Abstract
The structural stability of calmodulin (CaM) has been investigated previously by chemical and thermal methods. The calcium-loaded form of CaM has been found to be exceptionally stable, because it can be exposed to temperatures >90°C or to a 9 M urea solution without a marked change in its tertiary structure, and is therefore not experimentally accessible for unfolding studies using conventional analytical methods. In this study, we have developed a system for measuring the force for mechanically unfolding CaM using an atomic force microscope (AFM) by stretching the protein from its N- and C-terminal residues; we have been successful in obtaining force versus extension (F–E) curves for both apo and holo forms of CaM. In our experiment, distinguishable F–E curves have been obtained upon stretching of apoCaM and holoCaM to their full extensions. A very low force observed upon stretching of apoCaM indicated a relatively high flexibility of the apo form. On the contrary, a relatively high unfolding force and the appearance of a characteristic force peak were noted during full stretching of holoCaM. The F–E curve of the latter form of CaM most likely reflects a more rigid and probably more organized conformation of holoCaM than that of apoCaM. These experiments confirmed that the AFM is able to clearly distinguish two functionally distinct forms of CaM in terms of their mechanical properties.
Keywords: Calmodulin, unfolding mechanics, AFM, force, extension curve
Various spectroscopic techniques have been applied to study the stability of calmodulin (CaM) under different conditions. Studies using proton NMR (Guerini and Krebs 1983) and CD spectroscopy (Brzeska et al. 1983) have revealed that the presence of Ca2+ stabilizes the structural backbone of the native CaM, and even at 90°C, holoCaM retains most of its native conformation. Calcium ions also confer a structural stability to CaM in the presence of 9 M urea (Walace et al. 1980). It has therefore been difficult to study the unfolding mechanism of holoCaM. These findings encouraged us to find other techniques to study unfolding processes that would be applicable to thermally and chemically stable proteins like CaM.
One of the alternative techniques that have recently been used to study protein unfolding is the force spectroscopy method based on the atomic force microscopy (AFM; Mitsui et al. 1996; Zlatanova et al. 2000). This novel technique uses an extremely sensitive force sensor of AFM that is able to detect forces in the range of pico- to nanonewtons. In this technique, a protein molecule is sandwiched between a solid substrate and an AFM tip. The protein may either be immobilized by physical adsorption (Rief et al. 1997) or covalently linked to a solid substrate on one end and to the AFM tip on the other (Wang and Ikai 1999; Idiris et al. 2000). The sample stage is then lowered to increase the tip–sample distance by stretching the sandwiched protein molecule from its two ends up to its full contour length. As a result, the relationship between the tensile forces versus the extension of the protein molecule is obtained in terms of a force–extension (F–E) curve. The force required to stretch a target protein leading to a complete breakdown of its native conformation into an unfolded one is then analyzed from the F–E curve.
Unfolding mechanics of CaM have previously been studied by AFM using its tandem oligomers (Carrion-Vazquez et al. 2000; Idiris et al. 2000). The unfolding force spectrum of the CaM tetramer obtained by Carrion-Vazquez et al. (2000) was very similar to that of a theoretical random-coil chain either in the presence or absence of Ca2+. Idiris et al. performed a force spectroscopy on an engineered CaM dimer wherein two cysteine residues were added to both N and C ends of the protein; the force to unfold a Ca2+-saturated CaM dimer was found to be higher than the force to unfold a random coil, but the force spectrum was complex and difficult to interpret because the two monomeric units in a dimer appeared to behave as a single compound unit. The presence of a polyethylene glycol (PEG) spacer in the cross-linker used in their study had only a loosely defined length, and its use should be avoided if possible.
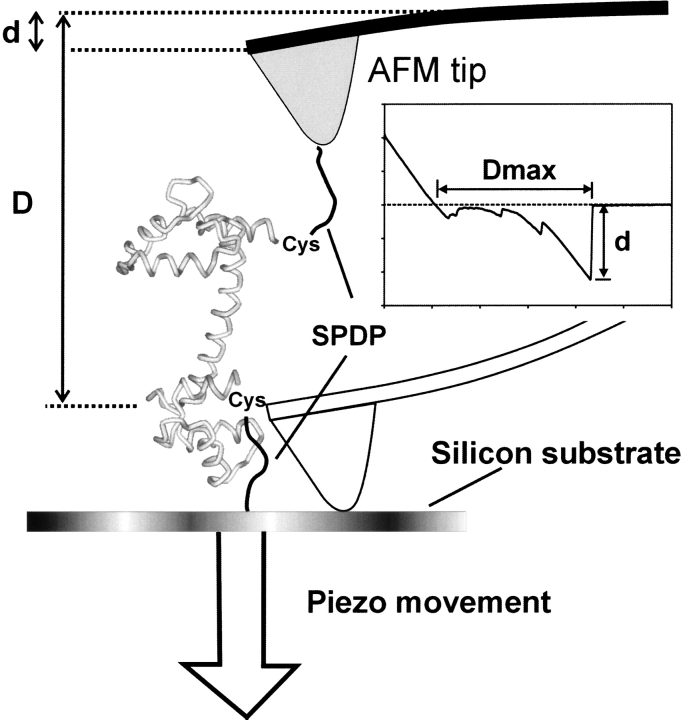
To obtain a detailed insight of the unfolding mechanics of a single molecule of CaM and to improve on previous techniques, we used a CaM monomer with additional cysteine residues next to its C and N termini and studied its forced unfolding in the presence and absence of Ca2+ ions. In our experiment, the engineered CaM was sandwiched between a functionalized substrate and an AFM tip, both coated with bifunctional cross-linkers without the PEG spacer (Fig. 1 ▶). By using this system, we have been able to show that the AFM-based force spectroscopy can distinguish two conformations of CaM based on their mechanical properties.
Fig. 1.
Schematic view of the unfolding mechanics experiment on calmodulin (CaM). CaM was covalently sandwiched through the cross-linking reactions between a Cys residue on one of its ends and a SPDP cross-linker on the silicon substrate and the AFM tip. The sandwiched protein is then mechanically unfolded by lowering the sample stage. The extension of the sandwiched protein (E) is determined by subtracting the cantilever deflection (d) from the distance covered by the piezo movement (Dmax).The tensile force is calculated by multiplying the cantilever deflection (d) by the cantilever spring constant (k) and is expressed as a function of the elongation (E), giving rise to an F–E curve.
Results
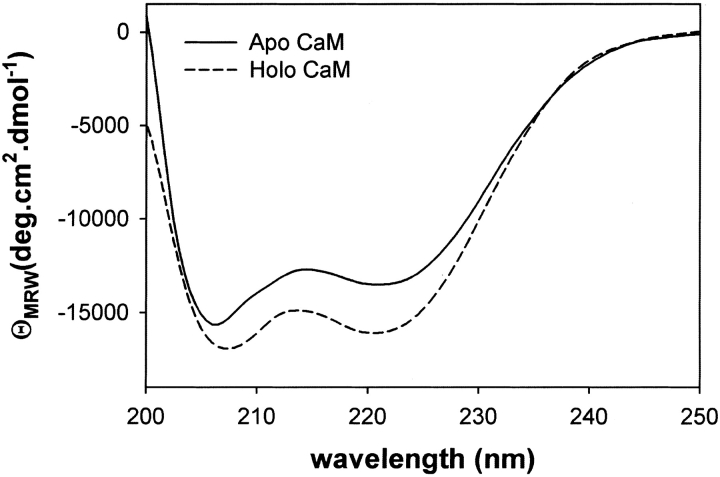
First we confirmed from the CD spectra that both apo and holo forms of the engineered CaM have similar secondary structures to those of corresponding forms of the wild-type protein. The CD spectra given in Figure 2 ▶ predict helical contents of 40% and 34% for holo and apo forms, respectively, and both values are in agreement with a previously published result for holo (40%–45%) and apo (30%–35%) forms (Klee 1977).
Fig. 2.
CD spectra of engineered CaM. The result confirmed that the engineered CaM has similar secondary structures in both holo and apo forms to those of the wild-type protein.
Before the start of stretching experiments, the spring constant of the cantilevers to be used was determined in air by the thermal fluctuation method as previously described (Hutter and Bechhoefer 1993). The mean spring constant of the long and soft V-shape NP cantilever with the nominal spring constant of 0.06 N/m (Digital Instrument) was 0.056 ± 0.004 N/m (N = 10). This mean value was used for the whole calculation in this paper.
Force–extension curve profiles for apo and holo calmodulin
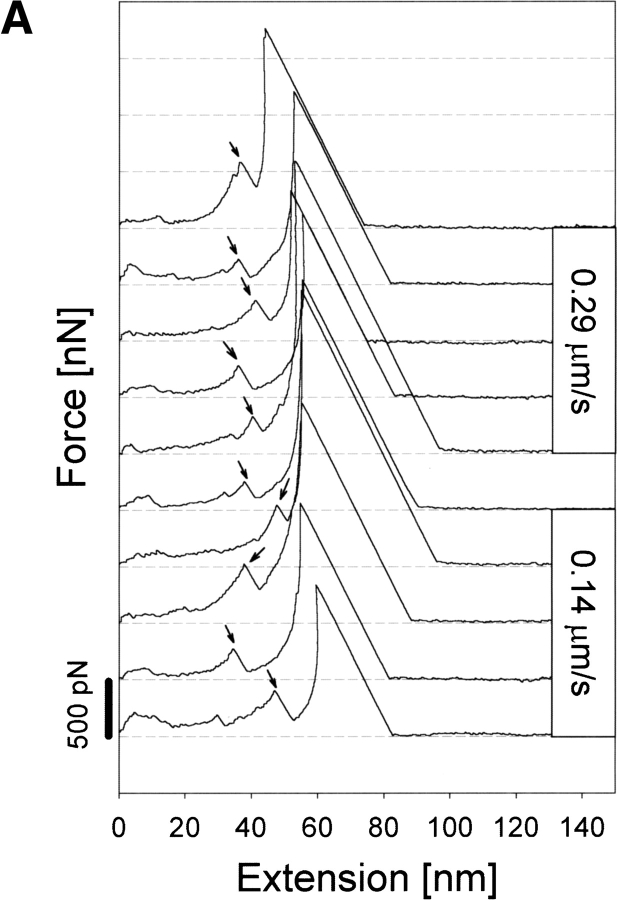
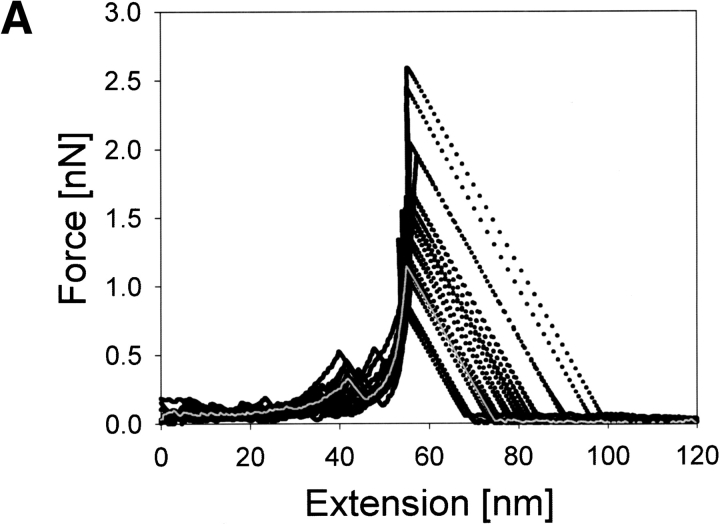
Figure 3A ▶ shows representative force (F)–extension (E) curves of holoCaM resulting from two different pulling speeds of 0.14 μm/sec and 0.29 μm/sec, showing the extension of the molecule almost to its full length of 55 nm before the final rupture of the system with a force >1.5 nN. As evident in the figure, an increase in E was generally accompanied by a somewhat nonlinear increase in F, but there was, unfailingly, at least one large force peak during stretching of holoCaM as indicated by the arrow on each curve. The top five F–E curves in Figure 3A ▶ were obtained with a pulling speed of 0.29 μm/sec, and the bottom five curves with 0.14 μm/sec. Both show the characteristic force peak at ∼35–45 nm in extension, with the peak force ranging from 0.2 to 0.6 nN. The following part of each F–E curve with a negative slope corresponds to a jump of the cantilever, thus containing no meaningful data. We think the intermediate force peak appeared as the result of the breakdown of a specific structure in holoCaM. Other smaller and not quite reproducible force peaks appearing in the initial part of extension were not considered as meaningful peaks at present because they might have resulted from nonspecific adsorptions of the protein to the substrate.
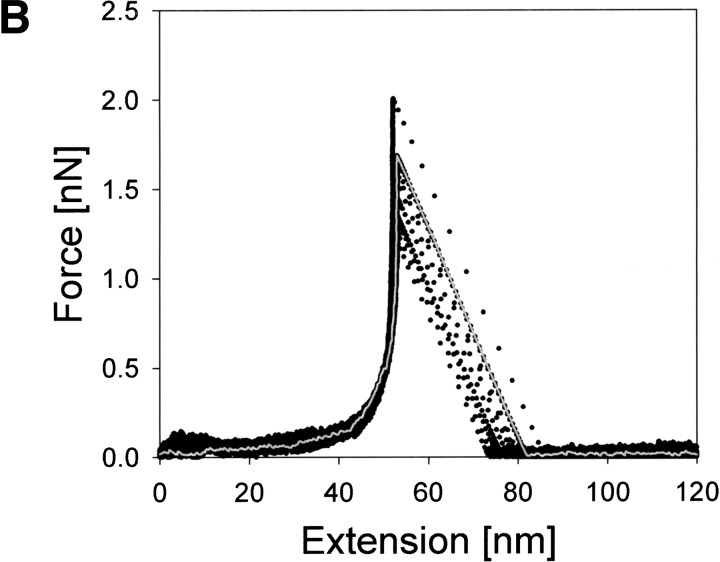
Fig. 3.
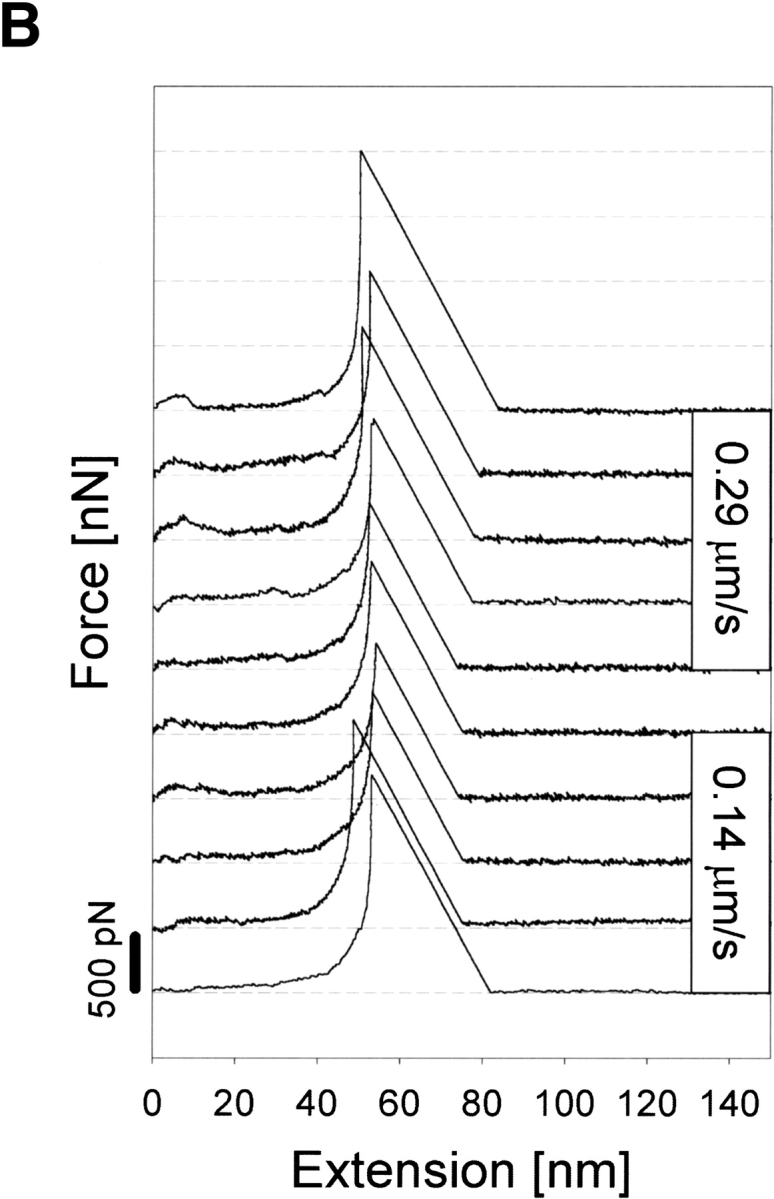
F–E curves of holoCaM and apoCaM obtained at two different pulling speeds of 0.14 μm/sec and 0.29 μm/sec. (A) Stretching of holoCaM produced force–extension curves having a single characteristic force peak marked by the single arrow at its reproducible position. Those parts in the F–E curves with negative slopes correspond to jumps of the cantilever and do not contain actual data. (B) F–E curves obtained from the stretching of apoCaM, also at two different pulling speeds.
In contrast to the peaked F–E curves of holoCaM explained above, typical F–E curves of apoCaM (Fig. 3B ▶) were of monotonously, though nonlinearly, increasing type without conspicuous force peaks in intermediate extension ranges. The slope of the F–E curves was less than those for holoCaM. Very similar F–E curves were obtained at two different pulling speeds of 0.14 and 0.29 μm/sec.
Distribution of final extension and intermediate force
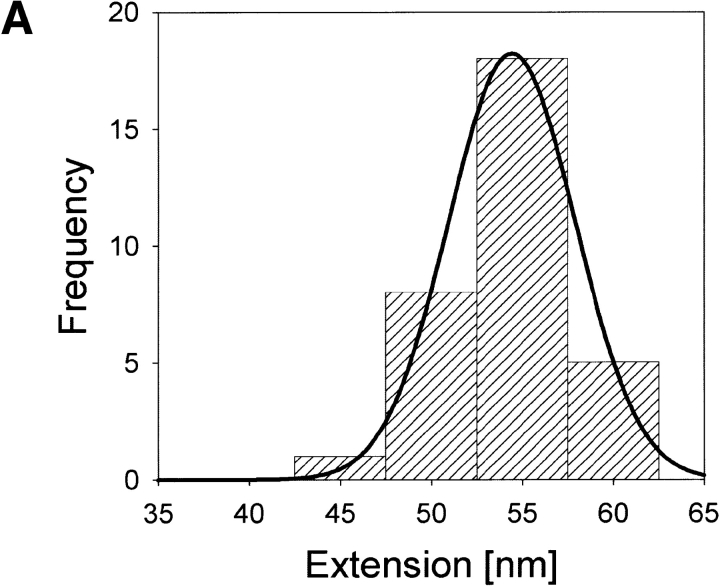
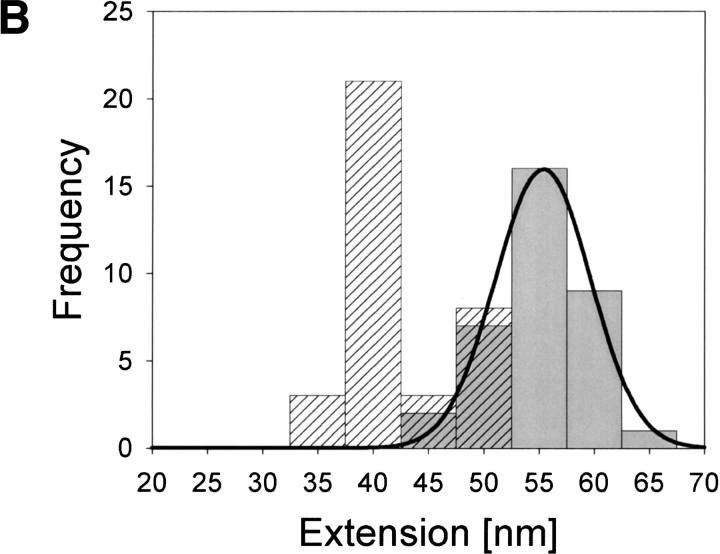
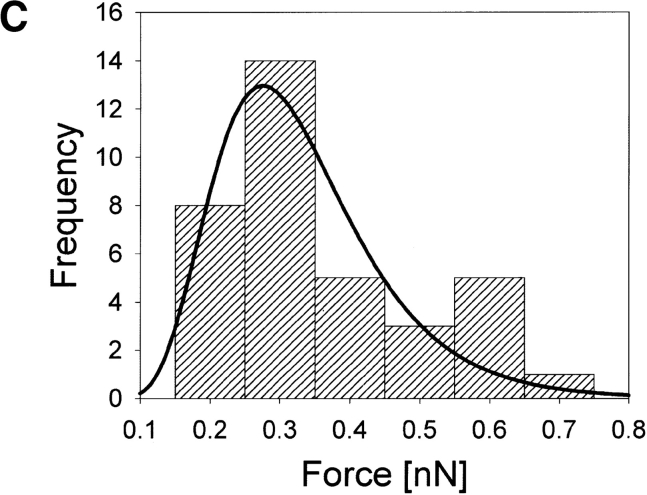
Histograms in Figure 4A,B ▶ give distributions of the overall extensions for apoCaM and holoCaM, respectively, as calculated according to the method described in Figure 1 ▶. As the histograms show, extension lengths are concentrated at ∼55 nm for both apoCaM and holoCaM. The mean values of extension for apoCaM and holoCaM obtained from a Gaussian fit are 54.4 ± 3.5 nm (N = 32) and 55.4 ± 4.3 nm (N = 35), respectively. An extension of 55 nm corresponds to the contour length of a polypeptide consisting of ∼150 amino acid residues if we take 0.37 nm for the length of the all-trans form of one amino acid residue (Schulz and Schrimer 1979). Because the engineered CaM used in the study had 150 amino acid residues, the result indicated that both apoCaM and holoCaM were extended to their full lengths most of the time. In the histogram of Figure 4B ▶, the distribution of the positions of the intermediate force peak is given. The intermediate force peak was most frequently observed when holoCaM was extended to ∼40 nm, or ∼70% of its total length. The probability of finding the intermediate peak at 37.5–42.5 nm was >55%, indicating that its appearance was not owing to nonspecific adsorptions but to a breakdown of some specific structure in holoCaM. The distribution of the height of the intermediate peak is presented by the histogram in Figure 4C ▶. The mean force calculated by a log-normal fit is 0.28 ± 0.15 nN (N = 35). We think the large standard error reflects a high variability in the structural rigidity of holoCaM molecules in solution.
Fig. 4.
Histograms of the distributions of the extension lengths for apoCaM (A) and holoCaM (B) and the force distribution of the intermediate peak (C). The distribution of the intermediate peaks position is also given in B (hatched column). The solid lines in A and B are the Gaussian fit, whereas the one in C is the log-normal fit.
Pulling speed dependence
As shown in Figure 3A ▶, the position and shape of the intermediate peak in the F–E curves of holoCaM were somewhat variable even at the same pulling speed. In order to see the pulling speed dependency and also to get the average F–E curves for further analysis, we superimposed 21 F–E curves by normalizing them to the same extension length (Fig. 5A ▶). Because one of the F–E curves (drawn by the gray lines in Fig. 5A ▶) was found to run close to the center of the superimposed curves according to our visual inspection, it was chosen as a representative F–E curve for the further analysis. It is important to note that despite variations in the shape and height of the intermediate peak, the F–E curves have similar slopes in the intermediate extension range. In conclusion, doubling of the pulling speed did not have a significant effect on the F–E curve, but the speed must be changed by at least a factor of 10 in the future. A representative F–E curve for apoCaM was also chosen in a similar way in which 10 normalized F–E curves were superimposed (Fig. 5B ▶). Dependence on the pulling speed was much less obvious in apoCaM compared with holoCaM. A representative curve (gray line in Fig. 5B ▶) was chosen from among the 10 curves and will be used for the further analysis of the F–E curves.
Fig. 5.
The superimposed F–E curves of holoCaM (A) and apoCaM (B). The gray line is a representative F–E curve chosen from actual data that were close to the center of the superimposed curves.
Worm-like chain and poly glutamic acid analysis
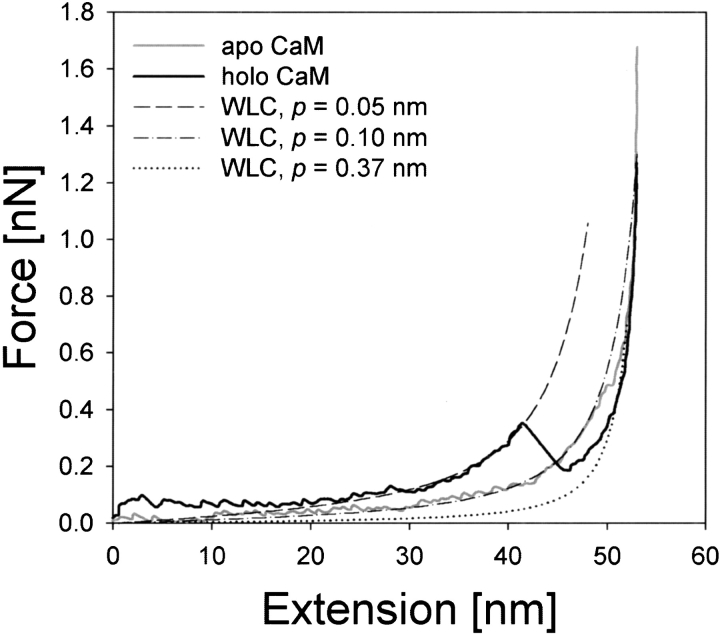
To see if the experimental F–E curves of CaM could be described by the entropic elasticity of a polypeptide chain or not, we adopted the WLC model (equation 2 below) as a purely entropic chain with the persistence length, p, as a freely adjustable parameter. Initially, p was set to the theoretical length of a single amino acid residue in the crystal structure, that is, 0.37 nm (Schulz and Schrimer 1979). As shown in Figure 6 ▶, neither of the representative experimental curves for apo and holoCaM could be fitted to a WLC model with p = 0.37 nm. To obtain better fittings of representative F–E curves of apoCaM and holoCaM to the WLC model, we lowered the value of p to <0.37 nm and found that the p values of 0.1 and 0.05 nm gave approximate fittings with major parts of the F–E curves of apoCaM and holoCaM, respectively. However, such p values are unrealistically small, being even less than the length of C—C or that of C—N bonds. As a result, we concluded that the F–E curves of both apoCaM and holoCaM cannot be fitted with a purely entropic model of polymer elasticity.
Fig. 6.
Fitting of the F–E curves of apoCaM and holoCaM to the WLC model. The WLC model was fitted to the average F–E curves of apoCaM and holoCaM by adjusting the persistence length (p).
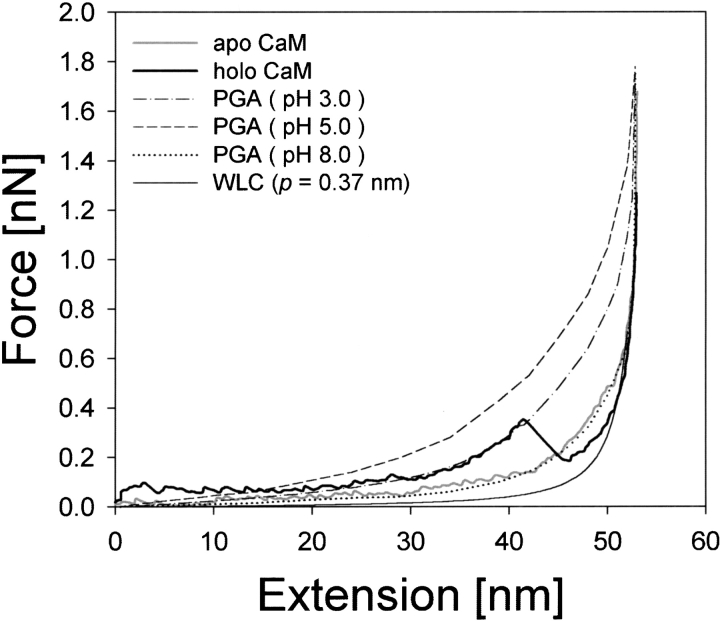
As an alternative way to analyze the experimental data, we compared them with the F–E curves of poly-L-glutamic acid (PGA) obtained by Idiris et al. (2000). The F–E curves of PGA at pH 3.0, 5.0, and 8.0 corresponding to the helical content of 80%, 40%, and 0%, respectively, were taken as reference curves and are given in Figure 7 ▶ together with the F–E curves of apoCaM and holoCaM. The F–E curve of apoCaM fits well with that of PGA at pH 8.0, whereas for holoCaM, the PGA curve at pH 5.0 could be fitted to its F–E curve up to the intermediate peak, and after the peak, a WLC curve with p = 0.37 nm became a closer fit. The result of this comparison for holoCaM up to the intermediate force peak was reasonable based on the helical content determined by CD spectroscopy but not so for apoCaM. We ascribe the presence of an intermediate force peak in the F–E curve of holoCaM to the breakdown of the residual tertiary structure in the molecule as stated above, and its absence in PGA. A good fit observed between the F–E curves of apoCaM and PGA at pH 8.0 could represent one of the following alternatives. First, PGA in a solution at pH 8.0 is not purely random coil but has a certain amount of secondary structure, which cannot be detected by a CD experiment. Second, apoCaM has 34% helix, but its helical structure is mechanically so weak that it is indistinguishable from a randomly coiled PGA in our mechanical measurement. We need further experimental work and theoretical considerations to elucidate the real reason for the observed similarities between the F–E curves of apoCaM and PGA at pH 8.0.
Fig. 7.
Comparison of the F–E curves of holoCaM and apoCaM with those of poly-L-glutamic acids (PGA) at different pHs (Idiris et al. 2000).
Discussion
The result of our mechanical stretching experiment on CaM monomers described in this paper is different from the previously reported ones for a CaM dimer (Idiris et al. 2000) or for a tetramer (Carrion-Vazcuez et al. 2000). The method of the attachment of protein molecules to the surface of the substrate and the choice of cross-linkers when used for immobilization of protein samples to the substrate may be among important factors that influenced the results of the three researches. Attaching protein molecules directly to a solid surface, for example, a gold surface, without using any cross-linkers (Carrion-Vazcuez et al. 2000) would promote nonspecific adsorption of the protein molecules to the solid surface. The use of a cross-linker having a PEG spacer largely prevented protein adsorption to the substrate, but the length of the PEG spacer had a certain distribution, which gave rise to a difficulty in the estimation of the net contour length of extended proteins (Idiris et al. 2000). In our system, we used SPDP as a cross-linker with a fixed length of 6.8 Å in full extension, which allowed us to evaluate the length of protein extension with confidence. Force curves accompanied with nonspecific force peaks were sometimes observed, but most of them were easily detected for their irregular and nonreproducible shape and therefore were discarded.
In addition to the use of the short cross-linker with a well-defined length, the choice of the monomeric form of CaM as our experimental sample gave us more informative results compared with the multimer systems used in the previous studies. Our experimental data obtained for both holoCaM and apoCaM could not be explained by the WLC model of a random coil, contrary to the previous report on the CaM tetramer in which apo and holo forms were mechanically indistinguishable from each other and were fitted well to the WLC model (Carrion-Vazcuez et al. 2000). In our case, apoCaMs and holoCaMs produced significantly different F–E curves: first, curves for the latter were for the most part higher in force than those for the former at corresponding lengths; and second, a conspicuous intermediate force peak was always noted for holoCaM, as presented in Figure 6 ▶.
The second interesting aspect in the experimental data is the presence of an intermediate force peak before the final rupture. In the previous work on tandemly repeated Ig domains of titin by Rief et al. (1997), several intermediate force peaks before the final rupture were found. They interpreted such peaks as representing sequential breakdowns of Ig domains, which are structurally almost totally independent from each other. In our experiment, we found a reproducible force peak in the F–E curve of single monomeric holoCaM between 30 and 50 nm in extension with a force range from 0.2 to 0.5 nN. Although the position and the height of the peak were somewhat variable, its unfailing appearance for holoCaM and its total absence for apoCaM strongly indicate that its origin was not in nonspecific adsorption of the protein to the substrate but was caused by a sudden breakdown of a structural element that was present only in holoCaM. Another important point to be noticed in the F–E curve of holoCaM is that, after passing the intermediate force peak, the force dropped to a lower level than for apoCaM at the same extension length, and then followed the curve for the WLC model with p = 0.37 nm (Fig. 7 ▶). A better fitting of the last part of the curve to the WLC model rather than that of apoCaM stresses the important point that a cooperative destruction of the residual structure reduced the polypeptide to a totally randomly coiled chain. Similar observations have been made on carbonic anhydrase (Wang and Ikai 1999), titin (Rief et al. 1997), spectrin domains (Rief et al. 1999), and tandemly repeated lysozyme (Yang et al. 2000).
Our experiment could not give direct information on the structural element responsible for the appearance of the intermediate force peak in the F–E curve of holoCaM. However, a previous thermodynamics study investigating the internal stability of holoCaM found that its C-terminal domain was thermodynamically more stable than the N-terminal domain (Masino et al. 2000). They stated that the C-terminal domain has a significantly greater affinity for calcium ions, hence a higher stability than other domains. For a further analysis, the use of various structural mutants of CaM combined with computer simulation studies of the stretching experiment will enable us to better explain the F–E curves of holoCaM and apoCaM.
The use of the PGA model for a more quantitative analysis of the data revealed that the F–E curves for holo and apoCaMs were fitted well to those of PGA containing 40% (pH 5.0) and 0% (pH 8.0) helix, respectively. The observed fit of the apoCaM F–E curve to the 0% helix curve of PGA was rather surprising, but this model is useful for the comparison of mechanical properties of helices of CaM and PGA, and the apparent discrepancy should be explained in the future in an extended study of protein nanomechanics.
Much more effort to refine the quality of the F–E curve is still required. The search for better cross-linkers and improvement in the methods of attachment of protein molecules to the substrate are necessary for future investigations. In spite of the problems associated with the present method, our success in mechanically identifying two distinct forms of CaM will open a new prospect for the application of AFM as a force spectroscopy.
Materials and methods
Protein and chemicals
A derivatized CaM with two cysteines at its N and C termini and with 6 His residues on the N termini was prepared according to a standard method of protein engineering (Sambrook and Russell 2001). A pRCaM plasmid containing cDNA of CaM and cysteine codons on both termini was transformed to Escherichia coli BL21 (DE3). The transformants were expressed by growing them at 37°C overnight in an LB medium containing 100 μg/mL ampicillin, and diluted at 1:100 into one volume of LB + ampicillin. IPTG (0.5 mM) was added to induce an expression when the optical density at 600 nm was 0.5. After shaking at 37°C for 3 h, the cells were harvested by centrifugation and resuspended in 10 mM Tris-HCl (pH 7.5). The expressed protein was purified from cytosolic supernatant by loading it onto an Ni-NTA column. The bound protein was eluted with a Tris-HCl buffer containing 150 mM imidazole. The protein was stored in the presence of a 100× excess of 1,4-dithiothreitol (DTT) at 4°C. These fractions were further purified by gel filtration on a Superdex 75 column using an elution buffer consisting of 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl.
Circular dichroism
Circular dichroism spectra were recorded at 20°C on a J-720WI spectropolarimeter (JASCO) using 1-mm quartz cells. The α-helicities of the solutes were calculated from the following equation:
 |
(1) |
where [θ]222 represents the mean residue ellipticity at 222 nm (Alder et al. 1973).
Sample preparation
Silicon wafers were purchased from Shin-Etsu Silicon and cut into small square pieces of 1 cm × 1 cm and used as substrates in AFM experiments. Commercially available silicon nitride cantilevers (NP; Digital Instruments) and the silicon substrates were treated with 3-aminopropryltriethoxysilane (APTES) after cleaning and oxidation (Idiris et al. 2000). Both tips and substrates were then treated with the heterobifunctional cross-linker N-succinimidyl 3-(2-pyridyl-dithio) propionate (SPDP; Pierce). An aliquot of CaM solution was dropped onto an active substrate treated with SPDP and incubated at room temperature for 1 h before measurement by AFM.
Force measurement
Force measurements were carried out on a Nanoscope III multiprobe AFM (Digital Instruments). Force curves were recorded at two piezo extension velocities of 0.14–0.29 μm/sec. Cantilevers were calibrated for their spring constants using a thermal fluctuations method (Hutter and Bechhoefer 1993).
Figure 1 ▶ depicts a schematic view of the force spectroscopy experiment performed on CaM. The protein was immobilized on a functionalized substrate with SPDP through the cross-linking reaction between one of the terminal cysteine residues and the cross-linker. The AFM tip, which was also treated with the same cross-linker, was carefully brought into contact with the protein-coated surface applying a loading force of <1 nN. During a brief contact time between the tip and the substrate, covalent bonds were occasionally formed between cross-linkers on the tip and the cysteine residues on the other terminus of CaM. The sandwiched protein was then pulled from its two ends when the sample stage was lowered by the movement of the piezomotor. The tensile force inflicted on the protein was calculated by multiplying the deflection of the cantilever by its force constant, and the extension of the sample stage was controlled by a piezo movement. When the probability of obtaining successful protein stretching curves was kept <10% of the total trials, we obtained quantitatively reproducible results, ensuring that they represented those of single molecule stretching.
All experiments were performed under aqueous conditions and at room temperature. Experiments on apoCaM and holoCaM were carried out in 50 mM Tris buffer (pH 7.5) containing 50 mM NaCl (ionic strength, I = 0.05 M) and in the presence of 1 mM EGTA and 1 mM CaCl2, respectively.
Worm-like chain (WLC) model
To estimate the structural flexibility of apoCaM and holoCaM from their force spectra and to obtain quantitative information on the extension of polypeptide length upon protein stretching, the following interpolation formula of the WLC model (Bustamante 1994) was used:
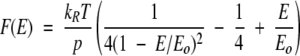 |
(2) |
This formula describes the relationship between the tensile force (F) and the extension (E) of an ideal entropic chain of a total length E0 and a persistence length of p, with kB and T representing the Boltzmann constant and the temperature in kelvins, respectively. The persistence length reflects the polymer stiffness.
Acknowledgments
This work was supported in part by grants-in-aid to A.I. from the Japan Society for the Promotion of Science (Research for the Future Program #99R16701) and from the Japanese Ministry of Education, Science, Culture and Sports (Scientific Research on Priority Areas [B] #11226202).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.3600102.
References
- Alder, A.J., Greenfield, N.J., and Fasman, G.D. 1973. Methods Enzymol. 27: 675–735. [DOI] [PubMed] [Google Scholar]
- Brzeska, H., Venyaminov, S.V., Grabarek, Z., and Drabikowski, W. 1983. Comparative studies on thermostability of calmodulin, skeletal muscle troponin C and their tryptic fragments. FEBS Lett. 153: 169–173. [DOI] [PubMed] [Google Scholar]
- Bustamante, C. 1994. Entropic elasticity of λ-phage DNA. Science 265: 1599–1600. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., Oberhauser, A.F., Fisher, T.E. Marszalek, P.E., Li, H., and Fernandez, J.M. 2000. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. & Mol. Biol. 74: 63–91. [DOI] [PubMed] [Google Scholar]
- Guerini, D. and Krebs, J. 1983. Influence of temperature and denaturing agents on the structural stability of calmodulin. FEBS Lett. 164: 105–110. [DOI] [PubMed] [Google Scholar]
- Hutter, J.L. and Bechhoefer, J. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64: 1868–1873. [Google Scholar]
- Idiris, A., Alam, M.T., and Ikai, A. 2000. Spring mechanics of α-helical polypeptide. Prot. Eng. 13: 763–770. [DOI] [PubMed] [Google Scholar]
- Klee, C.B. 1977. Conformational transition accompanying the binding of Ca2+ to the protein activator of 3`,5`-cyclic adenosine monophosphate phosphodiesterase. Biochemistry 16: 1017–1024. [DOI] [PubMed] [Google Scholar]
- Masino, L., Martin, S.R., and Bayley, P.M. 2000. Ligand binding and thermodynamics stability of a multidomain protein, calmodulin. Protein Sci. 9: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui, K., Hara, M., and Ikai, A. 1996. Mechanical unfolding of α-2-macroglobulin with atomic force microscope. FEBS Lett. 385: 29–33. [DOI] [PubMed] [Google Scholar]
- Rief, M., Gautel, M., Oesterhelt, F., Fernandez, J.M., and Gaub, H.E. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276: 1109–1112. [DOI] [PubMed] [Google Scholar]
- Rief, M., Pascual, J., Saraste, M., and Gaub, H. 1999. Single molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J. Mol. Biol. 286: 553–561. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schulz, G.E. and Schirmer, R.H. 1979. Principles of protein structure, pp. 108–130. Academic Press, New York.
- Walace, R.W., Talant, E.A., and Cheung, W.Y. 1980. Assay, preparation, and properties of calmodulin. In Calcium and cell function, Vol. 1, pp. 13–40. Academic Press, New York.
- Wang, T. and Ikai, A. 1999. Protein stretching III: Force-extension curves of tethered bovine carbonic anhydrase B to the silicon substrate under native, intermediate and denaturating conditions. Jpn. J. Appl. Phys. 38: 3912–3917. [Google Scholar]
- Yang, G., Cecconi, C., Baase, W., Vetter, I., Breyer, W., Haack, J., Matthews, B., Dahlquist, F., and Bustamante, C. 2000. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc. Natl. Acad. Sci. 97: 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova, J., Lindsay, S.M., and Leuba, S. H., 2000. Single molecule force spectroscopy in biology using the atomic force microscope. Prog. Biophys. Mol. Biol. 74: 37–61. [DOI] [PubMed] [Google Scholar]