Abstract
Autonomously folding β-hairpins (two-strand antiparallel β-sheets) have become increasingly valuable tools for probing the forces that control peptide and protein conformational preferences. We examine the effects of variations in sequence and solvent on the stability of a previously designed 12-residue peptide (1). This peptide adopts a β-hairpin conformation containing a two-residue loop (D-Pro-Gly) and a four-residue interstrand sidechain cluster that is observed in the natural protein GB1. We show that the conformational propensity of the loop segment plays an important role in β-hairpin stability by comparing 1 with DP→ N mutant 2. In addition, we show that the sidechain cluster contributes both to conformational stability and to folding cooperativity by comparing 1 with mutant 3, in which two of the four cluster residues have been changed to serine. Thermodynamic analysis suggests that the high loop-forming propensity of the DPG segment decreases the entropic cost of β-hairpin formation relative to the more flexible NG segment, but that the conformational rigidity of DPG may prevent optimal contacts between the sidechains of the GB1-derived cluster. The enthalpic favorability of folding in these designed β-hairpins suggests that they are excellent scaffolds for studying the fundamental mechanisms by which amino acid sidechains interact with one another in folded proteins.
Keywords: β-sheet, β-hairpin, protein design, hydrophobic cluster
It has recently become possible to design short peptides that adopt β-sheet conformations in aqueous solution (Nesloney and Kelly 1996; de Alba et al. 1997a,b; Gellman 1998; Andersen et al. 1999; Carulla et al. 2000; Serrano 2000; Cochran et al. 2001a; Searle 2001). These model systems are valuable tools for probing the factors that determine β-sheet stability without interference from a specific tertiary structural context. The hairpin motif is critical for generating small, autonomously folding β-sheets. β-hairpins are common substructures within folded proteins in which two antiparallel strands are connected via a short loop (Sibanda and Thornton 1985,Sibanda and Thornton 1993; Sibanda et al. 1989; Gunasekaran et al. 1997). There are now many reports of peptides containing 9–16 residues that fold autonomously to β-hairpin conformations in aqueous solution (for leading references, see Serrano 2000; Searle 2001). Three-stranded antiparallel β-sheets have been created in aqueous solution by interweaving two hairpins (Schenck and Gellman 1998; de Alba et al. 1999; Koepf et al. 1999; Griffiths-Jones and Searle 2000; Lopez de la Paz et al. 2001), and a self-associating four-stranded β-sheet has been described (Mayo and Ilyina 1998). Autonomously folding parallel β-sheet secondary structure can be generated in aqueous solution if a peptide-like diamine unit is used to connect strands via its C-termini (Fisk and Gellman 2001). These model systems are beginning to yield insights on β-sheet secondary structure analogous to those that have been obtained for α-helical secondary structure with short, autonomously folding peptides (Chakrabartty and Baldwin 1995; Baldwin and Rose 1999; Bolin and Millhauser 1999).
β-hairpin model systems have been used to identify several factors that are crucial to antiparallel β-sheet stability, including the conformational propensity of the loop-forming segment (de Alba et al. 1997a,b; Haque and Gellman 1997; Ramírez-Alvarado et al. 1997; Syud et al. 1999) and sidechain–sidechain interactions between neighboring strands (Ramírez-Alvarado et al. 1996; Maynard et al. 1998; Kobayashi et al. 2000; Russell and Cochran 2000; Santiveri et al. 2000; Espinosa et al. 2001; Syud et al. 2001). Here, we explore the stabilizing effects of loop propensity and interstrand sidechain interactions in the context of a designed β-hairpin that contains a hydrophobic cluster from the protein GB1. We show that altering the rigidity of the loop segment influences β-hairpin formation along the entire length of the strands, which highlights the cooperativity of β-hairpin formation in these short peptides. In addition, we show that favorable interstrand sidechain–sidechain contacts are essential for this cooperativity.
Results
Design
We have examined the effects of altering both the solvent and the peptide sequence to probe the forces that influence the stability of the β-hairpin conformation adopted by 1 (Scheme 1 ▶). Peptide 1 contains two five-residue strand segments (RWQYV and KFTVQ) connected via a D-Pro-Gly loop, which is a strong promoter of antiparallel β-sheet interactions between flanking residues (Haque et al. 1994,Haque et al. 1996; Haque and Gellman 1997; Ragothama et al. 1998; Stanger and Gellman 1998). The strand segments of 1 contain four hydrophobic residues, Trp-2, Tyr-4, Phe-9, and Val-11, which have been borrowed from the C-terminal β-hairpin of the small protein GB1 (Gallagher et al. 1994; Gronenborn et al. 1991). The C-terminal fragment of GB1, residues 41–56, has been shown to form a native-like β-hairpin in aqueous solution (Blanco et al. 1994; Honda et al. 2000). In peptide 1, as in the C-terminal β-hairpin of GB1, the first pair and the second pair of hydrophobic residues are arranged in i,i+2 fashion. In addition, these four residues in 1 are positioned so that they can form a native-like cluster if the DPG segment induces the expected β-hairpin conformation. Previously, we have shown that 1 adopts the intended β-hairpin conformation in aqueous solution (Espinosa and Gellman 2000). The folded-state population for 1 at 2°C is ∼61%.
Scheme 1.
In the present study, we have examined the effects of both conformation-stabilizing (methanol and 2,2,2-trifluoroethanol [TFE]) and conformation-destabilizing (urea) additives on the β-hairpin population of 1. We have also analyzed β-hairpin stability in two variants of 1, peptides 2 and 3 (Scheme 1 ▶). (Our discussion of stability as a function of conditions or sequence follows the approach of others who have studied autonomously folding β-hairpins; when we write that one peptide is more stable than another, we mean that the β-hairpin population of the former is higher than the β-hairpin of the latter at 3°C.) Peptide 2 contains L-asparagine in place of the D-proline residue of 1. L-Asn-Gly (NG) is the most common sequence for residues i+1 and i+2 of a type I` β-turn in natural proteins of known structure (Hutchinson and Thornton 1994), and type I` turns commonly form two-residue loops in β-hairpins of proteins (Sibanda and Thornton 1985,Sibanda and Thornton 1993; Sibanda et al. 1989). For this reason, the NG loop segment has been used in several β-hairpin designs (Ramírez-Alvarado et al. 1996; de Alba et al. 1997a; Maynard et al. 1998). A previous comparison, in a different peptide context, showed that a DPG loop induces a higher β-hairpin population than an NG loop (Stanger and Gellman 1998; Syud et al. 1999). Here, we provide a detailed thermodynamic comparison of DPG and NG as β-hairpin loops. Peptide 3 contains serine in place of the two hydrophobic residues in the C-terminal strand of 1, F9 and V11. Comparison of 1 and 3 allows us to assess the impact on β-hairpin stability of interstrand interactions among hydrophobic sidechains.
Peptide 1 forms a β-hairpin with a native-like hydrophobic sidechain cluster
As previously reported, 1 in aqueous solution at 3°C displays numerous NOEs between nonadjacent residues (Espinosa and Gellman 2000); all of these NOEs are consistent with adoption of a well-defined β-hairpin conformation. Figure 1A ▶ shows the characteristic CαH–CαH NOEs observed for 1 between W2 and V11 and between Y4 and F9. These and other NOEs involving nonadjacent residues were used as restraints for determination of the solution structure of 1 with the program DYANA (Guntert et al. 1997). Figure 2A ▶ shows an overlay of the 10 best structures from this analysis, backbone-heavy atoms only, for residues 2–11 (the two terminal residues were highly disordered). Root mean square deviation (RMSD) among the 20 best DYANA structures is 0.58 ± 0.16 Å for backbone heavy atoms; inclusion of sidechain heavy atoms leads to an RMSD of 1.25 ± 0.22 Å. Although no hydrogen-bonding restraints were used in the DYANA calculations, most of the best structures display four interstrand C=O–H-N hydrogen bonds, between V5 and K8 and between Q3 and T10.
Fig. 1.
Selected regions of ROESY spectra for peptides 1–3 (D2O at pH 3.8 and 3°C and 200 msec mixing time). (A) CαH–CαH NOEs for 1.6 mM 1. (B) CαH–CαH NOEs for 2.0 mM 2. (C) CαH–CαH NOEs for 3.1 mM 3.
Fig. 2.
(A) Superposition of the best 10 structures (backbone atoms only) for peptide 1 calculated based on NMR data using the program DYANA. (B) Comparison between the best NMR-derived structure of peptide 1 (lower) and the structure of residues 41–56 in the NMR structure of protein GB1 (upper). (C) Superposition of the best 10 structures (backbone atoms only) for cyclic peptide 5 calculated based on NMR data. (D) Superposition of the best 10 structures (backbone atoms only) for peptide 2 calculated based on NMR data.
Figure 2B ▶ compares the best DYANA structure of 1 with the C-terminal β-hairpin of GB1, on which the design of 1 was based (the GB1 fragment is taken from an NMR structure of the entire protein [Gronenborn et al. 1991; Protein Data Bank code 2GB1]). The hydrophobic cluster residues of 1, W2, Y4, F9, and V11 correspond to residues W43, Y45, F52, and V54 of GB1; only these four sidechains are shown in Figure 2B ▶. The two hairpins differ in that the loop segment is longer in GB1 than in 1 (DATK vs. DPG). It is evident from Figure 2B ▶ that the hairpin conformation of 1 is more highly twisted than the hairpin conformation of GB1 41–56 (most β-sheets in proteins are twisted [Chothia 1973]). Nevertheless, the spatial disposition of the four hydrophobic sidechains is similar in the two structures. In a flexible peptide like 1, the sidechains may explore several alternative spatial arrangements in the folded state, and the image of 1 in Figure 2B ▶ should not be interpreted to imply that there is necessarily a unique structure. The similarity between the sidechain cluster shown for 1 and that shown for the C-terminal β-hairpin of GB1, however, indicates that the unnatural backbone of 1 allows a native-like arrangement of sidechains.
β-hairpin population analysis: Identification of a reference peptide for the fully folded state
Evaluation of sequence-stability relationships among β-hairpins requires the ability to estimate β-hairpin population in solution. A number of NMR- and CD-based approaches have been reported for β-hairpin population analysis (for leading references, see Gellman 1998). We have focused on the chemical shifts of α-protons (δHα) for this purpose, because these NMR parameters are known to be sensitive to secondary structure (Wishart et al. 1991,Wishart et al. 1992). Because interconversion between unfolded and β-hairpin states of short peptides is rapid on the NMR timescale, chemical shift data represent a population-weighted average. We have used specifically designed reference peptides to provide δHα values for the fully unfolded (δU) and fully folded (δF) states (Syud et al. 1999, 2001). For 1, the unfolded reference is the diastereomer in which D-Pro is replaced by L-Pro (peptide 4); this change abolishes β-hairpin formation in a number of systems (Haque and Gellman 1997; Ragothama et al. 1998; Stanger and Gellman 1998). For 4, we detect none of the interstrand NOEs observed for D-Pro diastereomer 1. Our folded reference is a cyclic peptide, 5, in which the terminal residues of 1 are linked by a second DPG segment.
NMR evidence shows that a two-stranded antiparallel β-sheet conformation is very highly populated for 5, as required if this cyclic peptide is to represent the 100% β-hairpin conformation of linear peptide 1. Figure 3 ▶ summarizes the numerous NOEs observed between nonadjacent residues of 5 in aqueous solution. Figure 3A ▶ illustrates NOEs involving backbone protons (NH and/or CαH); this figure includes the two i,i+1 NOEs that indicate the reverse turns between G7 NH and K8 NH and between G14 NH and R1 NH (the numbering is analogous to that of the linear peptides; the "extra" loop residues are numbered 13 [D-Pro] and 14 [Gly]). Figure 3B ▶ shows NOEs involving sidechain protons. All of these NOEs are consistent with a two-stranded β-sheet that is closed off at each end by a DPG loop. Figure 2C ▶ shows an overlay of the 10 best structures resulting from NOE-restrained molecular dynamics analysis of 5 with DYANA. RMSD among the 20 best structures is 0.73 ± 0.23 Å for backbone heavy atoms only and 1.58 ± 0.29 Å for all heavy atoms.
Fig. 3.
Summary of NOE interactions involving nonadjacent residues, plus adjacent residue NOEs that define the turn segment, for cyclic peptide 5. (A) Backbone NOEs. (B) NOEs involving sidechains.
We estimated the β-sheet population of 5 based on the intensity of the W2 CαH–V11 CαH NOE, calibrated against the intraresidue CαH–CαH of G7. We assumed a W2 CαH–V11 CαH spacing of 2.32 Å in the β-sheet conformation, as observed for canonical antiparallel β-sheet in proteins. In addition, we assumed that the W2 CαH–V11 CαH spacing in any non-β-sheet conformation of 5 would be too large to give rise to an NOE. This analysis indicated that the β-sheet conformation of 5 is 100% populated. (It was not possible to carry out an independent analysis based on the Y4 CαH–F9 CαH NOE of 5, because this NOE was too close to the residual water signal and the diagonal for accurate integration.)
The high population of the β-sheet conformation of 5 was supported by qualitative H/D (hydrogen/deuterium) exchange studies. When 5 was dissolved in D2O (100 mM sodium acetate at pD 3.8 [uncorrected] and 3°C), the amide protons expected to be exposed to solvent in the β-sheet conformation (W2, Y4, G7, F9, V11, and G14) exchanged rapidly as detected by NMR. In contrast, the amide protons of the residues expected to be engaged in cross-strand hydrogen bonds in the β-sheet conformation (R1, Q3, V5, K8, T10, and Q12) were all visible in the NMR spectrum after 48 h in D2O. (All amide proton resonances of L-Pro peptide 4 disappeared from the spectrum within 10 min of dissolution in D2O.) The high stability of the β-sheet conformation of cyclic peptide 5 implied by these NMR and H/D exchange data are consistent with our previous analysis of similar cyclic peptides containing two DPG loops (Syud et al. 1999, 2001).
The β-hairpin population of 1 was estimated by interpolating α-proton chemical shifts (δHα) measured for 1 between the δHα values for the same residues in the unfolded reference peptide (L-Pro diastereomer 4) and the folded reference peptide (cyclic 5). We showed previously that not all residues of 1 are suitable for this analysis (Espinosa and Gellman 2000). Hydrogen-bonded strand residues Q3, V5, K8, and T10 were found to be optimal for population determination, apparently because the α-protons of these residues are oriented toward solvent rather than toward the other strand. Results for residues that are not hydrogen bonded to the opposite strand (W2, Y4, F9, and V11) displayed large and irregular variations, which we attributed to anisotropic effects of aromatic sidechains on the opposite strand. This distinction between hydrogen-bonded and nonhydrogen-bonded strand residues in terms of folded population analysis has been documented for a different designed β-hairpin (Syud et al. 1999, 2001), and the distinction may, therefore, prove to be a common feature, particularly among β-hairpins and β-sheets with sidechain clusters containing aromatic sidechains.
We estimated the β-hairpin population of 1 at 2°C by using summed δHα values for Q3, V5, K8, and T10 (∑δHα(1)) and for the corresponding residues of reference peptides 4 and 5 (equation 1); this approach
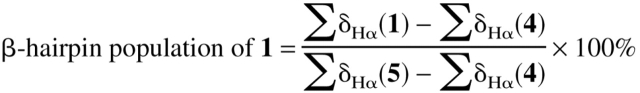 |
(1) |
follows that used by Searle et al. (1999) for a different designed β-hairpin. A β-hairpin population of 61% is deduced for 1 under these conditions. This conclusion is consistent with the results of population analysis of 1 based on the W2 CαH–V11 CαH NOE intensity, on the Y4 CαH–F9 CαH NOE intensity, or on the chemical shift difference between the diastereotopic α-protons of G7, as previously reported (Espinosa and Gellman 2000). Blanco et al. (1994) used a δHα approach to estimate that the β-hairpin population of the GB1 41–56 peptide is 42% populated at 5°C. (In this case, averaged δHα values from disordered regions of protein NMR structures were used as the 0% β-hairpin reference, and the δHα values from residues 41–56 in the NMR structure of the folded GB1 protein were used as the 100% reference.) This population estimate for GB1 41–56 has recently been confirmed by independent measurements (Cochran et al. 2001a). Thus, the β-hairpin conformation of peptide 1 appears to be somewhat more stable than the β-hairpin conformation of GB1 41–56.
Effects of solvent additives on β-hairpin stability
δHα data for residues Q3, V5, K8, and T10 were used to monitor qualitatively the effects of additives on the stability of the β-hairpin conformation of 1 in solution. We selected additives that have been widely used to stabilize (methanol or TFE) or destabilize (urea) folded protein and peptide conformations. Figure 4 ▶ summarizes the effect of these additives on α-proton chemical shifts of the indicator residues; the data are presented as ΔδHα = δHα(1) − δHα(4), where L-Pro diastereomer 4 in the appropriate solvent represents the fully unfolded state of 1. (ΔδHα values are usually calculated by using unfolded δHα data obtained from short peptides or from statistical analysis of NMR protein structures [Wishart et al. 1991ΔδHα values are usually calculated by using unfolded δHα data obtained from short peptides or from statistical analysis of NMR protein structures [Wishart et al. 1992]; use of a specific reference peptide like 4 should be more accurate because this approach accounts for local sequence effects. No NOEs between non-adjacent residues are observed for 4 in mixed water-alcohol solvents, indicating that 4 remains a good reference for the unfolded state in these solvents.) Also shown are ΔδHα = δHα(5) − δHα(4) data for the appropriate residues of cyclic peptide 5 in water, which approximates the fully folded state of 1 in water.
Fig. 4.
Chemical Shift Index, ΔδHα = δHα(observed) − δHα(unfolded reference peptide 4), for the inner hydrogen-bonded strand residues of peptide 1 in 9:1 H2O:D2O and 100 mM sodium deuteroacetate buffer at pH 3.8 (uncorrected) and 3°C with various solvent additives (solid bars: 6 M urea; open bars: no additive; horizontal stripes: 30% (v/v) TFE; vertical stripes: 50% (v/v) methanol), and for the same residues of cyclic peptide 5 (dotted bars, no additive). In each case, the reference data for 4 were obtained in the appropriate solvent system.
The ΔδHα data in Figure 4 ▶ for alcohol additives were obtained in 50% (v/v) methanol and in 30% (v/v) TFE. Incremental addition of methanol to an aqueous solution of 1 led to an increase in β-hairpin population, which appeared to reach a maximum at 40–50% (v/v) methanol based on NMR and CD measurements. These observations are consistent with prior reports of β-hairpin stabilization by alcohol cosolvents and a plateau in β-hairpin population near 1:1 water:cosolvent (Blanco et al. 1994; Searle et al. 1995; Ramírez-Alvarado et al. 1996; Maynard et al. 1998; Sharman and Searle 1998). Similar effects of alcohol cosolvents have been observed for α-helix formation by short peptides, and Luo and Baldwin (1997) have concluded that maximal α-helix populations detected in 40–50% (v/v) aqueous TFE do not necessarily correspond to 100% helix formation. In light of this conclusion for α-helices, it seems possible that the maximum extent of β-hairpin formation in cosolvent titrations may not represent 100% β-hairpin population. To probe this possibility in our system, we used ΔδHα data for 1 in 50% (v/v) methanol to represent the fully folded state in the calculation of β-hairpin population for 1 in aqueous solution (equation 1). The resulting population (77%) is significantly higher than the population calculated when ΔδHα data for cyclic peptide 5 in water are used to represent the fully folded state of 1 (61%). The population obtained using ΔδHα data for 5 is consistent with the average of two independent CαH–CαH NOE measurements (Espinosa and Gellman 2000); therefore, the higher population deduced for 1 in water when 1 in 50% (v/v) methanol is assumed to be fully folded suggests that this assumption may not be correct. We have recently observed comparable behavior for a different linear β-hairpin-forming peptide (Syud et al. 2001), i.e., use of ΔδHα data for a cosolvent-stabilized state of the linear peptide to represent the fully folded state in water implied significantly higher β-hairpin population than did use of ΔδHα data from a cyclic peptide in water to represent the fully folded state of the linear peptide in water. These results suggest that care should be taken when using NMR data for a linear peptide in a mixed solvent to represent the fully folded state of that peptide in water.
The data in Figure 4 ▶ show that addition of 6 M urea causes a decrease in β-hairpin population for 1. This observation is consistent with the denaturing effects exerted by urea on many other peptides and proteins. Analysis via equation 1 suggests 43% β-hairpin population for 1 in 6 M urea at 3°C, which suggests that the denaturant has only a modest effect on the stability of this β-hairpin. Maynard et al. (1998) have reported a similar resistance to urea-induced unfolding by a different designed β-hairpin.
Effect of loop sequence: Thermodynamic differences between β-hairpins with DPG and NG loops
NMR data show that 2 displays a significant population of the expected β-hairpin conformation, with a two-residue loop at NG in aqueous solution. Figure 5 ▶ summarizes NOEs between nonadjacent residues observed for 2. Figure 5A ▶ shows NOEs between backbone protons (NH and/or CαH; selected ROESY data are shown in Figure 1B ▶). Both of the CαH–CαH NOEs, consistent with the expected β-hairpin conformation, W2–V11 and Y4–F9, are observed, as is an NH–NH NOE between V5 and K8. In addition, a set of sequential NH–NH NOEs is observed around the expected loop segment (V5–N6, N6–G7, and G7–K8). Figure 5B ▶ summarizes nonadjacent NOEs involving protons on sidechains. Numerous interstand NOEs are observed, and all are consistent with a well-defined β-hairpin conformation. The nonadjacent residue NOEs and the sequential NOEs along the turn segment (a total of 22) were used as restraints for molecular dynamics with DYANA. Figure 2D ▶ shows an overlay of the 10 best structures from this analysis, backbone atoms only, residues 2–11. RMSD among the 20 best structures for these residues was 1.13 ± 0.39 Å for backbone heavy atoms only and 2.09 ± 0.48 Å for all heavy atoms.
Fig. 5.
Summary of NOE interactions involving nonadjacent residues, plus adjacent residue NOEs that define the turn segment, for peptide 2. (A) Backbone NOEs. (B) NOEs involving sidechains.
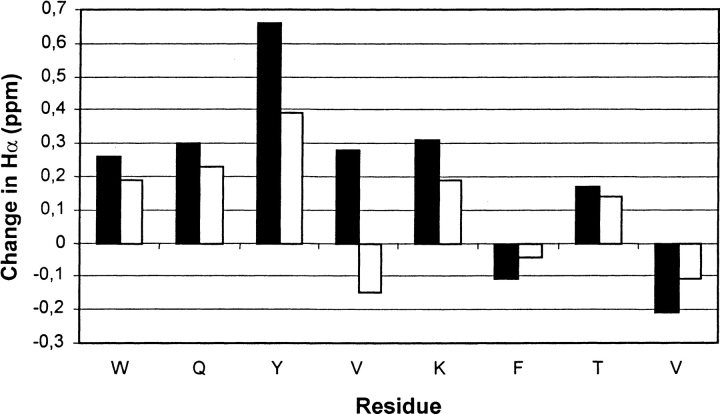
Figure 6 ▶ compares ΔδHα data for the nonterminal strand residues of 1 and 2 in aqueous solution at 3°C. The unfolded reference δHα values were obtained from L-Pro peptide 4. These data suggest qualitatively that the β-hairpin population of NG peptide 2 is smaller than the β-hairpin population of DPG peptide 1. β-Sheet secondary structure is generally indicated by ΔδHα > +0.1 (Wishart et al. 1992), and six of the eight strand residues of 1 meet this criterion. For F9 and V11 of 1, ΔδHα < 0, which we attribute to the magnetic anisotropy of nearby sidechains of W2 and Y4 in the β-hairpin conformation. For both of these residues, ΔδHα is closer to zero for 2 than for 1.
Fig. 6.
Chemical Shift Index, ΔδHα = δHα(observed) − δHα(unfolded reference peptide 4), for the inner hydrogen-bonded strand residues of peptides 1 (solid bars) and 2 (open bars) in 9:1 H2O:D2O and 100 mM sodium deuteroacetate buffer at pH 3.8 (uncorrected) and 3°C.
Quantitative analysis confirms that the β-hairpin population is lower for 2 than for 1. Analysis based on δHα data for indicator residues Q3, V5, K8, and T10 of 2 using equation 1 (δU values from L-Pro peptide 4 and δF values from cyclic peptide 5) suggests 41% β-hairpin population at 2°C for NG peptide 2. This conclusion is in excellent agreement with population determinations based on the intensities of the W2 CαH–V11 CαH NOE (39%) and the Y4 CαH–F9 CαH NOE (45%). Recall that the β-hairpin of DPG peptide 1 is ∼61% populated under these conditions. The observation that changing D-Pro to Asn exerts a consistent effect on β-hairpin population, as indicated by multiple parameters, supports our conclusion that β-hairpin formation in both 1 and 2 is cooperative.
We have previously used a nonlinear fitting method developed by Searle et al. (1999) to carry out van't Hoff thermodynamic analysis of β-hairpin formation by 1, based on changes in ∑ΔδHα for the indicator residues as a function of temperature (Espinosa and Gellman 2000). This approach has now been applied to 2. Figure 7 ▶ shows that the ΔδHα values of three of the four indicator residues decrease in a consistent manner over the temperature range (2°– 42°C), which suggests that 2 displays two-state behavior (fully unfolded vs. fully folded), as previously shown for 1 (Espinosa and Gellman 2000). (V5 could not be used to quantify the population of 2, because this residue is followed by proline in the reference peptides but by asparagine in 2; proline exerts unique conformational effects on immediately preceding residues [MacArthur and Thornton 1991].) Figure 8 ▶ shows ∑ΔδHα for both 1 and 2, along with the theoretical curves derived from fitting method of Searle et al. (1999). Table 1 summarizes the deduced thermodynamic parameters for β-hairpin formation by 2 at 25°C and provides the corresponding data for 1. In both peptides, β-hairpin formation is enthalphically favored and entropically disfavored at 25°C, although the balance between these two factors differs between the DPG and NG loops.
Fig. 7.
Relative population of the β-hairpin conformation of peptide 2 at 42°C vs. 2°C, as determined from δHα data at three hydrogen-bonded strand residues: Gln-3, Lys-8, and Thr-10. (As discussed in the text, data for Val-5 could not be used in this analysis.)
Fig. 8.
∑ΔδHα (defined in text) as a function of temperature for peptides 1 (circles) and 2 (crosses). The curves were generated via the fitting method of Searle et al. (1999), as described in the text.
Table 1.
Thermodynamic parameters (van't Hoff) for β-hairpin formation at 25°C (kcal/mole)
| Peptide | ΔH (kcal/mole) | ΔS (cal/mole deg) | ΔCp (cal/mole deg) |
| 1a | −3.2 ± 0.1 | −10 ± 0.2 | −98 ± 8 |
| 2 | −4.7 ± 0.1 | −18 ± 0.3 | −120 ± 10 |
a Espinosa and Gellman 2000.
Importance of the hydrophobic sidechain cluster to β-hairpin stability
We assessed the contribution of interstrand sidechain–sidechain interactions among the four residues corresponding to the hydrophobic cluster in GB1 by comparing 1 with double mutant 3, in which F9 and V11 have been replaced by serine. Serine was chosen because the sidechain is so short that contacts with sidechains on an adjacent strand are expected to provide little or no stabilization to the β-hairpin conformation. In addition, serine is reported to have a high β-sheet propensity in an edge strand, i.e., a strand that forms β-sheet interactions to only one side (Minor and Kim 1994); therefore, replacement of Phe and Val residues in 1 by Ser in 3 should not diminish the net β-sheet propensity of the strand residues.
NOE analysis indicates that 3 folds partially into a β-hairpin conformation, but that the β-hairpin population is lower for 3 than for analogs 1 and 2. CαH–CαH NOEs were observed between W2 and V11 and between Y4 and F9 (data in Figure 1C ▶), and an NH–NH NOE was observed between V5 and K8. There were too few interstrand NOEs for structure determination with DYANA. β-hairpin population analysis was performed based on the intensities W2 CαH–V11 CαH NOE (23%) and the Y4 CαH–F9 CαH NOE (45%). The difference between these two values suggests that the folding of 3 does not conform to a two-state model, i.e., that the conformational ensemble experienced by this peptide may include partial β-hairpin conformations. This conclusion was supported by observation that ΔδHα values for the indicator residues of 3 do not change in a coordinated way as a function of temperature (data not shown). For this reason, thermodynamic analysis of the folding of 3 was not performed.
Discussion
D-Pro-Gly vs. L-Asn-Gly as β-hairpin promoters
The data we have obtained for peptides 1 and 2 show that for the sequences RWQYVXGKFTVQ-NH2 β-hairpin population is greater when X = D-Pro than when X = L-Asn. This finding appears to represent a general trend, because we earlier observed similar behavior for a different designed β-hairpin series, RYVEVXGOKILQ-NH2 (O = ornithine) (Stanger and Gellman 1998; Syud et al. 1999). Cochran et al. (2001b) have made comparable observations with yet another set of β-hairpins.
We have compared van't Hoff thermodynamic parameters for β-hairpin formation by 1 and 2 to elucidate the difference in structure-promoting effects between DPG and NG (Table 1). One must be cautious when trying to interpret net thermodynamic parameters for a folding process in terms of detailed molecular phenomena. Nevertheless, we note that the observation that ΔS at 25°C for β-hairpin formation by 1 is less unfavorable than ΔS for β-hairpin formation by 2 is consistent with our design premise that DPG displays a greater conformational propensity than does NG to adopt the correct local conformation for β-hairpin folding. Interestingly, the entropic advantage of the DPG segment is balanced to some extent by an enthalpic advantage of the NG segment. This trend may indicate that the flexible NG segment allows the strands to achieve more energetically favorable contacts with one another than does the rigid DPG segment. We return to this point below.
Role of interstrand sidechain–sidechain contacts in β-hairpin stabilization
The behavior of double serine mutant peptide 3 suggests that the formation of an interstrand cluster of nonpolar sidechains contributes significantly to the stability of the β-hairpin conformation adopted by 1. The loss of favorable interstrand sidechain–sidechain interactions in 3 relative to 1 leads not only to a decrease in β-hairpin population, but also to a loss of two-state behavior. Thus, cooperative β-hairpin folding by 1 (and 2) results from an interplay between loop and strand segments. It seems likely that the favorable interstrand interactions occur between the sidechains themselves. Interstrand backbone–backbone hydrogen bonds have been proposed to stabilize β-hairpin conformations relative to the unfolded state (Constantine et al. 1995); however, interstrand hydrogen bonding should be comparable in the β-hairpin conformations of 1 and 3, unless larger sidechains (Val and Phe in 1 vs. Ser in 3) affect backbone solvation.
What is the nature of the interactions among the nonpolar GB1-derived sidechains in the β-hairpin conformations of 1 and 2? A negative change in heat capacity is characteristic of hydrophobically favorable processes (Baldwin 1986; Murphy et al. 1990; Livingstone et al. 1991). Thus, the observation that ΔCp < 0 for β-hairpin formation by 1 and 2 is consistent with an expectation that β-hairpin formation allows the nonpolar sidechains on each strand to shield poorly hydrated surfaces from the aqueous solvent by contact with nonpolar sidechains on the opposite strand. However, the observation that ΔH at 25°C is substantial and favorable for β-hairpin formation by 1, and even more favorable for β-hairpin formation by 2, suggests that dehydration of the GB1 sidechains in the β-hairpin conformation (i.e., a classical hydrophobic effect) is not the only contribution to β-hairpin stability resulting from interstrand sidechain clustering. At room temperature, the thermodynamic signature of a hydrophobically driven process (e.g., transfer of a simple hydrocarbon from aqueous solution to the pure liquid phase) is entropically favorable and enthalpically neutral, with a large negative change in heat capacity (Privalov and Gill 1989, and references therein). Indeed, this signature has been observed by Maynard et al. (1998) for the folding of a different designed β-hairpin, and these investigators proposed that the hydrophobic effect was the primary driving force for folding in that case. For 1 and 2, directly favorable interactions between nonpolar sidechains on opposite strands may contribute to β-hairpin stability along with dehydration effects (desolvation may be viewed as an indirect benefit of sidechain–sidechain contact). The fact that three of the four GB1 sidechains are aromatic may be significant in this regard, because it has been suggested (Makhatadze and Privalov 1994) that aromatic hydrocarbons are more prone to intrinsically favorable interactions (e.g., dispersion) than are aliphatic hydrocarbons. Griffiths-Jones and Searle (2000) have reached similar conclusions regarding aromatic sidechain interactions from thermodynamic analysis of the N-terminal β-hairpin in a designed three-stranded β-sheet. The enhancement of the favorable enthalpy of β-hairpin formation we observe on replacing the DPG loop with NG is consistent with the idea that the GB1 sidechains engage in enthalpically favorable interstrand contacts, because the more flexible NG loop may allow sidechains on opposite strands to optimize their contact geometries.
Comparison with related studies
The β-hairpin formed by GB1 residues 41–56 has inspired considerable interest. This segment was the first β-hairpin from a natural protein shown to fold autonomously in aqueous solution, i.e., when removed from the native protein (Blanco et al. 1994). This peptide has been examined by at least four different research groups (Blanco et al. 1994; Muñoz et al. 1997; Honda et al. 2000; Cochran et al. 2001a), and there has been disagreement on the extent of folding in solution. The original and most recent reports on GB1 41–56 suggest that the β-hairpin is ∼40% populated at low temperature in water; higher population estimates from other investigators under comparable conditions may have resulted from the incorrect assumption that ΔCp = 0 for folding. Interestingly, the extents of folding are similar when the GB1 cluster is combined with an NG loop (peptide 2) and when the GB1 cluster is in its natural context (GB1 41–56), even though the loop in GB1 (DATK) is longer and presumably more flexible. Kobayashi et al. (2000) have examined the effect on β-hairpin formation by GB1 41–56 of several alanine mutations. These investigators found that loss of the Tyr or Phe sidechain caused a substantial decrease in β-hairpin stability. Loss of the Trp sidechain caused a smaller decrease in β-hairpin stability, but loss of the Val sidechain had little effect. These results are consistent with our analysis of the double serine mutant of 1.
Cochran et al. (2001a) have recently described a set of short β-hairpin-forming peptides loosely related to GB1 41–56. In some of these peptides, the DATK loop from GB1 has been replaced by two-residue loops, including GN, NG, and DPN, which are comparable to the short loops we employ in 1–3. A major innovation in the design of Cochran et al. (2001a) is the use of Trp for all four of the nonpolar sidechains (these peptides were dubbed "tryptophan zippers"). The "trpzip" β-hairpins display remarkable behavior: higher conformational stability and higher folding cooperativity than any other autonomously folding β-hairpin or β-sheet. Several conclusions from the trpzip study are complementary to conclusions reached here. For example, Cochran et al. (2001a) show that decreasing the number of Trp sidechains in the nonpolar cluster leads to a decrease in β-hairpin stability and folding cooperativity, which is comparable to our findings on converting two of the nonpolar sidechains to serine (1 vs. 3). Cochran et al. (2001a) compare the thermodynamics of β-hairpin folding for NG and DPN turns, which is related to our comparison of NG and DPG turns (1 vs. 2). The folding of the more flexible NG trpzip peptide is less favorable entropically but more favorable enthalpically than folding of the DPN trpzip peptide (at the respective melting temperatures), and we find an analogous difference between 2 and 1 (at 25°C, Table 1). There are some contrasts between our results and those of Cochran et al. (2001a). At or slightly below room temperature, the NG and DPN trpzip peptides show very similar β-hairpin populations, whereas our DPG peptide (1) shows substantially greater β-hairpin population in this temperature range than does our NG peptide (2). When Cochran et al. (2001a) replaced the tetra-Trp cluster with the GB1 cluster (Trp, Tyr, Phe, and Val) in a 12-residue trpzip peptide with a GN loop, they found no evidence of β-hairpin formation. The investigators therefore concluded that the native cluster "is not sufficient to maintain a significant hairpin population without additional stabilizing elements." In contrast, we observe substantial β-hairpin folding for 12-residue 2, which contains the native cluster and a flexible NG loop. In addition, we have recently reported that β-hairpin formation can be observed in a designed 20-residue peptide in which the GB1 cluster is separated by five-residue segments from a DPG loop (Espinosa et al. 2001). The similarities and contrasts between our results and those of Cochran et al. (2001a) illustrate the value of analyzing and comparing related systems.
Conclusion
The effects of sequence variation and solvent variation on β-hairpin stability in peptides 1–3 provide insights on the forces that control antiparallel β-sheet stability in proteins and on the use of the β-hairpin strategy for modeling this common secondary structure. Comparisons among 1–3 in aqueous solution show that the loop segment and the interstrand clustering of sidechains both play key roles in β-hairpin formation, which is consistent with results of previous studies that considered both factors simultaneously. Our results show that the nonproteinogenic DPG loop is a very strong promoter of antiparallel β-sheet interactions between attached segments, leading to detectable β-hairpin formation even when interstrand sidechain–sidechain contacts are minimal (peptide 3) or when these contacts are compromised by 6 M urea. However, favorable sidechain–sidechain interactions are required for propagation of β-sheet secondary structure beyond the loop, and the more flexible NG loop may be superior to D-Pro-containing loops in terms of allowing optimal sidechain–sidechain contacts. The strong enthalpic drive we find for folding when interstrand clusters contain multiple aromatic groups, and the contrast between our findings and the behavior of a system that lacks an aromatic cluster (Maynard et al. 1998), suggest that β-hairpins may be ideal scaffolds for characterizing the fundamental mechanisms by which amino acid sidechains are attracted to one another in folded protein conformations.
Materials and methods
Materials
N-9-Fluorenylmethyloxycarbonyl amino acids were obtained from Advanced Chemtech. The arginine sidechain was protected with the 4-methoxy-2,3,6-trimethylbenzenesulfonyl group. Glutamine and asparagine were protected as the γ-triphenylmethyl derivatives. Lysine was tert-butoxycarbonyl protected. Tyrosine, threonine, and serine were protected as t-butyl ethers. Water used for HPLC was Millipore grade. HPLC grade acetonitrile, trifluoroacetic acid (TFA), D2O, and d4-acetic acid (CD3CO2D) were obtained from Aldrich. Diisopropylethylamine (DIEA), 2–(1-H-benzotriazolyl-1-yl)-N,N,N`,N`-tetramethyluronium hexaflurophosphate (HBTU), and piperidine were purchased from Applied Biosystems. The reference used for NMR proton chemical shifts was 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), which was obtained from Merck.
Peptide synthesis
All peptides were synthesized by standard solid-phase techniques with N-Fmoc amino acids on 2,4-dimethoxybenzhydrylamine resin (Rink amide resin), which provides an amide terminal group on cleavage. Couplings were performed on a 25-μmole scale on a Synergy 432A solid-phase peptide synthesizer (Applied Biosystems). For each coupling step, three equivalents of amino acid were used in DMF at room temperature. The coupling reagents were HBTU and DIEA, delivered at 80 μmole per cycle. Coupling times were ∼25 min. This time was doubled for couplings that are more difficult. The coupling reactions were monitored by the conductivity of the solution. Following each coupling reaction, the N-terminal Fmoc-protected amine was deprotected by treatment with 20% piperidine in DMF at room temperature. After the last residue was added, but before cleavage, the resin was rinsed with methanol and dried under a stream of nitrogen. The dried resin was transferred into a 10-mL round-bottom flask for the cleavage reaction. Peptides were cleaved from the resin by stirring for 3 h in 2 mL TFA with 100 mL thioanisol and 50 mL ethanedithiol. The resin beads were filtered off using glass wool, followed by rinsing with TFA. The TFA filtrate was concentrated to a volume of 1 mL and precipitated with 20 mL of anhydrous ether. The mixture was centrifuged to pellet the precipitate, and the ether was decanted off. The pellet was resuspended in cold ether and centrifuged again. This process was repeated three times. Millipore water was added to the tube during the final wash, and the mixture was centrifuged again. The aqueous layer, containing the crude peptide, was transferred to a round-bottom flask for lyophilization. Peptides were purified by reverse-phase semipreparative HPLC using a Vydac 214TP510 C4-silica column at a flow rate of 2 mL/min. The typical solvent system was 0.1% (v/v) TFA in H2O (solvent A) and 80% CH3CN/20% H2O/0.1% TFA (solvent B). The chromatograms were monitored by UV at 220 and 280 nm. Peptides were purified to ≥95% purity, as indicated by analytical HPLC (Vydac 214TP54 C4-silica analytical column). Peptides were checked by mass spectroscopy with a Bruker Reflex II MALDI-TOF mass spectrometer and by the complete assignment of the NMR spectra. Solid-phase synthesis of the cyclic peptide (Kates et al. 1993) began with attachment of a glutamic acid derivative to Rink amide resin via the sidechain carboxyl group. The α-carboxyl of this residue had previously been protected as an allyl ester and the α-amino group with Fmoc. Linear chain extension proceeded in standard fashion until all 14 residues were added. At this point, the α-carboxyl group of the Glu was deprotected by treatment with Pd(PPh3)4, the α-amino group of the last residues was depro-tected with 20% piperidine in DMF, and the peptide was cyclized with O-(-7-Azabenzotriazol-1-yl)-N,N,N`,N`-tetramethyluronium hexaflurophosphate (HATU) and 1-Hydroxy-7-azabenzotriazole (HOAt). Side chain deprotection, resin cleavage and purification were performed as outlined above for linear peptides.
NMR spectroscopy
NMR spectra were acquired on either a Varian INOVA 500 or a Bruker DRX 750 spectrometer. Two different solvents were used for NMR experiments: 9:1 H2O:D2O and D2O buffered to pH 3.8 (uncorrected) using 100 mM CD3COOD. The temperature of the NMR probe was calibrated using a methanol sample. DSS was used as internal reference. Peptide concentrations for NMR experiments were 1–3 mM. 1H spectra were recorded using 32K data points, which were zero-filled to 64K data points before Fourier transformation. Phase-sensitive COSY (Aue et al. 1976), TOCSY (Braunschweiler and Ernst 1983), NOESY (Jeener et al. 1979), ROESY (Bothner-By et al. 1984) spectra were recorded by standard techniques using presaturation or the WATERGATE pulse sequence (Piotto et al. 1992) to suppress the water signal. The number of increments was usually 512, and each transient contained 2048 data points. All spectra were zero-filled and baseline-corrected in both dimensions. Typically, a sine-squared window function with the corresponding shift optimized for every spectrum was applied. Mixing times of 200 msec were used for NOESY and ROESY spectra. TOCSY spectra were recorded using an 80-msec MLEV spin-lock. NOE intensities were measured by integration of the Hα-Hα NOEs characteristic of each hairpin in the ROESY spectra, taking the intraresidue Hα-Hα Gly NOE as the reference, as previously described (Searle et al. 1995; Ramírez-Alvarado et al. 1996).
Analytical ultracentrifugation
All peptides were shown via sedimentation equilibrium analysis to be monomeric under NMR conditions. A Beckman Optima XL-A analytical ultracentrifuge was used for the centrifugation experiments. Single-sample cells were assembled using 0.3- or 1.2-cm spacers, and each well contained 38 or 100 μL of sample, respectively. Samples were referenced against a blank corresponding to the solvent without the peptide. The density of the buffer solution was determined experimentally to be 1.011 g/mL. The centrifugation cells were placed in a four-bucket rotor. A vacuum was applied, and the temperature was set to 4°C. Sedimentation was monitored at 2-h intervals until the distribution was unchanged, indicating that the samples had equilibrated. Observed molecular weights were determined from plots of (distance)2 versus ln(absorbance) at two rotor speeds using the program Current XLA developed by Dr. D. McCaslin (University of Wisconsin-Madison Biophysics Instrumentation Facility).
Structure calculations
Medium- and long-range NOE cross peaks were qualitatively evaluated according to their intensities and classified as strong, medium, weak, and very weak and translated into upper-limit distance restraints. Pseudoatom corrections were added when necessary. φ angles were restricted to the range of 0° to −180° except for Gly and Asn. For those residues with 3JHα-NH coupling constants > 8 Hz, φ angles were restricted to the range −80° to −160° as described previously (de Alba et al. 1997a). The 3JHα-NH coupling constants for the inner strand residues (2–5 and 8–11) of peptides 1–5 are provided in the supplementary material (available online at www.proteinscience.org). Structures were calculated with the program DYANA (Guntert et al. 1997) using a modified library to include D-Pro residues.
Thermodynamic analysis
The thermodynamic analysis is based on the nonlinear fitting method described by Maynard et al. (1998). In short, assuming a two-state model, the equilibrium constant for folding (K) is given by
 |
where v is the fraction of the folded peptide and is related to the chemical deviation from random coil values (ΔδHα). Therefore, v can be expressed as shown by
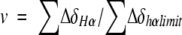 |
where ∑ΔδHα is the sum of the α-proton chemical shifts for the four-strand residues in hydrogen-bonded positions for the equilibrating peptides minus the analogous sum for unfolded reference 4 at the corresponding temperature, and ∑ΔδHαlimit is the sum of the α-proton chemical shifts for the same residues for the totally folded cyclic reference peptide 5 minus the analogous sum for 4. Using standard thermodynamic expressions
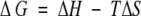 |
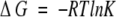 |
where
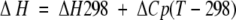 |
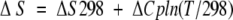 |
substitution and rearrangement yield
 |
where
 |
The experimental data were fitted iteratively to this final equation to determine the thermodynamic parameters. The final errors were calculated from the fitting routine.
Acknowledgments
This research was supported by the NIH (GM61238). J.F.E. was supported in part by a fellowship from the Ministerio de Educacion y Cultura (Spain) and the Fulbright Commission. The Biophysics Instrumentation Facility was supported by NSF; we thank BIF Director Dr. Darrell McCaslin for assistance. NMR instruments in the Department of Chemistry were supported in part by grants from NIH and NSF, and NMR instruments in NMRFAM were supported in part by NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.4140102.
References
- Andersen, N.H., Dyer, R.B., Fesinmeyer, R.M., Gai F., Liu, Z.H., Neidigh, J.W., and Tong, H. 1999. Effect of hexafluoroisopropanol on the thermodynamics of peptide secondary structure formation. J. Am. Chem. Soc. 121: 9879–9880. [Google Scholar]
- Aue, W.P., Bartholdi, E., and Ernst, R.R. 1976. Two-dimensional spectroscopy. Application to nuclear magnetic resonance. J. Chem. Phys. 64: 2229–2246. [Google Scholar]
- Baldwin, R.L. 1986. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. 83: 8069–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, R.L. and Rose, G.D. 1999. Is protein folding hierarchic? I. Local structure and peptide folding. Trends Biochem. Sci. 24: 26–33. [DOI] [PubMed] [Google Scholar]
- Blanco, F.J., Rivas, G., and Serrano, L. 1994. A short linear peptide that folds into a native stable β-hairpin in aqueous solution. Nat. Struct. Biol. 1: 584–590. [DOI] [PubMed] [Google Scholar]
- Bolin, K.A. and Millhauser, G.L. 1999. Alpha and 3(10): The split personality of polypeptide helices. Acc. Chem. Res. 32: 1027–1033. [Google Scholar]
- Bothner-by, A.A., Stephens, R.L., Lee, J.M., Warren, C.D., and Jeanloz, R.W. 1984. Structure determination of a tetrassacharide: Transient nuclear Overhauser effect in the rotating frame. J. Am. Chem. Soc. 106: 811–813. [Google Scholar]
- Braunschweiler, L. and Ernst, R.R. 1983. Coherence transfer by isotropic mixing - application to proton correlation spectroscopy. J. Magn. Reson. 53: 521–528. [Google Scholar]
- Carulla, N., Woodward, C., and Barany, G. 2000. Synthesis and characterization of a β-hairpin peptide that represents a `core module' of bovine pancreatic trypsin inhibitor (BPTI). Biochemistry 39: 7927–7937. [DOI] [PubMed] [Google Scholar]
- Chakrabartty, A. and Baldwin, R.L. 1995. Stability of α-helices. Adv. Protein Chem. 46: 141–176. [PubMed] [Google Scholar]
- Chothia, C. 1973. Conformation of twisted β-pleated sheets in proteins. J. Mol. Biol. 75: 295–302. [DOI] [PubMed] [Google Scholar]
- Cochran, A.G., Skelton, N.J., and Starovasnik, M.A. 2001a. Tryptophan zippers: Stable, monomeric β-hairpins. Proc. Natl. Acad. Sci. 98: 5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran, A.G., Tong, R.T., Starovasnik, M.A., Park, E.J., McDowell, R.S., Theaker, J.E., and Skelton, N.J. 2001b. A minimal peptide scaffold for β-turn display: Optimizing a strand position in disulfide-cyclized β-hairpins. J. Am. Chem. Soc. 123: 625–632. [DOI] [PubMed] [Google Scholar]
- Constantine, K.L., Mueller, L., Andersen, N.H., Tong, H., Wandler, C.F., Friedrichs, M.S., and Bruccoleri, R.E. 1995. Structural and dynamic properties of a β-hairpin-forming linear peptide. 1. Modeling using ensemble-averaged constraints. J. Am. Chem. Soc. 117: 10841–10854. [Google Scholar]
- de Alba, E., Jimenez, M.A., and Rico, M. 1997a. Turn residue sequence determines β-hairpin conformation in designed peptides. J. Am. Chem. Soc. 119: 175–183. [Google Scholar]
- ———. 1997b. Cross-strand side-chain interactions versus turn conformation in β-hairpins. Protein Sci. 6: 2548–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alba, E., Santoro, J., Rico, M., and Jiminez, M.A. 1999. De novo design of a monomeric three-stranded β-sheet protein. Protein Sci. 8: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa, J.F. and Gellman, S.H. 2000. A designed β-hairpin containing a natural hydrophobic cluster. Angew. Chem. Int. Ed. Engl. 39: 2330–2333. [DOI] [PubMed] [Google Scholar]
- Espinosa, J.F., Muñoz, V., and Gellman, S.H. 2001. Interplay between hydrophobic cluster and loop propensity in β-hairpin formation. J. Mol. Biol. 306: 397–402. [DOI] [PubMed] [Google Scholar]
- Fisk, J.D. and Gellman, S.H. 2001. A parallel β-sheet model system that folds in water. J. Am. Chem. Soc. 123: 343–344. [DOI] [PubMed] [Google Scholar]
- Gallagher, T., Alexander, P., Bryan, P., and Gilliland, G.L. 1994. 2 crystal-structures of the B1 immunoglobulin-binding domain of streptococcal protein-G and comparison with NMR. Biochemistry 33: 4721–4729. [PubMed] [Google Scholar]
- Gellman, S.H. 1998. Minimal model systems for β-sheet secondary structure in proteins. Curr. Opin. Chem. Biol. 2: 717–724. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S.R. and Searle, M.S. 2000. Structure, folding, and energetics of cooperative interactions between the β-strands of a de novo designed three-stranded antiparallel β-sheet peptide. J. Am. Chem. Soc. 122: 8350–8356. [Google Scholar]
- Gronenborn, A.M., Filpula, D.R., Essig, N.Z., Achari, A., Whitlow, M., Wingfield, P.T., and Clore, G.M. 1991. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein-G. Science 253: 657–661. [DOI] [PubMed] [Google Scholar]
- Gunasekaran, K., Ramakrishnan, C., and Balaram, P. 1997. β-Hairpins in proteins revisited: Lessons for de novo design. Protein Eng. 10: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Guntert, P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Haque, T.S. and Gellman, S.H. 1997. Insights on β-hairpin stability in aqueous solution from peptides with enforced type I` and type II` β-turns. J. Am. Chem. Soc. 119: 2303–2304. [Google Scholar]
- Haque, T.S., Little, J.C., and Gellman, S.H. 1994. `Mirror image' reverse turns promote β-hairpin formation. J. Am. Chem. Soc. 116: 4105–4106. [Google Scholar]
- ———1996. Stereochemical requirements for β-hairpin formation: Model studies with four-residue peptides and depsipeptides. J. Am. Chem. Soc. 118: 6975–6985. [Google Scholar]
- Honda, S., Kobayashi, N., and Munekata, E. 2000. Thermodynamics of a β-hairpin structure: Evidence for cooperative formation of folding nucleus. J. Mol. Biol. 295: 269–278. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.G. and Thornton, J.M. 1994. A revised set of potentials for β-turn formation in proteins. Protein Sci. 3: 2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeener, J., Meier, B.H., Bachmann, P., and Ernst, R.R. 1997. Investigation of exchange processes by two dimensional NMR spectroscopy. J. Phys. Chem. 71: 4546–4553. [Google Scholar]
- Kates, S.A., Sole, N.A., Johnson, C.R., Hudson, D., Barany, G., and Albericio, F. 1993. A novel, convenient, 3-dimensional orthogonal strategy for solid-phase synthesis of cyclic-peptides. Tetrahedron Lett. 34: 1549–1552. [Google Scholar]
- Kobayashi, N., Honda, S., Yoshii, H., and Munekata, I. 2000. Role of side-chains in the cooperative β-hairpin folding of the short C-terminal fragment derived from streptococcal protein G. Biochemistry 39: 6564–6571. [DOI] [PubMed] [Google Scholar]
- Koepf, E.K., Petrassi, H.M., Ratnaswamy, G., Huff, M.E., Sudol, M., and Kelly, J.W. 1999. Characterization of the structure and function of W→F WW domain variants: Identification of a native unfolded protein that folds upon ligand binding. Biochemistry 38: 14338–14351. [DOI] [PubMed] [Google Scholar]
- Livingstone, J.R., Spolar, R.S., and Record, M.T. 1991. Contribution to the thermodynamics of protein folding from the reduction in water-accessible nonpolar surface area. Biochemistry 30: 4237–4244. [DOI] [PubMed] [Google Scholar]
- Lopez de la Paz, M., Lacroix, E., Ramírez-Alvarado, M., and Serrano, L. 2001. Computer-aided design of β-sheet peptides. J. Mol. Biol. 312: 229–246. [DOI] [PubMed] [Google Scholar]
- Luo, P. and Baldwin, R.L. 1997. Mechanism of helix induction by trifluoroethanol: A framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry 36: 8413–8421. [DOI] [PubMed] [Google Scholar]
- MacArthur, M.W. and Thornton, J.M. 1991. Influence of proline residues on protein conformation. J. Mol. Biol. 218: 397–412. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I. and Privalov, P.L. 1994. Energetics of interactions of aromatic hydrocarbons with water. Biophys. Chem. 50: 285–291. [DOI] [PubMed] [Google Scholar]
- Maynard, A.J., Sharman, G.J., and Searle, M.S. 1998. Origin of β-hairpin stability in solution: Structural and thermodynamic analysis of the folding of a model peptide supports hydrophobic stabilization in water. J. Am. Chem. Soc. 120: 1996–2007. [Google Scholar]
- Mayo, K.H. and Ilyina, E. 1998. A folding pathway for βpep-4 peptide 33mer: From unfolded monomers to β-sheet sandwich dimers to well-structured tetramers. Protein Sci. 7: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor, D.L. and Kim, P.S. 1994. Context is a major determinant of β-sheet propensity. Nature 371: 264–267. [DOI] [PubMed] [Google Scholar]
- Muñoz, V., Thompson, P.A., Hofrichter, J., and Eaton, W.A. (1997) Folding dynamics and mechanism of β-hairpin formation. Nature 390; 196–198. [DOI] [PubMed]
- Murphy, K.P., Privalov, P.L., and Gill, S.J. 1990. Common features of protein unfolding and dissolution of hydrophobic compounds. Science 247: 559–561. [DOI] [PubMed] [Google Scholar]
- Nesloney, C.L. and Kelly, J.W. 1996. Progress toward understanding β-sheet structure. Bioorg. Med. Chem. 4: 739–766. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions. J. Biomol. NMR 2: 661–665. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L. and Gill, S.J. 1989. The hydrophobic effect: a reappraisal. Pure Appl. Chem. 61: 1097–1104. [Google Scholar]
- Ragothama, S.R., Awasthi, S.K., and Balaram, P. 1998. β-Hairpin nucleation by Pro-Gly β-turns. Comparison of D-Pro-Gly and L-Pro-Gly sequences in an apolar octapeptide. J. Chem. Soc. Perk. T. 2 137–143.
- Ramírez-Alvarado, M., Blanco, F.J., and Serrano, L. 1996. De novo design and structural analysis of a model β-hairpin peptide system. Nat. Struct. Biol. 3: 604–612. [DOI] [PubMed] [Google Scholar]
- Ramírez-Alvarado, M., Blanco, F.J., Niemann, H., and Serrano, L. 1997. Role of β-turn residues in β-hairpin formation and stability in designed peptides. J. Mol. Biol. 273: 898–912. [DOI] [PubMed] [Google Scholar]
- Russell, S.J. and Cochran, A.G. 2000. Designing stable β-hairpins: Energetic contributions from cross-strand residues. J. Am. Chem. Soc. 122: 12600–12601. [Google Scholar]
- Santiveri, C.M., Rico, M., and Jimenez, M.A. 2000. Position effect of cross-strand side-chain interactions on β-hairpin formation. Protein Sci. 9: 2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck, H.L. and Gellman, S.H. 1998. Use of a designed triple-stranded antiparallel β-sheet to probe β-sheet cooperativity in aqueous solution. J. Am. Chem. Soc. 120: 4869–4870. [Google Scholar]
- Searle, M.S. 2001. Peptide models of protein β-sheets: Design, folding and insights in stabilising weak interactions. J. Chem. Soc. Perk. T. 2 1011–1020.
- Searle, M.S, Williams, D.H., and Rackman, L.C. 1995. A short linear peptide derived from the N-terminal sequence of ubiquitin folds into a water-stable non-native β-hairpin. Nat. Struct. Biol. 2: 999–1006. [DOI] [PubMed] [Google Scholar]
- Searle, M.S., Griffiths-Jones, S.R., and Skinner-Smit, H. 1999. Energetics of weak interactions in a β-hairpin peptide: Electrostatic and hydrophobic contributions to stability from lysine salt bridges. J. Am. Chem. Soc. 121: 11615–11620. [Google Scholar]
- Serrano, L. 2000. The relationship between sequence and structure in elementary folding units. Adv. Protein Chem. 53: 49–85. [DOI] [PubMed] [Google Scholar]
- Sibanda, B.L. and Thornton, J.M. 1985. β-Hairpin families in globular proteins. Nature 316: 170–174. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Accommodating sequence changes in β-hairpins in proteins. J. Mol. Biol. 229: 428–447. [DOI] [PubMed] [Google Scholar]
- Sibanda B.L., Blundell, T.L., and Thornton, J.M. 1989. Conformation of β-hairpins in protein structures. J. Mol. Biol. 206: 759–777. [DOI] [PubMed] [Google Scholar]
- Stanger, H.E. and Gellman, S.H. 1998. Rules for antiparallel β-sheet design: D-Pro-Gly is superior to L-Asn-Gly for β-hairpin nucleation. J. Am. Chem. Soc. 120: 4236–4237. [Google Scholar]
- Syud, F.A., Espinosa, J.F., and Gellman, S.H. 1999. NMR-based quantification of β-sheet populations in aqueous solution through use of reference peptides for the folded and unfolded states. J. Am. Chem. Soc. 121: 11577–11578. [Google Scholar]
- Syud, F.A., Stanger, H.E., and Gellman, S.H. 2001. Analysis of interstrand sidechain-sidechain contacts in a designed β-hairpin: Significance of both lateral and diagonal interactions. J. Am. Chem. Soc. 123: 8667–8677. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Sykes, B.D., and Richards, F.M. 1991. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222: 311–333. [DOI] [PubMed] [Google Scholar]
- ———. 1992. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31: 1647–1651. [DOI] [PubMed] [Google Scholar]