence of the scavengers anisole (10% v/v) and 1,2 ethanedithiol (EDT) (1% v/v), magnetically stirred for 90 min in a sodium chloride (NaCl) waterbath at 4°C. The resin was then washed three times with cold diethyl ether for the removal of scavengers and amino acid protecting groups. Subsequent resin extraction with glacial acetic acid and overnight lyophilization yields the crude peptide.
Crude peptides were purified by reversed-phase chromatography (RPC) (reviewed in Mant et al. 1997) on a Zorbax semipreparative 300 SB-C8 column (250 × 9.4 mm I.D., 5 μm particle size, 300 Å pore size; Agilent Technologies) by linear AB gradient elution (0.2% acetonitrile/min) at a flow rate of 2 mL/min, where eluent A is 0.05% aqueous TFA and eluent B is 0.05% TFA in acetonitrile. The purification was carried out at RT. The purity of the peptide was verified by analytical RPC on a Zorbax analytical 300 SB-C8 column (150 × 2.1 mm I.D., 5 μm, 300 Å), by quantitative amino acid analysis (Beckman Model 6300 amino acid analyzer), and by electrospray mass spectrometry on a Fisons Quattro (Fisons, Pointe-Claire, Quebec, Canada).
Disulfide-bridge formation
The disulfide-bridged peptide was formed by air oxidation of purified peptide in its reduced state in 100 mM NH4HCO3 buffer (pH 8.5) at RT. The reaction was allowed to proceed with stirring overnight. Oxidation was terminated by neutralization with dilute acetic acid, and the desired disulfide-bridged peptide was purified by RPC (described above).
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy was performed on a Jasco-720 spectropolarimeter (Jason, MD, USA) interfaced with an Epson Equity 386/25 computer running the Jasco DP-500/PS2 system software (version 1.33a). A Lauda model circulating water bath (Brinkman Instruments, Ontario) was used to control the temperature of the optic cell chamber. For wavelength scan analysis, a 1.0 mM stock solution of each disulfide-bridged peptide in 100 mM KCl, 50 mM PO4 (K2HPO4/KH2PO4) buffer, pH 7, was diluted 20-fold, loaded into a silica CD cell of 0.02 cm path length, and its ellipticity subsequently scanned from 190 to 250 nm. Each peptide was scanned in 100 mM KCl, 50 mM PO4, pH 7.0 in the absence or presence of 50% trifluoroethanol (TFE) (v/v) at 25°C. CD data were presented as mean residue molar ellipticity [θ] (degrees • cm2 • dmol−1), calculated from the equation
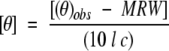 |
where (θ)obs is the observed ellipticity in millidegrees, MRW is the mean residue molecular weight (molecular weight of the peptide divided by its number of residues), l is the optical path length of the CD cell in cm, and c is the peptide concentration (mg/mL), determined by quantitative amino acid analysis. Each peptide spectrum was the average of ten scans collected at 0.1 nm intervals. The uncertainty in the molar ellipticity values is +/− 300 degrees • cm2 • dmol−1.
For chemical denaturation experiments, approximately 1.0 mM stock peptide solutions were similarly diluted with appropriate volumes of buffer (100 mM KCl, 50 mM PO4, pH 7.0) and 8.0 M guanidinium hydrochloride (GdnHCl) in buffer to a give a series of increasing denaturant concentrations. The concentration of peptides ranged from 87 to 104μM. These peptides and denaturant mixtures were left to equilibrate overnight at RT and were scanned the following day at 222 nm to monitor the unfolding of peptide in the presence of GdnHCl. To ensure accuracy and reproducibility of the denaturation curves, selected analogs were rescanned to ascertain proper denaturant equilibration.
Calculation of the difference in free energy of unfolding, ΔΔGu
A two-stage unfolding model was used to derive peptide stability values from GdnHCl denaturation results. The ellipticity readings were normalized to the fraction of peptide folded, ff and fraction unfolded, fu
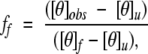 |
where [θ ]f and [θ ]u represent the mean residue molar ellipticity for the fully folded and fully unfolded species, respectively, and the [θ]obs is the observed ellipticity at a given denaturant concentration. The free energy of unfolding was derived from the equation:
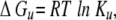 |
where Ku is the equilibrium constant of the unfolding process. In the case of disulfide-bridged peptides, where the unfolding process is concentration-independent,
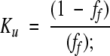 |
thus,
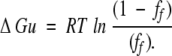 |
Estimates of the free energy of unfolding in the absence of denaturant ΔGu(H20) and slope term m can be obtained by linear extrapolation to zero by plotting
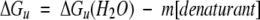 |
(Pace 1986; Shortle 1989), where m is the slope term associated with unfolding. Because small errors in the slope term m may lead to large errors in the extrapolated ΔGu(H20) value (Serrano et al. 1990), the change in free energy of unfolding between peptide analogs (ΔΔGu values) was calculated using the equation
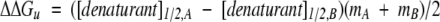 |
(Serrano et al. 1990) because the concentration of denaturant at 50% unfolding, [denaturant]1/2, was found by Kellis et al. (1989) to be the most reproducible quantity from repetitive experiments over a period of time and experimental methods. Thus, the relative difference in stability between two analogs is most accurately measured using this equation. It has also been reported that ΔΔGu (in denaturant) closely approximates ΔΔG(H20) if the slope terms of the analogs are similar (Serrano et al. 1990). The m values of the different analogs in this study were similar (1.66 +/− 0.07). We determined the slope term, m, by linear intrapolation of the ΔGu values in the range of +/− 0.5 M units of denaturant about the transition midpoint. We compared this method to that of Santoro and Bolen (1988), who used a nonlinear least square fitting protocol with pre- and postdenaturation baselines fitting into the sigmoidal around the transition midpoint. The linear correlation of m values obtained by the two methods correlated with high confidence (r2 = 0.95).
Numerical analysis of secondary structural specificity determinants
A search of the NN dipeptide motif was performed using SEQSEE and VADAR software (Wishart et al. 1994, 1997) on a nonredundant version of the Brookhaven Protein Database generated with 1021 protein sequences.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research Group in Protein Structure and Function (R.S.H.), the University of Colorado Health Sciences Center (R.S.H.), an NIH grant to R.S.H. (R01GM 61855), and studentships from the National Science and Engineering Research Council of Canada and Alberta Heritage Foundation for Medical Research (S.C.K.). We thank Kim Oikawa, Bob Luty, Les Hicks, Lorne Burke, and Paul Semchuk for technical expertise.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110ps.4170102.
References
- Argos, P. 1987. Analysis of sequence-similar pentapeptides in unrelated protein tertiary structures. Strategies for protein folding and a guide for site-directed mutagenesis. J. Mol. Biol. 197 331–348. [DOI] [PubMed] [Google Scholar]
- Aurora, R. and Rose, G.D. 1998. Helix capping. Protein Sci. 7 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg, J., Jensen, M.H., Sneppen, K., and Tiana, G. 2001. Hydrogen bonds in polymer folding. Phys. Rev. Lett. 86 1031–1033. [DOI] [PubMed] [Google Scholar]
- Chakrabartty, A., Kortemme, T., and Baldwin, R.L. 1994. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 3 843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-H., Yang, J.T. and Chau, K.H. 1974. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13 3350–3359. [DOI] [PubMed] [Google Scholar]
- Chou, P.Y. and Fasman, G.D. 1974. Pedictions of protein conformation. Biochemistry 13 222–245. [DOI] [PubMed] [Google Scholar]
- Chou, P.Y. and Fasman, G.D. 1978. Empirical predictions of protein conformation. Annu Rev Biochem 47 251–276. [DOI] [PubMed] [Google Scholar]
- Cohen, F.E. 1999. Protein misfolding and prion diseases. J. Mol. Biol. 293 313–320. [DOI] [PubMed] [Google Scholar]
- Cooper, T.M. and Woody, R.W. 1990. The effect of conformation on the CD of interacting helices: A theoretical study of tropomyosin. Biopolymers 30 657–676. [DOI] [PubMed] [Google Scholar]
- Cregut, D., Civera, C., Macias, M.J., Wallon, G., and Serrano, L. 1999. A tale of two secondary structure elements: when a β-hairpin becomes an α-helix. J. Mol. Biol. 292 389–401. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. 1999. Polymer principles and protein folding. Protein Sci. 8 1166–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman, R., Chao, H.G., Mueller, L., Lavoie, T.B., Shen, L., Novotny, J., and Matsueda, G.R. 1995. Characterization of a new four-chain coiled-coil: Influence of chain length on stability. Protein Sci. 4 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot, J.D., Ball, J.M., Miller, M.A., David, C.M., and Montelaro, R.C. 1991. A survey of potential problems and quality control in peptide synthesis by the fluorenylmethoxycarbonyl procedure. J. Pep. Res. 1 19–25. [PubMed] [Google Scholar]
- Harbury, P.B., Zhang, T., Kim, P.S., and Alber, T. 1993. A switch between two-, three- and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262 1401–1407. [DOI] [PubMed] [Google Scholar]
- Hodges, R.S. 1996. Boehringer Mannheim award lecture 1995. La conference Boehringer Mannheim 1995. De novo design of alpha-helical proteins: basic research to medical applications. Biochem. Cell. Biol. 74 133–154. [DOI] [PubMed] [Google Scholar]
- Hodges, R.S., Saund, A.K., Chong, P.C.S., St-Pierre, S.A., and Reid, R.E. 1981. Synthetic model for two-stranded α-helical coiled-coils. J. Biol. Chem. 256 1214–1224. [PubMed] [Google Scholar]
- Hodges, R.S., Semchuk, P.D., Taneja, A.K., Kay, C.M., Parker, J.M.R., and Mant, C.T. 1988. Pept. Res. 3 123–137. [Google Scholar]
- Houston, M.E., Jr., Campbell, A.P., Lix, B., Kay, C.M., Sykes, B.D., and Hodges, R.S. 1996. Lactam bridges stabilization of alpha-helices: the role of hydrophobicity in controlling dimeric versus monomeric alpha-helices. Biochemistry 35 10041–10050. [DOI] [PubMed] [Google Scholar]
- Kammerer, R.A., Schulthess, T., Landwehr, R., Engel, J., Aebi, U., and Steinmetetz, M.O. 1998. An autonomous folding unit mediates the assembly of two-stranded coiled coils. Proc. Natl. Acad. Sci. 95(23): 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, P.A. 1997. Hydrophobicity regained. Protein Sci. 6 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, J.T., Jr., Nyberg, K., and Fersht, A.R. 1989. Energetics of complementary side-chain packing in a protein hydrophobic core. Biochemistry 28 4914–4922. [DOI] [PubMed] [Google Scholar]
- Kohn, W.D., Mant, C.T., and Hodges, R.S. 1997. α-Helical protein assembly motifs. J. Biol. Chem. 272 2583–2586. [DOI] [PubMed] [Google Scholar]
- Kwok, S.C., Tripet, B., Man, J.H., Chana, M.S., Lavigne, P., Mant, C.T., and Hodges, R.S. 1998a. Structural cassette mutagenesis in a de novo designed protein: proof of a novel concept for examining protein folding and stability. Biopolymers (Pept. Sci.) 47 101–123. [DOI] [PubMed] [Google Scholar]
- Kwok, S.C., Mant, C.T., and Hodges, R.S. 1998b. Effect of α-helical and β-sheet propensities of amino acids on protein stability. Peptides 1998: Proc. 25th European Peptide Symposium, pp.34–35.
- Kwok, S.C., Mant, C.T., and Hodges, R.S. 2001. Using conformationally-restricted lactam-bridged peptides to examine the effects of α-helical propensity of solvent-exposed residues. Peptides: the wave of the future: Proc. 2nd International and 17th American Peptide Symposium, (accepted).
- Lattman, E.E. and Rose, G.D. 1993. Protein folding—What's the question? Proc. Natl. Acad. Sci. 90 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S.Y.M., Taneja, A.K., and Hodges, R.S. 1984. Synthesis of a model protein of defined secondary and quaternary structure: Effect of chain length on the stabilization and formation of two-stranded α-helical coiled-coils. J. Biol. Chem. 259 13253–13261. [PubMed] [Google Scholar]
- Lee, D.L., Lavigne, P., and Hodges, R.S. 2001. Are trigger sequences essential in the folding of two-stranded α-helical coiled-coils? J. Mol. Biol. 306 539–553. [DOI] [PubMed] [Google Scholar]
- Lehmann, M., Pasamontes, L., Lassen, S.F., and Wyss, M. 2000. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta. 1543 408–415. [DOI] [PubMed] [Google Scholar]
- Lu, M., Shu, W., Ji, H., Spek, E., Wang, L., and Kallenbach, N.R. 1999. Helix capping in the GCN4 leucine zipper. J. Mol. Biol. 288 743–752. [DOI] [PubMed] [Google Scholar]
- Lyu, P.C., Liff, M.I., Marky, L.A., and Kallenbach, N.R. 1990. Side chain contributions to the stability of alpha-helical structure in peptides. Science 250 669–673. [DOI] [PubMed] [Google Scholar]
- Mant, C.T, Kondejewski, L.H., Cachia, P.J., Monera, O.D., and Hodges, R.S. 1997. Practical aspects of analysis of synthetic peptides by high-performance liquid chromatography. Meth. Enzymol. 289 426–469. [DOI] [PubMed] [Google Scholar]
- Meizi, M. 1998. Chameleon sequences in the PDB. Protein Eng. 11 411–414. [DOI] [PubMed] [Google Scholar]
- Merrifield B. 1997. Concept and early development of solid-phase peptide synthesis. Meth. Enzymol. 289 3–13. [DOI] [PubMed] [Google Scholar]
- Micklatcher, C. and Chmielewski, J. 1999. Helical peptide and protein design. Curr. Opin. Chem. Biol. 3 724–729. [DOI] [PubMed] [Google Scholar]
- Minor, D.L., Jr. and Kim, P.S. 1994. Measurement of the beta-sheet-forming propensities of amino acids. Nature 367 660–663. [DOI] [PubMed] [Google Scholar]
- Minor, D.L., Jr. and Kim, P.S. 1996. Context-dependent secondary structure formation of a designed protein sequence. Nature 380 730–734. [DOI] [PubMed] [Google Scholar]
- Monera, O.D., Sereda, T.J., Zhou, N.E., Kay, C.M., and Hodges, R.S. 1995. Relationship of side-chain hydrophobicity and α-helical propensity on the stability of single-stranded amphipathic α-helix. J. Pep. Res. 1 312–329. [DOI] [PubMed] [Google Scholar]
- Monne, M., Hermansson, M., and von Heijne, G. 1999. A turn propensity scale for transmembrane helices. J. Mol. Biol. 288 141–145. [DOI] [PubMed] [Google Scholar]
- O'Neil, K.T. and Degrado, W.F. 1990. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250 646–651. [DOI] [PubMed] [Google Scholar]
- Ohmura, T., Ueda, T., Hashimoto, Y., and Imoto, T. 2001. Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Eng. 14 421–425. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. 1986. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131 266–280. [DOI] [PubMed] [Google Scholar]
- Penel, S. and Doig, A.J. 2001. Rotamer strain energy in protein helices quantification of a major force opposing protein folding. J. Mol. Biol. 305 961–968. [DOI] [PubMed] [Google Scholar]
- Prusiner, S.B. 1997. Prion diseases and the BSE crisis. Science 278 245–251. [DOI] [PubMed] [Google Scholar]
- Santoro, M.M. and Bolen, D.W. 1988. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenymethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27 8063–8068. [DOI] [PubMed] [Google Scholar]
- Sereda, T.J., Mant, C.T., Quinn, A.M., and Hodges, R.S. 1993. Effect of the α-amino group on peptide retention behavior in reversed-phase chromatography. Determination of the pKa values of the α-amino group of 19 different N-terminal amino acid residues. J. Chromatogr. 646 17–30. [DOI] [PubMed] [Google Scholar]
- Serrano, L., Horovitz, A., Avron, B., Mycroft, M., and Fersht, A.R. 1990. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry 29 9343–9352. [DOI] [PubMed] [Google Scholar]
- Shortle, D. 1989 Probing the determinants of protein folding and stability with amino acid substitutions. J. Biol. Chem. 264 5313–5318. [PubMed]
- Sönnichsen, F.D., Van Eyk, J.E., Hodges, R.S., and Sykes, B.D. 1992. The effect of trifluoroethanol on protein secondary structure: An NMR and CD study using a synthetic actin peptide. Biochemistry 31 8790–8798. [DOI] [PubMed] [Google Scholar]
- Su, J.Y., Hodges, R.S., and Kay, C.M. 1994. Effects of chain length on the formation and stability of synthetic alpha-helical coiled coils. Biochemistry 33 15501–15510. [DOI] [PubMed] [Google Scholar]
- Tripet, B., Wagschal, K., Lavigne, P., Mant, C.T., and Hodges, R.S. 2000. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position "d". J. Mol. Biol. 300 377–402. [DOI] [PubMed] [Google Scholar]
- Wagschal, K., Tripet, B., Lavigne, P., Mant, C., and Hodges, R.S. 1999. The role of position "a" in determining the stability and oligomerization state of α-helical coiled coils: 20 amino acid stability coefficients in the hydrophobic core of proteins. Prot. Sci. 8 2312–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, W.Y. and Milner-White, E.J. 1999. A natural grouping of motifs with an aspartate or asparagine residue forming two hydrogen bonds to residues ahead in sequence: Their occurrence at α-helical N termini and in other situations. J. Mol. Biol. 286 1633–1649. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Fortin S., Woloschuk, D.R., Wong, W., Rosborough, T., van Domselaar, G., Schaeffer, J., and Szafron, D. 1997. A platform-independent graphical user interface for SEQSEE and XALIGN. Comput. Appl. Biosci. 13 561–562. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Boyko, R.F., Willard, L., Richards, F.M., and Sykes, B.D. 1994. SEQSEE: A comprehensive program suite for protein sequence analysis. Comput. Appl. Biosci. 10 121–132. [DOI] [PubMed] [Google Scholar]
- Xu, J., Baase, W.A., Baldwin, E., and Matthews, B.W. 1998. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 7 158–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B.Y., Lavigne, P., Kay, C.M., Hodges, R.S., and Privalov, P.L. 1999. Contribution of translational and rotational entropy to the unfolding of a dimeric coiled-coil. J. Phys. Chem. 103 2270–2278. [Google Scholar]
- Yu, Y., Monera, O.D., Hodges, R.S., and Privalov, P.L. 1996. Ion-pairs significantly stabilize coiled-coils in the absence of electrolyte. J. Mol. Biol. 255 367–372. [DOI] [PubMed] [Google Scholar]
- Zhou, N.E., Monera, O.D., Kay, C.M., and Hodges, R.S. 1994. α-Helical propensities of amino acids in the hydrophobic face of an amphipathic α-helix. Protein and Pept. Lett 1 114–119. [Google Scholar]
- Zhou, N.E., Zhu, B.Y., Kay, C.M., and Hodges R.S. 1992. The two-stranded α-helical coiled-coil is an ideal model for studying protein stability and subunit interactions. Biopolymers 32 419–426. [DOI] [PubMed] [Google Scholar]