Abstract
Nano-electrospray ionization time-of-flight mass spectrometry (ESI-MS) was used to study the conformational consequences of metal ion binding to the colicin E9 endonuclease (E9 DNase) by taking advantage of the unique capability of ESI-MS to allow simultaneous assessment of conformational heterogeneity and metal ion binding. Alterations of charge state distributions on metal ion binding/release were correlated with spectral changes observed in far- and near-UV circular dichroism (CD) and intrinsic tryptophan fluorescence. In addition, hydrogen/deuterium (H/D) exchange experiments were used to probe structural integrity. The present study shows that ESI-MS is sensitive to changes of the thermodynamic stability of E9 DNase as a result of metal ion binding/release in a manner consistent with that deduced from proteolysis and calorimetric experiments. Interestingly, acid-induced release of the metal ion from the E9 DNase causes dramatic conformational instability associated with a loss of fixed tertiary structure, but secondary structure is retained. Furthermore, ESI-MS enabled the direct observation of the noncovalent protein complex of E9 DNase bound to its cognate immunity protein Im9 in the presence and absence of Zn2+. Gas-phase dissociation experiments of the deuterium-labeled binary and ternary complexes revealed that metal ion binding, not Im9, results in a dramatic exchange protection of E9 DNase in the complex. In addition, our metal ion binding studies and gas-phase dissociation experiments of the ternary E9 DNase-Zn2+-Im9 complex have provided further evidence that electrostatic interactions govern the gas phase ion stability.
Keywords: Colicin E9, metal ion binding, protein conformation, hydrogen exchange, mass spectrometry
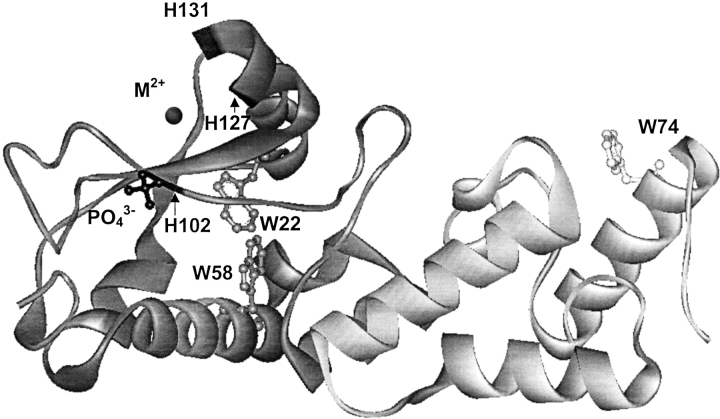
Colicins are bacterial protein antibiotics that are released in times of nutrient or environmental stress by Escherichia coli (James et al. 1996). The family of E-type colicins can be subdivided into two major groups: (1) pore-forming colicins such as E1 (Cramer et al. 1990) and (2) nuclease colicins with either RNase activity (E3, E5, and E6) or DNase activity (E2, E7, E8, and E9) (Lau et al. 1992; Masaki et al. 1992). Colicin E9 is a 60 kD endonuclease composed of three functional domains: the N-terminal translocation domain, the receptor binding domain, and the C-terminal cytotoxic domain (Kleanthous et al. 1998). Although the cell-killing mechanism is still not completely understood, it apparently involves three steps. Initial binding of the colicin E9 to the BtuB extracellular receptor for vitamin B12 is followed by translocation of the E9 DNase domain into the periplasm through the Tol system of proteins (a process that is possibly assisted by the porin OmpF) (James et al. 1996; Lazdunski et al. 1998). On entry of the DNase domain of colicin E9 into the cytoplasm, cell death is caused by cleavage of the chromosomal DNA (Schaller and Nomura 1976; Chak et al. 1991; Pommer et al. 1998). The E9 DNase domain has a high sequence homology with at least three other members of the DNase family (the colicins E2, E7, and E8) (Kleanthous et al. 1998). All these colicins share sequence identity with a larger group of enzymes known as HNH endonucleases (Gorbalenya 1994; Shub et al. 1994). Under normal conditions, colicin-producing bacteria protect themselves from suicide by coexpressing a cognate immunity protein (Kleanthous et al. 1998). Colicin E9 forms a tight heterodimeric complex with its cognate 9.5-kD immunity protein Im9 with a dissociation constant of ∼10−16 M (Wallis et al. 1992; 1995). Immunity proteins specific for other colicins (Im2, Im7, and Im8) also bind to the E9 DNase but with affinities that are six to eight orders of magnitude weaker than the cognate immunity protein Im9, highlighting that these complexes show both high affinity and a high degree of specificity (Wallis et al. 1995). Recent x-ray crystal structures of the E9 DNase domain-Im9 complex (Fig. 1 ▶) indicate that the E9 DNase is a metalloprotein containing a single transition metal within the active site located more than 10 Å away from the protein–protein interface. The HNH motif located at the C terminus spans 32 amino acids and wraps around the bound metal ion to form the core of the E9 DNase active site. It is noteworthy that the transition metal ion is not essential for the cleavage of double-stranded DNA; even so it is centrally located in the active site of the enzyme (Pommer et al. 1999). The crystallography data also revealed that the immunity protein does not bind directly at the active site of the DNase but at an adjacent exosite (Kleanthous et al. 1999; Ko et al. 1999; Kleanthous and Walker 2001). It is believed that bound Im9 sterically and electrostatically repels the putative substrate DNA and, thus, blocks the DNase activity (Kleanthous et al. 1999; Kuhlmann et al. 2000). An interesting conformational feature of the E9 DNase has been revealed by nuclear magnetic resonance (NMR) studies. The E9 DNase interchanges between two conformers with a forward and a backward rate constant of ∼1.6 s−1 and 1.1 s−1 (at 15°C), respectively (Whittaker et al. 1998; 1999), and this conformational equilibrium seems largely to be unaffected by the presence of Zn2+. No extensive conformational changes were observed on Zn2+-binding (Hannan et al. 2000).
Fig. 1.
X-ray structure of the E9 DNase (dark) bound to Im9 (light) (Brookhaven Protein accession code 1bxi). The His residues H102, H127, and H131 coordinating the metal ion are shown in black. The phosphate ion located at the active site is labeled. The tryptophan residues W22 and W58 of the E9 DNase are indicated, as well as W74 of Im9. The figure was constructed with WebLab ViewerPro.
In the present work, we report on the use of nano-electrospray ionization time-of-flight mass spectrometry (ESI-MS) to probe the effect of metal ion binding/release and Im9 binding on the conformational properties of colicin E9 DNase. The determination of composition of metalloproteins and peptides by ESI-MS is well documented (Loo 1997; Lei et al. 1998; Veenstra et al. 1998). ESI-MS is also increasingly recognized as a technique capable to report on protein conformational changes (Konermann and Douglas 1998; Vis et al. 1998; Maier et al. 1999; Krutchinsky et al. 2000). Here we take advantage of the unique capability of ESI-MS-based approaches to monitor conformational heterogeneity and metal ion binding simultaneously. In addition, hydrogen/deuterium (H/D) exchange monitored by MS is used to obtain information regarding the structural integrity, conformational heterogeneity, and dynamics of the metal ion protein interaction. Circular dichroism (CD) and intrinsic fluorescence spectroscopy are used as complementary probes. We show that ESI-MS is a capable approach to study in considerable detail the interaction of the colicin E9 DNase with metal ions, as well as with its cognate immunity protein Im9. Furthermore, we provide evidence that ESI-MS is sensitive toward changes of the thermodynamic stability of E9 DNase. We also point out some limitations of our MS-based approaches for studying conformational properties of proteins.
Results
ESI-MS enables simultaneous observation of different conformational states for apo- and holo-E9 DNase in solution
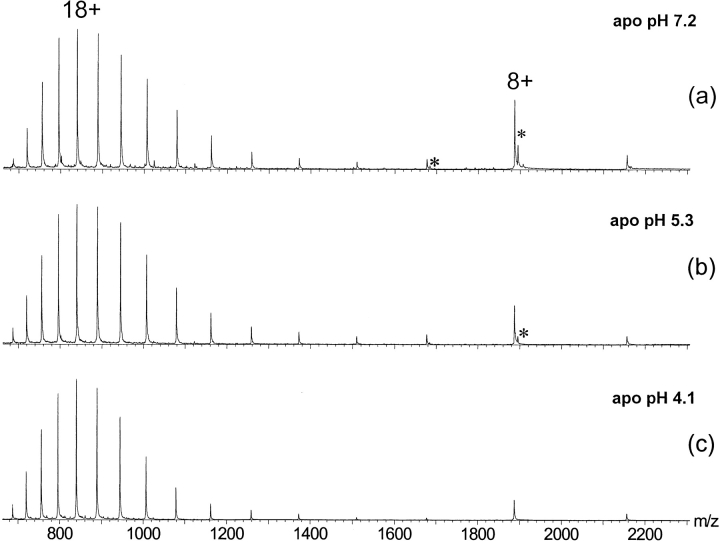
Initial efforts were directed toward establishing experimental conditions that allow the observation of the metal protein complex. The ESI mass spectrum of apo-E9 DNase, acquired in 50 mM ammonium acetate adjusted to pH 7.2, displayed a bimodal charge state distribution encompassing a broad distribution of charge states (with a maximum at 18+) and a second narrow distribution of approximately three charge states (with a maximum at 8+) (Fig. 2 ▶). The calculated masses of the protein, using the individual two charge distributions, were both identical to the expected mass of apo-E9 DNase (Table 1). Bimodal charge distributions of proteins in ESI mass spectra are not unprecedented, and it is assumed that protein populations with different conformational properties contribute to the appearance of the ESI mass spectrum (Chowdhury et al. 1990; Konermann and Douglas 1998; Maier et al. 1999). In the present case we assume that the charge states 7+, 8+, and 9+ originate primarily from more compact conformational states (termed F, folded), whereas the ions with charge states between 11+ and 22+ are considered to originate from more unfolded, loosely packed conformational states (termed U, unfolded). By using the total ion peak areas of the ion peaks termed F and U, respectively, an acid-induced unfolding curve for apo-E9 DNase was deduced by plotting the peak area ratio AF/(AU + AF) as a function of the pH of the solution (Fig. 3 ▶). On further acidification, no significant changes in the ESI spectra of apo-E9 DNase were observed, although the intensity of the highly charged ion peaks increased slightly (Fig. 2 ▶).
Fig. 2.
Nano-electrospray ionization (ESI) mass spectra of the apo-E9 DNase recorded at different pH values: (a) 7.2, (b) 5.3, and (c) 4.1. The ion series labeled with an * has an additional mass of 98 Da.
Table 1.
Summary of the calculated and measured molecular masses of E9 DNase, E9 DNase bound to divalent transition metal ions, Im9, and the immunity complexes
| Average mass (Da) | Apo-E9 DNase | E9-Zn2+ | E9-Ni2+ | E9-Co2+ | Im9 | E9Im9 | E9Im9-Zn |
| Calculated | 15,088.0 | 15,151.4a | 15,143.7a | 15,144.9a | 9582.5 | 24,670.5 | 24,733.4a |
| Measured | 15,088.2 ± 0.3 | 15,151.2 ± 0.3 | 15,142.8 ± 2.0 | 15,146.2 ± 5.0 | 9582.0 ± 0.8 | 24,668.0 ± 3.0 | 24,733.5 ± 0.7 |
a The average mass calculated for the holo-proteins is based on the formula (apo-protein + n Mem − n m H+), where n is the number of metal ions and m their charge (Lei et al. 1998).
Fig. 3.
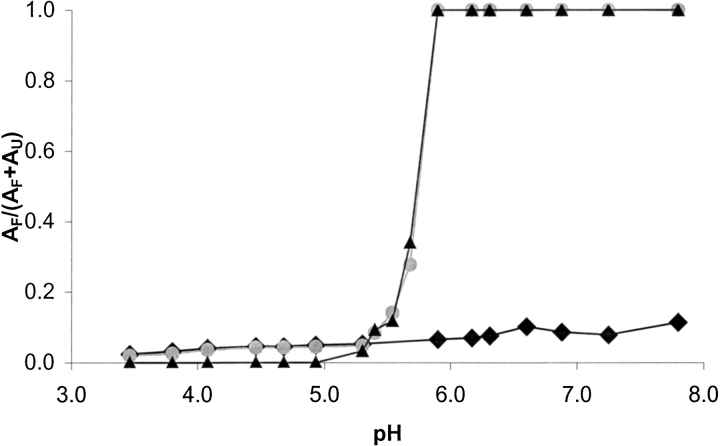
pH-induced unfolding curves for the E9 DNase were deduced by plotting the peak area ratios AF/(AU + AF) as a function of pH. Unfolding curves of holo-E9 DNase and apo-E9 DNase are labeled with (•) and (♦), respectively. Curve indicated with (▴) is deduced from AF Zn /(AF Zn + AF apo + AU); whereby (F) represents the folded conformer (from 9+ to 7+) and (U) the unfolded conformer (from 10+ to 22+) as illustrated in Figure 2 ▶. The data points for the unfolding curve of holo-E9 DNase were extracted from nano-ESI mass spectra of E9 DNase in the presence of an equimolar amount of Zn2+ in 50 mM ammonium acetate adjusted to different pH values. Some representative mass spectra are shown in Figure 6 ▶.
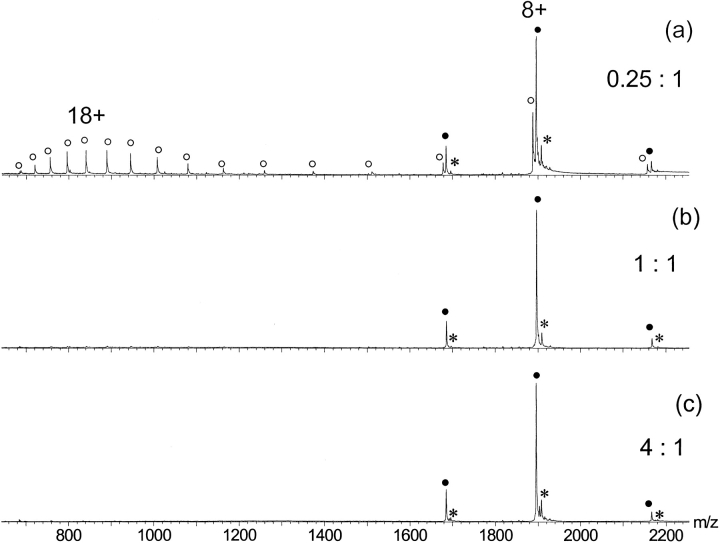
A particularly interesting structural feature of the E9 DNase domain is the so-called HNH motif, an amino acid sequence stretch that is shared with homing endonucleases (Gorbalenya 1994; Shub et al. 1994). Two histidine residues within the HNH motif (His102 and His127) and one additional His residue located at the end of α-helix 9 (His131) tetrahedrally coordinate the metal ion. A phosphate ion acts as a fourth ligand in the X-ray structure (Kleanthous et al. 1999; Kuhlmann et al. 2000). The transition metal ion Zn2+ binds with the highest affinity to the HNH motif of E9 DNase (Pommer et al. 1999). Thus, metal-binding studies were initiated by performing titration experiments with Zn2+. Adding Zn2+ in stoichiometric amounts to a 13-μM apo-DNase E9 solution at pH 7.2 resulted in mass spectra that showed a dramatically different appearance compared with those observed for apo-E9 DNase under identical conditions. Although for apo-E9 DNase, a bimodal charge state distribution was observed (Fig. 2 ▶), for Zn2+-bound E9 DNase a narrow charge state distribution encompassing exclusively charge states between 7+ and 9+ (Fig. 4 ▶) was observed. The measured mass of 15,151.2 ± 0.3 Da indicated that these ions originate from the E9 DNase protein bound to one single Zn2+-ion, reflecting the expected 1 : 1 stoichiometry. In the case of one equivalent of Zn2+, binding appears to be completely saturated (Fig. 4b ▶). Even with a large excess of Zn2+, the protein with one metal ion bound was the predominantly observed species (Fig. 4c ▶). Less than stoichiometric amounts of Zn2+ resulted in mass spectra displaying concurrently the bimodal charge state distribution typically observed for the apo-protein and the narrow charge state envelope of the holo-protein (Fig. 4a ▶).
Fig. 4.
Nano-ESI mass spectra of E9 DNase in the presence of increasing amounts of Zn2+. Molar ratio of zinc acetate to apo-E9 DNase: (a) 0.25 : 1, (b) 1 : 1, and (c) 4 : 1. Ion peaks of Zn2+-bound and metal-free E9 DNase are labeled with filled circles (•) and open circles (○), respectively. The ion series labeled with an * has an additional mass of 98 Da.
ESI-MS reports prevalence of E9 DNase for Zn2+
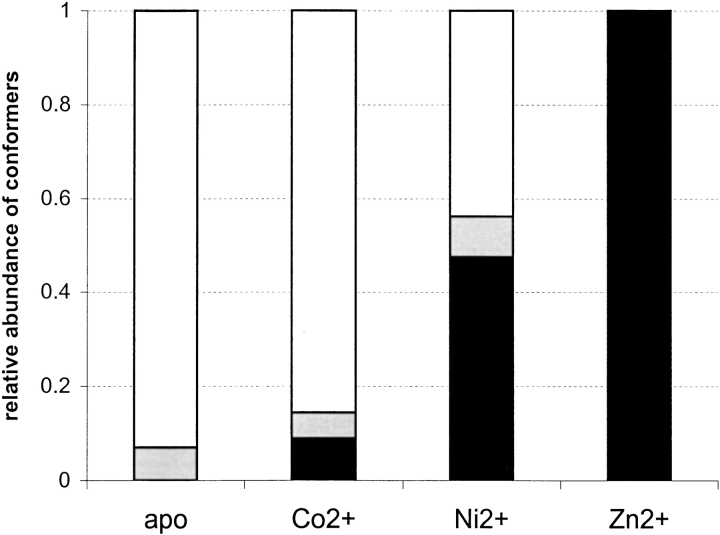
Zn2+ is reported as the metal ion having the strongest affinity for the HNH motif of E9 DNase (Kd in the low nM range), whereas the affinity for Co2+ and Ni2+ is ∼1000-fold lower (i.e., in the μM range) (Pommer et al. 1999). Thus, we were interested if metal ion affinities for the E9 DNase derived from ESI-MS would reflect the metal binding behavior in solution. Stoichiometric mixtures of the DNase and the two other transition metal ions, Co2+ and Ni2+, respectively, were subjected to analysis. MS analysis of a stoichiometric mixture of apo-E9 DNase and Co2+ yielded ion species resulting in an average mass of 15,088.2 ± 0.3 Da (i.e., metal-free E9 DNase) and 15,146.2 ± 5.0 Da (i.e., Co2+-bound E9 DNase), respectively, indicating the concomitant presence of the apo- and holo-protein. The Co2+-bound holo-protein displayed again only a single, narrow charge state distribution encompassing the charge states 7+ to 9+. The mass spectrum revealed further that metal binding is not yet saturated when using one equivalent of Co2+ (data not shown). A similar experiment using a 1 : 1 molar mixture of E9 DNase and Ni2+ provided an ESI mass spectrum similar to the one observed for Co2+, although the charge envelope for Ni2+-bound holo-protein (giving an experimental mass of 15,142.8 ± 2.0 Da) was relatively more abundant than the one observed for the Co2+-bound holo-protein (data not shown). Measured masses for the holo-proteins demonstrating 1 : 1 stoichiometry of transition metal ion binding are summarized in Table 1. From the ESI mass spectra (at pH 7.2) obtained for the 1 : 1 mixtures of protein and different transition metal ions, ratios of apo- to holo-ions were calculated by summing up the corresponding ion peak areas. These ratios are graphically presented in Figure 5 ▶. Assuming that the deduced apo- to holo-ion ratios reflect relative metal ion binding affinities, it is found that Zn2+ > Ni2+ > Co2+ is in qualitative agreement with literature data (Pommer et al. 1999). Earlier work indicated that Mg2+ does not bind to the protein at least in the absence of DNA, but that it is essential for the nonspecific endonuclease activity of E9 DNase, (Pommer et al. 1999). Thus, Mg2+ was used as a probe to test the specificity of our MS-based metal-binding studies. MS analysis of an E9 DNase solution containing a ninefold excess of Mg-acetate in a 50 mM ammonium acetate solution (pH 7.2) yielded exclusively the bimodal charge state distribution typically observed for the apo-protein. No charge states of Mg2+-bound E9 DNase were observed (data not shown), thus stressing the specificity of the MS-based metal-binding study.
Fig. 5.
The relative binding affinities of the E9 DNase for different divalent metal ions. Bars shown in white represent the nonmetal-containing unfolded conformer; bars shown in grey represent the nonmetal-containing folded conformer; and bars shown in black represent the folded metal-containing conformer.
Acid-induced metal release causes conformational destabilization of E9 DNase
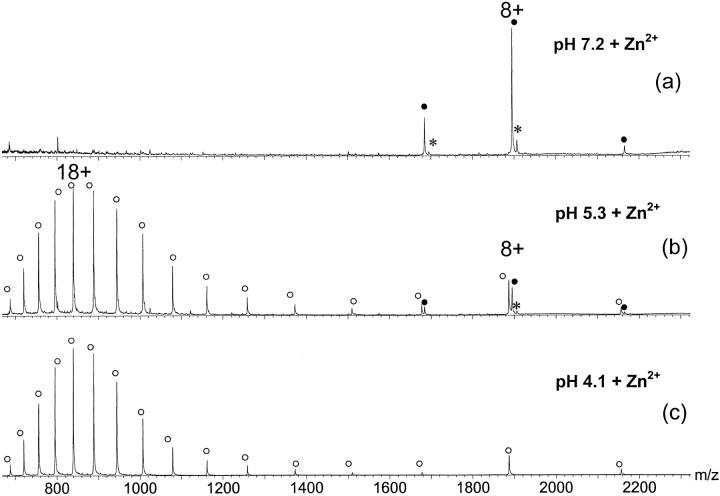
Pommer et al. (1999) have suggested previously that association of colicin E9 with the outer membrane receptor BtuB and the periplasmic Tol proteins causes dissociation of both Im9 and the bound metal ion in preparation for translocation of the cytotoxic DNase domain into the cell, possibly via a destabilized state (Pommer et al. 1999). Thus, we were interested to determine if the local acidic pH near the membrane surface of the target cell may trigger the release of the metal ion causing conformational destabilization of the DNase domain. ESI-MS is particularly well suited for this kind of conformational unfolding study because of its unique capability of monitoring simultaneously different conformational populations and metal ion binding. To study the pH-induced conformational transitions of the holo-E9 DNase, protein solutions with pH values between 3.5 and 7.8 were analyzed by ESI-MS. Three illustrative ESI mass spectra are shown in Figure 6 ▶. The mass spectrum of the holo-E9 DNase at pH 7.2 was dominated by the less charged peaks with the 8+-ion peak as the predominant one. All the ions observed originate from the E9 DNase protein bound to one Zn2+-ion. Decreasing the pH to 5.3 caused metal ion release accompanied with the appearance of highly charged ion peaks in the lower mass-to-charge (m/z) range. No Zn2+-binding was observed for these higher charge state ions that presumably originate from less compact conformational states. On further acidification, the abundance of the highly protonated charge states increased, but the maximum of the charge state envelopes remained at 18+ (Fig. 6 ▶). As described for the apo-protein, the AF/(AU + AF) ratios were calculated from the ESI mass spectra acquired at different pH values and gathered in Figure 3 ▶, illustrating a strong pH-dependent release of Zn2+ that is apparently accompanied by major conformational destabilization.
Fig. 6.
Nano-ESI mass spectra of E9 DNase in the presence of an equimolar amount of Zn2+ at pH 7.2 (a), 5.3 (b), and 4.1 (c) in 50 mM ammonium acetate. The filled circles (•) represent ion peaks of the Zn2+ bound; the open circles (○) represent ion peaks of the metal-free E9 DNase. The repeatedly observed ion series with an additional mass of 98 Da is labeled with an *.
MS characterization of E9 DNase in complex with its cognate immunity protein Im9
To investigate E9 DNase in complex with Im9, an equimolar amount of Im9 was added to a solution of E9 DNase in ammonium acetate adjusted to pH 7.2. The nano-ESI spectrum of the E9 DNase-Im9 complex showed a very narrow charge state distribution dominated by two charge states, that is, 9+ and 10+ at m/z 2743 and 2469, respectively (data not shown). The determined mass of 24,668.0 ± 3.0 Da agreed well with the expected mass of the E9 DNase-Im9 complex (Table 1). These narrow charge state distributions are frequently observed in studies of noncovalent complexes and are generally considered as an indication of compact structures that are preserved in the gas phase (Loo 2000). Interestingly, at pH 7.2 the most abundant charge states observed in the ESI mass spectra of E9 DNase and Im9 are 8+ and 5+, respectively. The apparent reduction in the degree of protonation observed for the complex may to some extent be caused by salt bridges formed on Im9 complexation linking Arg54 and Lys97 of E9 DNase with Glu30 and Glu41 of Im9, respectively (Kuhlmann et al. 2000).
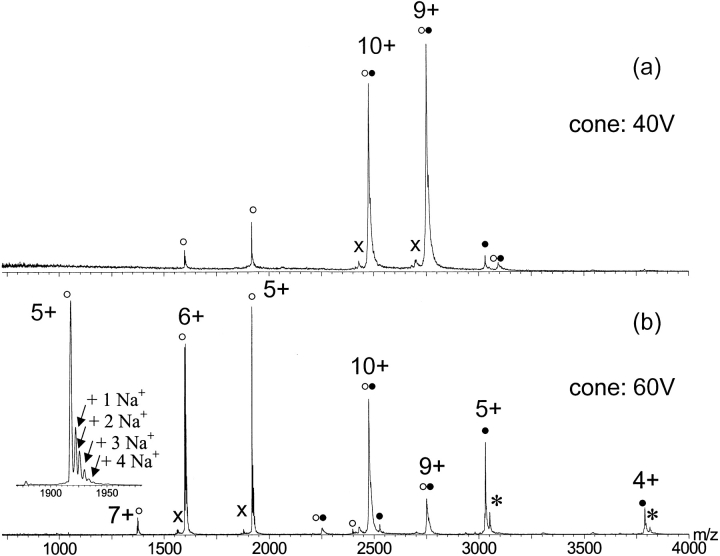
ESI-MS of the ternary complex obtained by adding zinc acetate to the preformed E9 DNase-Im9 complex (at pH 7.2) revealed a charge state distribution comparable to the one observed for the binary complex, encompassing the charge states 8+ to 10+ at m/z 2249, 2474, and 2749, respectively (Fig. 7 ▶). The observed mass of 24,733.5 ± 0.7 Da is consistent with the expected mass of the Zn-bound E9 DNase-Im9 ternary complex (Table 1).
Fig. 7.
Nano-ESI mass spectra of the E9 DNase-Im9-Zn complex in 50 mM ammonium acetate at pH 7.2 at cone voltages of 40 V (a) and 60 V (b). The filled circles (•) represent ion peaks of the E9 DNase containing the Zn2+ ion, and open circles (○) correspond to ion peaks of Im9 after cone voltage dissociation. The filled and open circles combined (○•) denote ion peaks representing the E9 DNase-Im9-Zn complex. The insert shows the 5+-ion peak of Im9 after cone voltage dissociation with up to four Na+ ions remaining. The ion series labeled with (x) is lacking a mass of 475 ± 3 Da, probably because of a truncated Im9 variant missing the C-terminal tetrapeptide FKQG.
The gas phase stability of noncovalent complexes can be evaluated by collision-induced dissociation- (CID) MS experiments (Potier et al. 1998; Versluis and Heck 2001). Dissociation of noncovalent complexes is achieved by increasing the internal energy of the ions entering the mass spectrometer. In our instrumental setup, the cone voltage can be used to incrementally increase the internal energy of the multiply charged ions until dissociation of the complex is achieved. Increasing the cone voltage above 35 V caused dissociation of the E9 DNase-Im9 binary complex, resulting in two additional charge state distributions in addition to the one of the binary complex. The charge state envelope below m/z 2400 encompassing the charge states 4+ to 7+ was assigned to Im9. A more detailed look at the individual charge states of Im9 revealed the presence of some sodium adduction. The second envelope with charges states +4, +5, and +6 at m/z 3774, 3019, and 2516, respectively, was attributed to apo-E9 DNase. Interestingly, apo-E9 DNase ion peaks did not show sodium adduction. We may speculate that the high number of acidic residues in Im9 (16 of 86) promotes alkali metal ion binding. Similar persistent sodium adduction had been encountered in ESI-MS studies of oligonucleotides (Liu et al. 1996) and DNA-binding proteins (Potier et al. 1998), and here it was assumed that the highly negatively charged phosphate backbone of the oligonucleotide causes very tight alkali metal ion binding (Donald et al. 2001). At higher cone voltages, very minor additional peaks, subsidiary to the predominant charge envelope of Im9, appeared that may indicate breakage of covalent bonds in Im9 (data not shown).
Likewise, gas-phase dissociation experiments of the ternary Zn2+-loaded E9 DNase-Im9 complex (Fig. 7 ▶) yielded mass spectra similar to those observed for the binary complex. However, a more detailed analysis revealed that the charge state distribution assigned to E9 DNase resulted in an average mass of 15,151.1 ± 0.4 consistent with one Zn2+-ion bound to E9 DNase. Despite the very high affinity of the E9 DNase-Im9 complex observed in solution, in the gas-phase disruption of the protein–protein interaction occurs before dissociation of the protein–metal ion interaction. This is in accordance with the emerging view that electrostatic interactions are enhanced, whereas the hydrophobic effect becomes less significant in the gas phase (Loo 1997; Smith et al. 1997a,b).
Assessing the conformational integrity of E9 DNase by H/D exchange
The E9 DNase domain possesses a total of 261 exchangeable hydrogens (123 backbone amide hydrogens, 135 side-chain hydrogens, and 3 hydrogens from both termini). For Im9, a total of 153 labile hydrogens were counted (82 backbone hydrogens, 68 side-chain hydrogens, and 3 hydrogens from the termini). ESI-MS was used to monitor directly H/D exchange of peptide amide and side-chain hydrogens. Consequently, multiply deuterated molecular ions, that is, (M + D)n+, rather than multiply protonated protein ions, that is, (M + H)n+, were observed. It is important to point out that all H/D exchange experiments were monitored by "normal" ESI-MS, and that the protein solutions were introduced into the ESI source via a syringe pump. With this setup, deuterium levels for fully deuterated E9 DNase and Im9 of (97 ± 1)% and (96 ± 1)%, respectively, were observed.
H/D exchange data for the E9 DNase were obtained from three independent measurements in the presence and absence of a sixfold excess of Zn2+-ions. An attempt was made to follow the H/D exchange kinetically. However, it was observed that deuterium incorporation had already reached a plateau level ∼2 min after diluting the protein solution into deuterating ammonium acetate solution. Because this time is about the earliest time point that was achieved with the current instrumental setup, we decided instead to focus on the number of deuterons present in the protein under equilibrium conditions, that is, an incubation period of 5 min. More extended incubation periods (up to 1 hr) did not result in substantially higher deuterium incorporation. The results of the H/D experiments are summarized in Table 2.
Table 2.
Levels of deuterium incorporation observed for E9 DNase, Im9, and the E9 DNase/Im9 complex in the presence and absence of Zn2+
| Deuterium incorporation after 5 min H/D exchange pD 7.2 | Deuterium incorporation fully deuterateda | ||||
| Complex | Protein | % | % | ||
| E9 | 246 ± 4 | 94 ± 2 | 253 ± 3 | 97 ± 1 | |
| E9-Zn2+ | 197 ± 2 | 75 ± 1 | n.m.c | n.m.c | |
| Im9 | 115 ± 5 | 75 ± 3 | 147 ± 2 | 96 ± 1 | |
| E9-Im9 | E9b | 243 ± 6 | 93 ± 2 | n.m.c | n.m.c |
| Im9b | 99 ± 6 | 65 ± 4 | n.m.c | n.m.c | |
| E9-Im9-Zn2+ | E9-Zn2+b | 188 ± 3 | 72 ± 1 | n.m.c | n.m.c |
| Im9b | 97 ± 6 | 63 ± 4 | n.m.c | n.m.c | |
Noted as well are the deuterium levels determined for E9 DNase and Im9 after collision-induced dissociation of the immunity complex.
a After 12 h at 37°C in pD 2.4.
b After cone voltage dissociation.
c No complex at pD 2.4.
For the apo-E9 protein at pD 7.2, H/D exchange resulted after 5 min in an average mass increase of 246 ± 4 Da corresponding to an exchange of (94 ± 2)% of all labile hydrogens to deuterons (Table 2). No difference in mass increase was observed for any of the charge states assigned to the bimodal charge state distribution typically observed for apo-E9 DNase. In contrast, holo-E9 DNase domain showed a significantly lower deuterium uptake, and the observed average mass increase was 197 ± 2 Da, indicating that (∼75 ± 1)% of all labile hydrogens experienced exchange (Table 2).
For the binary complex composed of apo-E9 DNase and Im9, a deuterium uptake of 342 ± 6 (83 ± 1)% was determined, whereas the ternary complex, that is, Zn2+-bound E9 and Im9, showed a drastically lower deuterium incorporation (i.e., 285 ± 6, (68 ± 1)%). A somewhat lower deuterium uptake as a consequence of complex formation is anticipated because previous solvent accessible labile hydrogens will become buried within the protein–protein interface. However, to further be able to rationalize the drastically different exchange behavior of the binary compared with the ternary complex, the labeled complexes were subjected to collision-induced dissociation experiments. Subsequently, the deuterium levels were determined from the mass shifts of the complex constituents observed. The deuterium level found for Im9 as a constituent of the binary complex was very similar to the one that was found for Im9 in the ternary complex, namely, ∼99 ± 6 in the binary complex and 97 ± 6 in the ternary complex (Table 2). In contrast, holo-E9 DNase when a constituent of the ternary complex showed significantly less deuterium incorporation, that is, 188 ± 3 versus the 243 ± 6 found for apo-E9 DNase in the binary complex (Table 2).
Conformational studies of E9 DNase by tryptophan fluorescence emission spectroscopy
To complement our MS experiments, we studied the consequence of metal binding/release on the conformational properties of E9 DNase by intrinsic tryptophan fluorescence emission spectroscopy. It has been shown previously that tryptophan fluorescence emission is a sensitive probe to study changes of the tertiary structure of the E9 DNase (Pommer et al. 1999). The nuclease contains two Trp residues, W22 and W58 (Fig. 1 ▶). Both Trp residues are part of helical regions and are buried in the protein core. The Trp residues are therefore sensitive probes to study the conformational transitions of E9 DNase on acid-induced metal ion release and complementing the MS-derived findings on changes in the tertiary structure of E9 DNase. The fluorescence spectrum of holo-E9 DNase at pH 7.4 in 50 mM ammonium acetate shows an emission maximum at 334 nm, indicating that the Trp residues are buried in the nonpolar protein core. The fluorescence spectrum of the apo-protein (under identical solution conditions) was similar to that recorded for the holo-protein, that is, λmax = 334 nm, but with a slight fluorescence enhancement. These data are in agreement with previously reported fluorescence data obtained in potassium phosphate buffer (Pommer et al. 1999), indicating that the absence of phosphate ions seems not to substantially affect the tertiary structural properties of E9 DNase. This is important because all MS experiments were performed in 50 mM ammonium acetate solutions (as preferred for the ESI process). Furthermore, the similarity of the fluorescence spectra for apo- and holo-protein is interesting because dramatically different ESI mass spectra were obtained for the holo- and apo-protein under corresponding solution conditions, that is, for the holo-E9 DNase a unimodal charge state distribution was observed, whereas the apo-protein showed a bimodal charge state distribution.
However, pH-induced unfolding experiments revealed differences in the conformational stability of apo- and Zn2+-bound E9 DNase. Illustrative fluorescence spectra of the apo- and holo-E9 DNase are shown in Figure 8 ▶. Fluorescence spectra of the holo-E9 DNase did not change significantly between pH 7.4 and 5.3, but further acidification caused red-shifting of the fluorescence maximum to about 353 nm, marking unfolding. In contrast, the fluorescence spectra of the apo-E9 DNase revealed a gradual shift of the fluorescence maximum with decreasing pH values. At pH 3.9 the fluorescence spectra of the apo- and holo-E9 DNase proteins were very similar, showing fluorescence maxima of 353 nm observed typically for unfolded states.
Fig. 8.
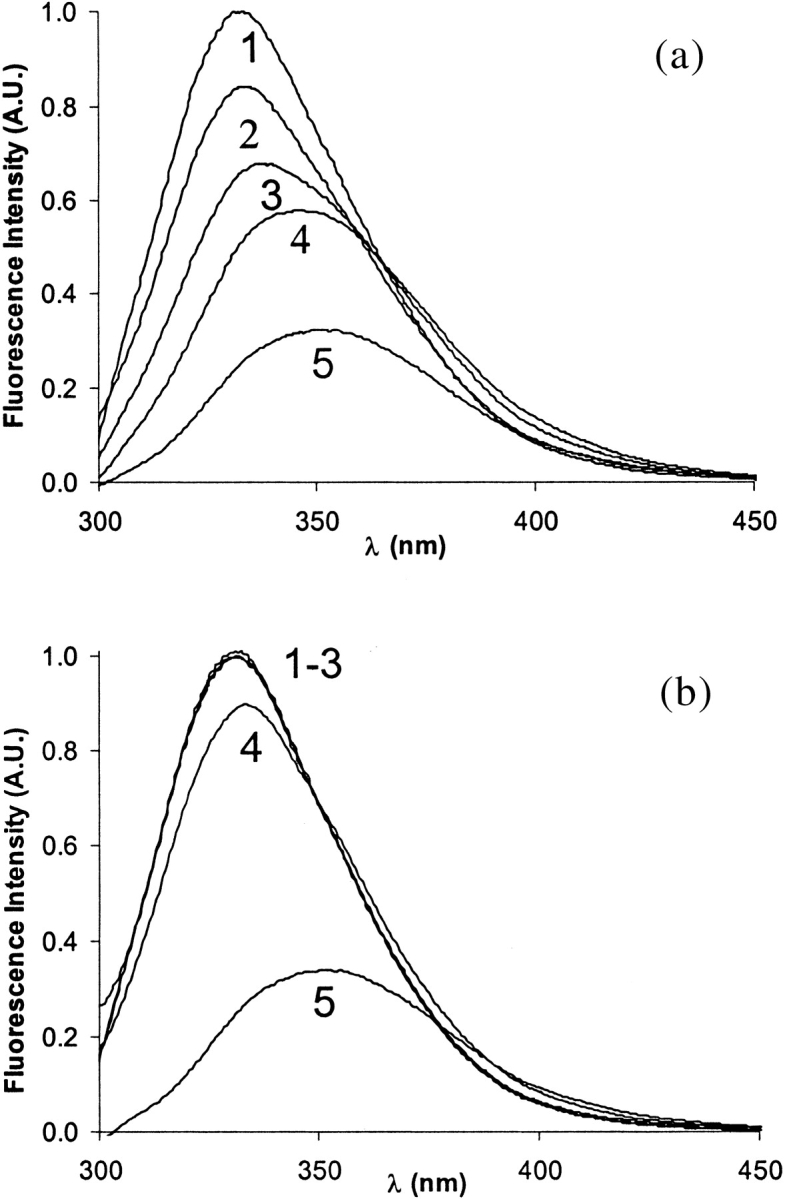
Changes in the tryptophan fluorescence emission spectra during acid-induced unfolding of E9 DNase (6 μM), Im9 (6 μM), and E9 DNase-Im9 complex (6 μM) in 50 mM ammonium acetate. (a) Curves 1–5 represent the apo-E9 DNase at pH 7.4, 5.9, 5.3, 4.9, and 3.9; (b) curves 1–5 represent E9 DNase in the presence of a sixfold molar excess of Zn2+ at pH 7.4, 5.9, 5.3, 4.9, and 3.9, respectively.
Acid-induced metal ion release probed by CD spectroscopy
In addition, CD spectroscopy was used to assess structural changes in E9 DNase on addition of acid in the presence and absence of Zn2+. Far-UV CD spectra of apo- (Fig. 9a ▶) and holo-protein (Fig. 9b ▶) in 50 mM ammonium acetate at neutral pH were similar to those previously reported (Pommer et al. 1999), confirming that apparently only minor changes in the secondary structure occur on metal binding/release. Interestingly, on acidification to pH 4 the negative ellipticity at 222 nm remained largely unaffected independent of the presence or absence of Zn2+. In contrast, a conformational transition on acid-induced metal release was observed in the ESI-MS, as well as in the fluorescence spectroscopic experiments.
Fig. 9.
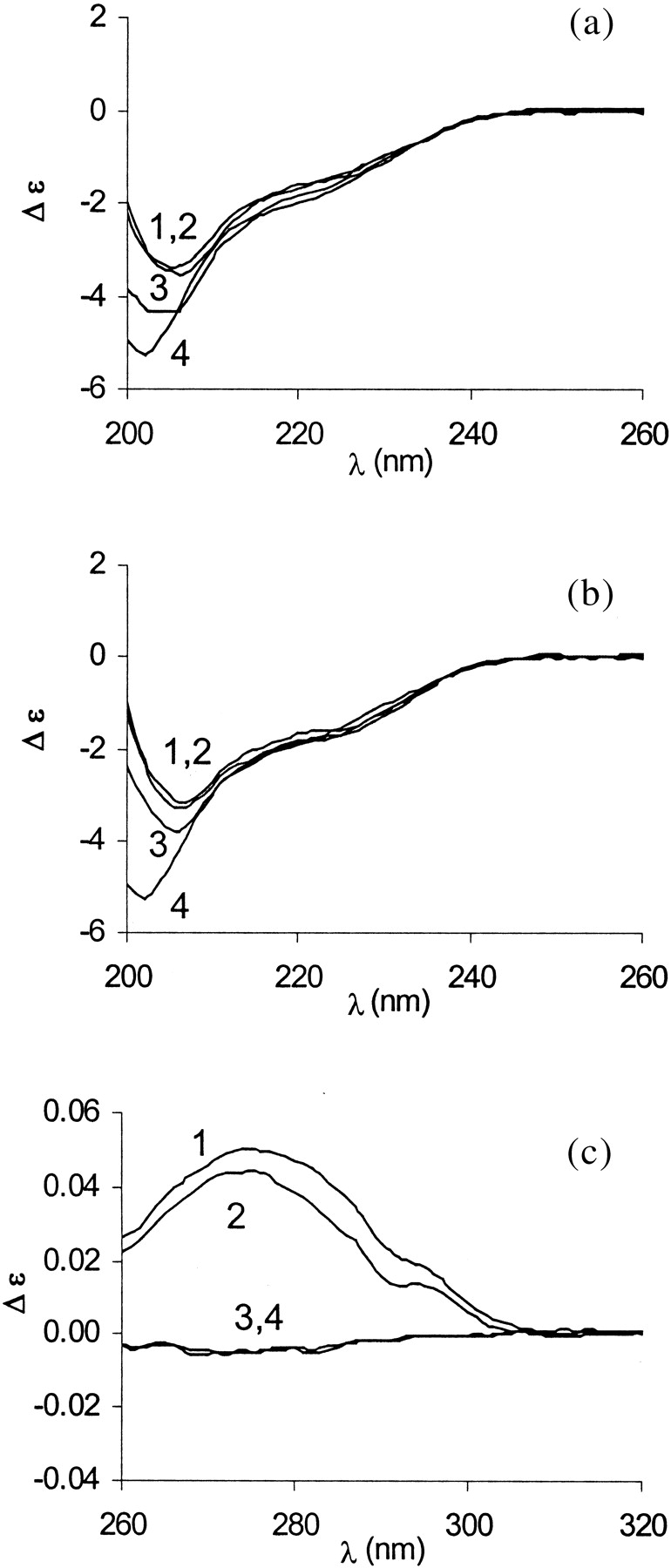
Far-UV (a, b) and near-UV (c) CD spectra of E9 DNase in 50 mM ammonium acetate. (a,b) Curves 1–4 represent pH 7.4, 5.9, 5.3, and 3.9 of the apo- and holo E9 DNase (8 μM), respectively. (c) Curves 1 and 2 denote the E9 DNase (50 μM) at pH 7.4 in the absence and presence of Zn2+. Curves 3 and 4 represent the E9 DNase at pH 3.9 in the absence and presence of Zn2+.
The near-UV CD spectrum of holo-E9 DNase at neutral pH shows a broad prominent positive band at 275 nm and a minor one at 295 nm (Fig. 9c ▶). Under identical solution conditions, the near-UV CD spectrum of the apo-protein has a very similar appearance (Fig. 9c ▶) stressing the fact that apo-E9 DNase has a defined tertiary structure even in the absence of metal ions. However, at pH 4, apo- and holo-E9 preparations are devoid of the near-UV CD signal, thus indicating loss of ordered structure (Fig. 9c ▶).
Discussion
ESI-MS is sensitive to metal-induced changes in the thermodynamic stability of E9 DNase
In this work, we evaluate the use of ESI-MS to probe the conformational properties of the metalloprotein colicin E9 DNase. In the ESI-MS process, multiple protonated protein ions (charge states) are generated and transferred intact into the gas phase of the mass spectrometer (Smith et al. 1990). It has been observed that ESI mass spectra of a protein analyzed from solutions causing protein unfolding experience a greater extent of charging in the ESI process when compared with the same protein analyzed from solutions that favor the folded, "native" conformation (Chowdhury et al. 1990; Przybylski and Glocker 1996; Konermann et al. 1997). The metal-binding stoichiometry in ESI mass spectra is reflected as corresponding mass shifts. Thus, these intrinsic properties of ESI-MS allow simultaneous assessment of the composition of the protein–ligand complex and the conformational heterogeneity. In contrast, the optical techniques commonly used to monitor conformational changes, such as CD (Swint and Robertson 1993) and fluorescence spectroscopy (Eftink 1994), measure the average properties of an ensemble but do not allow a direct observation of distinct conformational states.
The metal-free colicin E9 DNase domain showed bimodal charge state distributions in which the charge states centered around the 18-fold protonated ion peak were attributed to loosely packed, conformational states of metal-free E9 DNase and those centered around the 8-fold protonated ion peak to more compact, folded states. No intermediate charge states were observed. These results have been interpreted as showing that the apo-E9 DNase is significantly destabilized in solution with the ESI mass spectra able to reflect to the conformational states populated in solution, that is, more loosely packed, unfolded conformational states, collectively termed "U" states, in equilibrium with more compact, folded "F" states. The thermodynamic stability of E9 DNase increases substantially on metal ion binding (Pommer et al. 1999), thus shifting the thermodynamic equilibrium to the compact states. This shift of the conformational equilibrium is reflected in the ESI mass spectra by the absence of highly charged ion peaks. Hence, ESI-MS seems to be very responsive to changes of protein conformational stability.
However, the present work also provides evidence that ESI-MS overestimates somewhat the contribution of the loosely packed conformational states to the conformational heterogeneity in solution. For instance, the ion peak ratio AF/(AU + AF) is close to 1/9 at neutral pH (Fig. 3 ▶), but the fluorescence spectrum under the identical solution condition is clearly dominated by a strong intrinsic fluorescence emission at 334 nm, indicating that the two tryptophans in E9 DNase (W22 and W58) are in a nonpolar environment (Fig. 8 ▶). Likewise, apo-E9 DNase at neutral pH showed a strong near-UV CD signal (Fig. 9 ▶), also indicating dominance of folded conformational states with stable tertiary structure. To rationalize the pronounced bimodal charge state distribution observed for apo-E9 DNase, we have to recall some intrinsic features of ESI-MS. First, the relative abundance of conformer-specific ions in ESI mass spectra cannot simply be related to their fractional contribution in solution. Recent work by Cech and Enke (Cech and Enke 2000;2001) on tripeptides shows that the ionization efficiency depends critically on the tripeptide's relative affinity for the droplet surface. Tripeptides with higher nonpolar character and, thus, higher affinity for the droplet surface gave a higher ESI response than more hydrophilic ones. It can be speculated that in highly destabilized conformational states the increased exposure of hydrophobic patches enhances the affinity for the droplet surface, leading to relatively increased ionization rates and ESI response. Dobo and Kaltashov (2001) also observed an apparent higher ionization efficiency for structurally disordered protein conformational states (Dobo and Kaltashov 2001). Second, for highly charged ions, the ion detection efficiency is somewhat increased. Both aspects of ESI-MS contribute that ions originating from disordered conformational states show relatively increased signals in ESI-MS.
The intrinsic conformational heterogeneity of E9 DNase is not observed in ESI-MS experiments
NMR experiments have revealed that the E9 DNase slowly interconverts between two conformers. Two distinct regions showing this conformational heterogeneity have been identified, namely, the sequence stretches encompassing the residues 20–25 and 65–72 (Whittaker et al. 2000). Hannan et al. reported that on Zn2+-binding, the conformational equilibrium remains undisturbed, and that Zn2+-binding does not result in significant conformational changes in E9 DNase (Hannan et al. 2000). However, in our present MS experiments at pH 7, holo-E9 DNase yielded mass spectra showing a unimodal charge envelope with a charge maximum at 8+. Narrow charge envelopes are generally considered as an indication of compact gas-phase structures originating from compact conformational states. Hence, it has to be concluded that ESI-MS failed to report on the conformational heterogeneity described for the Zn2+-bound E9 DNase. In the X-ray structure, the conformationally heterogeneous regions are in spatial proximity and located at or near the protein surface. Thus, we may speculate that the conformational switch between the two conformers has to be rather localized and apparently does not result in changes of the accessibility of protonation sites. At present, a full understanding of the conformational heterogeneity of E9 DNase is still lacking. Apparently, Trp 22 plays a central role in the conformational dynamics (Whittaker et al. 2000). However, to further elucidate if a possible reorientation of the Trp residue is involved in the multiple conformations observed in NMR, we intend to apply time-resolved tryptophan fluorescence in combination with time-resolved anisotropy.
Acid-induced metal ion release studies underscore the role of the metal ion as a conformational clamp
ESI mass spectra of colicin E9 DNase in the presence of Zn2+ at pH values ranging from 4 to 5.5 enabled the observation of distinctive charge state distributions for the apo- and holo-protein at the same time, that is, the bimodal charge state distribution attributed to apo-E9 DNase and the narrow charge state distribution of the holo-E9 DNase are observed simultaneously. The lack of intermediate charge state distributions on acid-induced metal release is interpreted as an indication that partially folded conformational states are not significantly sampled. There was also no indication that partly unfolded holo-species were present, as observed in the study of the N-lobe of transferrin on acid-induced unfolding (Gumerov and Kaltashov 2001). In addition, our Zn2+-titration experiments provide further evidence that metal ion binding is exclusively associated with the folded state.
Interestingly, apo-E9 DNase at low pH had negative ellipticity at 222 nm, whereas the near-UV CD signal and the Trp fluorescence were lost. Thus, at least under acidic conditions, apo-E9 DNase seems to adopt features that are reminiscent of molten globule-like states, that is, states that lack a fixed tertiary structure but in which secondary structure is retained (Le et al. 1996; Chak et al. 1998). In this regard, it is intriguing to speculate that the local acidic pH at the membrane surface triggers metal ion release and concomitantly renders the DNase in a molten globule-like state competent for membrane translocation (Prats et al. 1986; McLaughlin 1989; Bychkova et al. 1996). Interestingly, Chak et al. have suggested that colicin E7 uptake into cells is aided by the acidic pH at the membrane surface and involves an unfolded intermediate (Chak et al. 1998). Furthermore, it is reported that insertion of the pore-forming domain of colicin A in the cytoplasmic membrane is preceded by a native to molten globule transition at acidic pH (Van der Goot et al. 1991).
Gas phase stability and, thus, ESI-MS detectability is governed by electrostatic interactions
E9 DNase is a metalloprotein that binds a single transition metal ion within the HNH motif. Isothermal titration calorimetry established that Zn2+ is by far the tightest binding metal ion compared with the other two tested transition metal ions, namely Co2+ and Ni2+. Zn2+ possesses Kd values in the low nanomolar range, whereas the Kd values for Co2+ and Ni2+ are in the micromolar range (Pommer et al. 1999). Although the presented metal ion-binding studies qualitatively indicate the prevalence of E9 DNase for Zn2+, they do not accurately reflect the solution-derived binding parameters. We believe this deviation is a strong indication that the stabilization of gas phase complexes by electrostatic interactions may have a dramatic influence on the ion species detected by ESI-MS. In the absence of an aqueous environment, van der Waals interactions become less important, whereas ionic and hydrogen bonding interactions are enhanced. As a consequence, the stability of metal ion–protein complexes derived from ESI-MS experiments do not necessarily reflect their solution stability.
Noticeably, our gas-phase dissociation experiments of the ternary E9 DNase-Zn-Im9 complex highlight the dominance of electrostatic interactions in influencing the gas phase stability of noncovalent complexes. The protein–protein complex of E9 DNase and its cognate immunity protein Im9 are considered as having one of the lowest equilibrium dissociation constants reported so far for a protein complex (Kd ∼ 10−16 M in the absence of salt) (Wallis et al. 1995). In fact, the difference in binding affinity between Zn2+ and Im9 for the E9 DNase is seven orders of magnitude. Nevertheless, in our gas-phase dissociation experiments, dissociation of the protein constituents occurs before the release of the metal ion. A detailed analysis of the protein–protein interface of the E9 DNase-Im9 complex (Kleanthous et al. 1999; Kuhlmann et al. 2000) shows that 14 residues from the E9 DNase and 17 residues from Im9 are directly involved in forming the interfacial contact. Key contributions in defining the stability and specificity of the complex are (1) a hydrophobic core with a stacking interaction between Phe86 of the E9 DNase and Tyr54 of Im9 in the center, (2) a hydrogen-bonding network of 12 hydrogen bonds including two salt bridges, Arg54 and Lys97, from the E9 DNase and Glu30 and Glu41 from Im9, respectively, and (3) five interfacial water molecules, two of which are completely buried. The interfacial water molecules are involved in an extended network of interprotein hydrogen bonds mediating stability and specificity of the interface. The mass spectra of the E9 DNase/Im9 complexes in the presence and absence of Zn2+ showed no indication of the presence of bound water molecules. Recent surveys of structural data on protein–protein and protein–DNA interface indicate that water molecules play a crucial role in mediating polar interactions that stabilize complexes (Janin 1999). Thus we may speculate that the diminishing complex stabilization in the gas phase by van der Waals forces and water-mediated interprotein hydrogen-bonding interactions may contribute, at least to some extent, to the apparent weakening of the protein–protein interaction.
Another interesting feature is noticeable in ESI mass spectra of holo- and apo-E9 DNase under solution conditions favoring the folded species. Under these conditions, we repeatedly observed that exclusively the charge states of the folded states showed a second minor charge state envelope with a mass difference of 98 Da (labeled with an * in Figs. 2, 4, and 6 ▶ ▶ ▶). Interestingly, in the x-ray structures of the E9 DNase-Im9 complex in the presence of Ni2+, a phosphate ion is seen within the active site cleft (Kleanthous et al. 1999) that is retained in the absence of a bound metal ion (Kuhlmann et al. 2000). The phosphate is thought to represent the binding site for the product of DNA hydrolysis (Pommer et al. 2001). We may speculate that the low abundant ion species observed may represent compact protein ions retaining a specifically bound phosphate ion in the gas phase. Nonspecific ion adduction is less likely because the ion peaks assigned to the "U" states do not show corresponding satellite peaks.
H/D exchange experiments highlight that metal ion binding, not Im9 binding, induces the conformational stabilization of the colicin E9 DNase
H/D exchange is a sensitive method to probe protein structures and folding/unfolding mechanisms (Miranker et al. 1993; Englander et al. 1996; Nemirovskiy et al. 1999; Engen and Smith 2001). In proteins, side-chain and backbone amide hydrogens are exchangeable, whereby the latter ones report on the protein structural properties. Before backbone amide hydrogen exchange in proteins can take place, hydrogen bonds have to be broken and exposed to solvent. In general, in H/D exchange experiments, more open protein conformational states with a weak hydrogen bonding network and/or increased solvent accessibility show more rapid exchange reaction than do stable, compact conformational states (Englander and Kallenbach 1983). The utility of MS for extracting exchange levels allows distinguishing of coexisting protein structural populations that experience different exchange rates. Our H/D exchange data obtained for apo-E9 DNase enabled us to conclude that (1) the interconversion between the population termed F and U has to be fast relative to the exchange rate because for both charge envelopes identical exchange-in rates were observed, and (2) the high deuterium incorporation indicated lack of persistent structure. In contrast, in Zn2+-bound E9 DNase, ∼19% of all labile hydrogen were found to be protected against exchange, indicating the presence of compact structural elements associated with hydrogen bonding and reduced surface accessibility, which is consistent with the increased thermodynamic stability of holo-E9 DNase.
Interestingly, on Im9-binding, apo-E9 DNase apparently still lacks, or at best possesses only to a minor extent, persistent structural elements as indicated by the high deuterium uptake. Recently reported NMR studies show that the slow conformational dynamics of the E9 DNase seem to persist after binding to its cognate immunity protein Im9, although a general decrease in the extent of the heterogeneity was observed (Whittaker et al. 2000). However, Zn2+-binding of Im9-bound E9 DNase apparently diminishes the conformational instability of E9 DNase, as evident from the reduced deuterium uptake (Table 2) and in line with the Zn2+-induced increase of conformational stability. On the other side, one can argue that the H/D exchange of the complex constituents can be rationalized by taking into account that transient dissociation of the complex is a prerequisite for global exchange. In the absence of Zn2+, the dissociated E9 unfolds, resulting in high deuterium incorporation, whereas in the presence of Zn2+ it remains folded and less deuterium incorporation is observed. However, under the conditions used, the Kd of the E9 DNase/Im9 complex is in the femtomolar range; thus the dissociation rate is very low. We therefore believe that the relatively elevated deuterium level for E9 DNase observed in the absence of Zn2+ rather relates to the intrinsic conformational mobility of apo-E9 DNase.
Conclusions
In summary, the strength of MS-based conformational studies of metalloproteins lies in the capability of concomitantly monitoring composition and metal-induced conformational changes. Complementing spectroscopic and NMR-based findings, we explored this unique feature of the ESI-MS-based approach for gaining additional insights into some of the mechanistic aspects of the effects of metal ion binding/release on the conformational properties of E9 DNase. Intriguing is the apparent adoption of molten globule-like properties of the apo-E9 DNase at low pH, a condition frequently discussed in the context of membrane translocation. However, the present study also reveals some limitations of MS-based conformational studies. ESI-MS allowed probing of global conformational changes induced by metal ion binding/release by monitoring alteration of the charge state distributions. However, the apparently more locally occurring intrinsic conformational heterogeneity observed for the E9 DNase in NMR solution experiments was not reported by ESI-MS. The present study substantiates that electrostatic interactions govern the stabilization of the gas phase structures and, consequently, the ion species that are detected by ESI-MS.
Materials and methods
Colicin E9 DNase and its immunity protein Im9 were expressed in E. coli and purified as previously described (Wallis et al. 1992; 1994). The concentrations for E9 DNase and Im9 were determined by measuring the UV absorption at 280 nm, using ɛ = 17,550 M−1 cm−1 and 11,400 M−1 cm−1, respectively (Wallis et al. 1995). Confirmation of the expressed E9 DNase and Im9 by ESI-MS under denaturing conditions, that is, 50% (v/v) acetonitrile containing 0.1% formic acid, yielded average masses of 15,088.3 ± 0.3 Da and 9582.0 ± 0.8 Da, respectively. Both experimentally determined masses agreed well with the calculated masses (Table 1). The final protein concentrations used were 13 μM unless stated otherwise. Zinc acetate, nickel chloride, and cobalt chloride (Merck, Darmstadt, Germany) were used without further purification. For the pH titration experiments, the ammonium acetate solutions were adjusted by adding acetic acid or ammonia. The pH measurements were taken with a standard combi-electrode (Metrohm Ltd., Herisau, Switzerland).
Preparation of deuterated proteins
The deuterium exchange experiments were preceded by prediluting the samples in 100 mM ammonium acetate (1 : 1) followed by a 1 : 25 dilution in D2O (>99.9% D Aldrich) containing 50 mM ammonium acetate (pD 7.2). The fully deuterated control samples were prepared by diluting the protein stock solutions (1 : 50) into D2O at pD 2.4 containing 0.1% formic acid and incubated overnight at 37°C.
Mass spectrometry
Electrospray ionization mass spectra were recorded on a Micromass LC-T TOF mass spectrometer (Manchester, U.K.) operating in the positive ion mode. Prior analysis on a 600–3000 m/z scale was calibrated with CsI (2 mg/mL) in isopropanol/water (1 : 1). Samples for charge state distribution analysis were introduced via a nanoflow electrospray source. Nano-electrospray needles were made from borosilicate glass capillaries (Kwik-Fil, World Precision Instruments, Sarasota, FL) on a P-97 puller (Sutter Instruments, Novato, CA). The needles were coated with a thin gold layer by using an Edwards Scancoat (Edwards Laboratories, Milpitas, CA) six Pirani 501 (at 40 mV, 1 kV, for 200 sec). In all experiments an aliquot (1–3 μL) of protein sample at a concentration of 13 μM was introduced into the electrospray needles. The nanospray needle potential was typically set to 1300 V and the cone voltage to 30 V. Source block temperature was set to 60°C. The temperature at the location of capillary tip was measured with a thermocouple and was ∼27°C ± 2°C. During individual titration experiments, all parameters of the mass spectrometer were kept constant. For in-source CID MSMS experiments, the cone voltage was gradually increased up to 80 V.
Analysis of acid-induced conformational changes of bimodal charge state distributions were performed by assuming that the compact conformational state of the E9-DNase includes the charge states 7+, 8+, and 9+ (termed F). For the more destabilized or loosely packed states, the charge states were 10+ to 24+ (termed U). The acid-induced folding curve as a function of pH was obtained by calculating the ion peak area ratio AF/(AF + AU) where AF and AU were obtained by summing the ion peak areas of charge states assigned to the compact conformational state (F) and the more destabilized or loosely packed state (U) (Mirza et al. 1993).
For the H/D exchange-in measurements, "normal" ESI-MS was used to prevent the fast exchanging deuterium atoms from back exchanging. Nitrogen gas was used as desolvation gas with a flow rate of 250 L/hr. The desolvation temperature was set at 110°C. The potential between the needle and the cone of the mass spectrometer was 3000 V; the cone voltage was set for 30 V. Samples were introduced into the ionization source at a flow rate of 5 μL/min via a syringe pump (kdScientific Inc., New Hope, PA). The syringe and transfer tubing were flushed prior to sample introduction with the same solvent system to ensure that traces of previous samples were removed. The degree of deuterium incorporation was deduced from the protein's mass shift during the course of an experiment. The raw data were processed using Masslynx software 3.5 with minimal smoothing.
Tryptophan fluorescence spectroscopy
Measurements of the intrinsic tryptophan fluorescence emission of E9 DNase, Im9, and the heterodimeric complex were performed on a Perkin Elmer LS50 luminescence spectrometer using an excitation wavelength of 295 nm. Emission spectra were recorded from 300–450 nm. The excitation bandwidth was set on 5 nm and the emission bandwidth on 10 nm. The protein concentrations were 6 μM each in 50 mM ammonium acetate at various pH values. A quartz cell with a 5-mm path length was used at 25°C. For the metal loaded protein experiment, measurements were performed with a sixfold excess (33 μM) of zinc acetate.
CD spectroscopy
A dual-beam DSM 1000 CD spectrophotometer (On-Line Instrument Systems, Bogart, GA) was used for CD measurements. The subtractive double-grating monochromator was equipped with a fixed disk, holographic gratings (2400 lines/mm, blaze wavelength 230 nm) and 1.24-mm slits. Samples were measured in a 1-mm path length cell. The E9 DNase concentration was 8 μM in 50 mM ammonium acetate. For the Zn2+-containing experiments, a threefold excess (24 μM) of zinc acetate was used. Denaturation was performed by diluting the DNase in 6 M guanidine hydrochloride (Merck). Far-UV CD spectra were recorded from 260–200 nm at 25°C.
The E9 DNase concentration in the near-UV CD measurements was 50 μM and was measured in a cuvette with a path length of 10 mm. The subtractive double-grating monochromator was equipped with 0.6-mm slits. For the Zn2+-containing E9 DNase measurements, a 10-fold excess of zinc acetate (500 μM) was used. Near-UV CD spectra were recorded from 320–260 nm at 25°C.
Each measurement was the average of at least six repeated scans (step resolution 1 nm, 1 sec each step) from which the corresponding background spectrum was subtracted. The measured signals were converted to molar absorbance difference on the basis of a mean residual weight of 112.6.
Acknowledgments
The present work was supported by the Center for Biomolecular Genetics and the Netherlands Organization for Scientific Research (NWO No. 9803 to A.J.R.H.). the colicin work of R.J., G.R.M., and C.K. in this paper was funded by the Wellcome Trust. We thank Christine Moore, Ann Reilly, and Nick Cull (UEA) for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ESI, electrospray ionization
MS, mass spectrometry
CD, circular dichroism
m/z, mass-to-charge
CsI, Cesium iodide
H/D, hydrogen/deuterium
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0200502.
References
- Bychkova, V.E., Dujsekina, A.E., Klenin, S.I., Tiktopulo, E.I., Uversky, V.N., and Ptitsyn, O.B. 1996. Molten globule-like state of cytochrome c under conditions simulating those near the membrane surface. Biochemistry 35 6058–6063. [DOI] [PubMed] [Google Scholar]
- Cech, N.B. and Enke, C.G. 2000. Relating electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 72 2717–2723. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Effect of affinity for droplet surfaces on the fraction of analyte molecules charged during electrospray droplet fission. Anal. Chem. 73 4632–4639. [DOI] [PubMed] [Google Scholar]
- Chak, K.F., Kuo, W.S., Lu, F.M., and James, R. 1991. Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol. 137 91–100. [DOI] [PubMed] [Google Scholar]
- Chak, K.-F., Hsieh, S.-Y., Lao, C.-C., and Kan, L.-S. 1998. Change of thermal stability of colicin E7 triggered by acidic pH suggests the existence of unfolded intermediate during the membrane-translocation phase. Proteins 32 17–25. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S.K., Katta, V., and Chait, B.T. 1990. Probing conformational changes in proteins by mass spectrometry. J. Am. Chem. Soc. 112 9012–9013. [Google Scholar]
- Cramer, W.A., Cohen, F.S., Merrill, A.R., and Song, H.Y. 1990. Structure and dynamics of the colicin E1 channel. Mol. Microbiol. 4 519–526. [DOI] [PubMed] [Google Scholar]
- Dobo, A. and Kaltashov, I.A. 2001. Detection of multiple protein conformational ensembles in solution via deconvolution of charge-state distributions in ESI MS. Anal. Chem. 73 4763–4773. [DOI] [PubMed] [Google Scholar]
- Donald, L.J., Hosfield, D.J., Cuvelier, S.L., Ens, W., Standing, K.G., and Duckworth, H.W. 2001. Mass spectrometric study of the Escherichia coli repressor proteins, IcIR and GcIR, and their complexes with DNA. Protein Sci. 10 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink, M.R. 1994. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 66 482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen, J.R. and Smith, D.L. 2001. Investigating protein structure and dynamics by hydrogen exchange MS. Anal. Chem. 73 256A–265A. [DOI] [PubMed] [Google Scholar]
- Englander, S.W. and Kallenbach, N.R. 1983. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16 521–655. [DOI] [PubMed] [Google Scholar]
- Englander, S.E., Sosnick, T.R., Enlander, J.J., and Mayne, L. 1996. Mechanisms and uses of hydrogen exchange. Curr. Opin. Struct. Biol. 6 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A.E. 1994. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 3 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumerov, D.R. and Kaltashov, I.A. 2001. Dynamics of iron release from transferrin N-lobe studied by electrospray ionization mass spectrometry. Anal. Chem. 73 2565–2570. [DOI] [PubMed] [Google Scholar]
- Hannan, J.P., Whittaker, S.B., Hemmings, A.M., James, R., Kleanthous, C., and Moore, G.R. 2000. NMR studies of metal ion binding to the Zn-finger-like HNH motif of colicin E9. J. Inorg. Biochem. 79 365–370. [DOI] [PubMed] [Google Scholar]
- James, R., Kleanthous, C., and Moore, G.R. 1996. The biology of E colicins: Paradigms and paradoxes. Microbiology 142 1569–1580. [DOI] [PubMed] [Google Scholar]
- Janin, J. 1999. Wet and dry interfaces: The role of solvent in protein-protein and protein-DNA recognition. Structure Fold. Des. 7 R277–279. [DOI] [PubMed] [Google Scholar]
- Kleanthous, C., Hemmings, A.M., Moore, G.R., and James, R. 1998. Immunity proteins and their specificity for endonuclease colicins: Telling right from wrong in protein-protein recognition. Mol. Microbiol. 28 227–233. [DOI] [PubMed] [Google Scholar]
- Kleanthous, C., Kuhlmann, U.C., Pommer, A.J., Ferguson, N., Radford, S.E., Moore, G.R., James, R., and Hemmings, A.M. 1999. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 6 243–252. [DOI] [PubMed] [Google Scholar]
- Kleanthous, C. and Walker, D. 2001. Immunity proteins: Enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 26 624–631. [DOI] [PubMed] [Google Scholar]
- Ko, T.P., Liao, C.C., Ku, W.Y., Chak, K.F., and Yuan, H.S. 1999. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure Fold. Des. 7 91–102. [DOI] [PubMed] [Google Scholar]
- Konermann, L., Collings, B.A., and Douglas, D.J. 1997. Cytochrome c folding kinetics studied by time-resolved electrospray ionization mass spectrometry. Biochemistry 36 5554–5559. [DOI] [PubMed] [Google Scholar]
- Konermann, L. and Douglas, D.J. 1998. Equilibrium unfolding of proteins monitored by electrospray ionization mass spectrometry: Distinguishing two-state from multi-state transitions. Rapid Commun. Mass Spectrom. 12 435–442. [DOI] [PubMed] [Google Scholar]
- Krutchinsky, A.N., Ayed, A., Donald, L.J., Ens, W., Duckworth, H.W., and Standing, K.G. 2000. Studies of noncovalent complexes in an electrospray ionization/time-of-flight mass spectrometer. Methods Mol. Biol. 146 239–249. [DOI] [PubMed] [Google Scholar]
- Kuhlmann, U.C., Pommer, A.J., Moore, G.R., James, R., and Kleanthous, C. 2000. Specificity in protein-protein interactions: The structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J. Mol. Biol. 301 1163–1178. [DOI] [PubMed] [Google Scholar]
- Lau, P.C.K., Parsons, M., and Uchimura, T. 1992. Molecular evolution of E colicin plasmids with emphasis on the endonuclease types. In Bacteriocins, microcins and lantibiotics. NATO ASI Series, (eds. R. James, C. Lazdunski, and F. Pattus), pp. 353–348. Berlin: Springer-Verlag
- Lazdunski, C.J., Bouveret, E., Rigal, A., Journet, L., Lloubes, R., and Benedetti, H. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180 4993–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, W.P., Yan, S.X., Zhang, Y.X., and Zhou, H.M. 1996. Acid-induced folding of yeast alcohol dehydrogenase under low pH conditions. J. Biochem. 119 674–679. [DOI] [PubMed] [Google Scholar]
- Lei, Q.P., Cui, X., Kurtz, D.M., Jr., Amster, I.J., Chernushevich, I.V., and Standing, K.G. 1998. Electrospray mass spectrometry studies of non-heme iron-containing proteins. Anal. Chem. 70 1838–1846. [DOI] [PubMed] [Google Scholar]
- Liu, C., Wu, Q., Harms, A.C., and Smith, R.D. 1996. On-line microdialysis sample cleanup for electrospray ionization mass spectrometry of nucleic acid samples. Anal. Chem. 68 3295–3299. [DOI] [PubMed] [Google Scholar]
- Loo, J.A. 1997. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 16 1–23. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Electrospray ionization mass spectrometry: A technology for studying noncovalent macromolecular complexes. Int. J. Mass Spectrom. 200 175–186. [Google Scholar]
- Maier, C.S., Schimerlik, M.I., and Deinzer, M.L. 1999. Thermal denaturation of Escherichia coli thioredoxin studied by hydrogen/deuterium exchange and electrospray ionization mass spectrometry: Monitoring a two-state protein unfolding transition. Biochemistry 38 1136–1143. [DOI] [PubMed] [Google Scholar]
- Masaki, H., Yajima, S., Akutsu-Koide, A., Ohta, T., and Uozumi, T. 1992. Immunity specificity and evolution of the nuclease-type E colicins. In Bacteriocins, microcins and lantibiotics (eds. R. James, C. Lazdunski, and F. Pattus), pp. 379–395 Berlin: Springer-Verlag
- McLaughlin, S. 1989. The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem. 18 113–136. [DOI] [PubMed] [Google Scholar]
- Miranker, A., Robinson, C.V., Radford, S.E., Aplin, R.T., and Dobson, C.M. 1993. Detection of transient protein folding populations by mass spectrometry. Science 262 896–900. [DOI] [PubMed] [Google Scholar]
- Mirza, U.A., Cohen, S.L., and Chait, B.T. 1993. Heat-induced conformational changes in proteins studied by electrospray ionization mass spectrometry. Anal. Chem. 65 1–6. [DOI] [PubMed] [Google Scholar]
- Nemirovskiy, O., Giblin, D.E., and Gross, M.L. 1999. Electrospray ionization mass spectrometry and hydrogen/deuterium exchange for probing the interaction of calmodulin with calcium. J. Am. Soc. Mass. Spectrom. 10 711–718. [DOI] [PubMed] [Google Scholar]
- Pommer, A.J., Wallis, R., Moore, G.R., James, R., and Kleanthous, C. 1998. Enzymological characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli. Biochem. J. 334 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommer, A.J., Kuhlmann, U.C., Cooper, A., Hemmings, A.M., Moore, G.R., James, R., and Kleanthous, C. 1999. Homing in on the role of transition metals in the HNH motif of colicin endonucleases. J. Biol. Chem. 274 27153–27160. [DOI] [PubMed] [Google Scholar]
- Pommer, A.J., Cal, S., Keeble, A.H., Walker, D., Evens, S.J., Kuhlman, U.C., Cooper, A., Connolly, B.A., Hemmings, A.M., Moore, G.R., et al. 2001. Mechanisms and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 314 735–749. [DOI] [PubMed] [Google Scholar]
- Potier, N., Donald, L.J., Chernushevich, I., Ayed, A., Ens, W., Arrowsmith, C.H., Standing, K.G., and Duckworth, H.W. 1998. Study of a noncovalent trp repressor: DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 7 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats, M., Teisié, J., and Tocanne, J.-F. 1986. Lateral proton conduction at lipid-water interfaces and its implications for the chemiosmotic-coupling hypothesis. Nature 322 756–758. [Google Scholar]
- Przybylski, M. and Glocker, M.O. 1996. Electrospray mass spectrometry of biomolecular complexes with noncovalent interactions: New analytical perspectives for supramolecular chemistry and molecular recognition processes. Angew. Chem. Int. Ed. Engl. 35 806–826. [Google Scholar]
- Schaller, K. and Nomura, M. 1976. Colicin E2 is DNA endonuclease. Proc. Natl. Acad. Sci. USA. 73 3989–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub, D.A., Goodrich-Blair, H., and Eddy, S.R. 1994. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 19 402–404. [DOI] [PubMed] [Google Scholar]
- Smith, R.D., Loo, J. A., Edmonds, C. G., Barinaga, C. J., and Udseth, H. R. 1990. New developments in biochemical mass spectrometry: Electrospray ionization. Anal. Chem. 62 882–899. [DOI] [PubMed] [Google Scholar]
- Smith, R.D., Bruce, J.E., Wu, Q.Y., and Lei, Q.P. 1997a. New mass spectrometric methods for the study of noncovalent associations of biopolymers. Chem. Soc. Rev. 26 191–202. [Google Scholar]
- Smith, D.L., Deng, Y., and Zhang, Z. 1997b. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J. Mass. Spectrom. 32 135–146. [DOI] [PubMed] [Google Scholar]
- Swint, L. and Robertson, A.D. 1993. Thermodynamics of unfolding for turkey ovomucoid third domain: Thermal and chemical denaturation. Protein Sci. 2 2037–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Goot, F.G., Gonzalez-Manas, J.M., Lakey, J.H., and Pattus, F. 1991. A `molten-globule' membrane-insertion intermediate of the pore-forming domain of colicin A. Nature 354 408–410. [DOI] [PubMed] [Google Scholar]
- Veenstra, T.D., Johnson, K.L., Tomlinson, A.J., Craig, T.A., Kumar, R., and Naylor, S. 1998. Zinc-induced conformational changes in the DNA-binding domain of the vitamin D receptor determined by electrospray ionization mass spectrometry. J. Am. Soc. Mass. Spectrom. 9 8–14. [DOI] [PubMed] [Google Scholar]
- Versluis, C. and Heck, A.J.R. 2001. Gas-phase dissociation of hemoglobin. Int. J. Mass Spectrom. 210 637–649. [Google Scholar]
- Vis, H., Heineman, H., Dobson, C.M., and Robinson, C.V. 1998. Detection of a monomeric intermediate associated with dimerization of protein HU by mass spectrometry. J. Am. Chem. Soc. 120 6427–6428. [Google Scholar]
- Wallis, R., Moore, G.R., Kleanthous, C., and James, R. 1992. Molecular analysis of the protein-protein interaction between the E9 immunity protein and colicin E9. Eur. J. Biochem. 210 923–930. [DOI] [PubMed] [Google Scholar]
- Wallis, R., Reilly, A., Barnes, K., Abell, C., Campbell, D.G., Moore, G.R., James, R., and Kleanthous, C. 1994. Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur. J. Biochem. 220 447–454. [DOI] [PubMed] [Google Scholar]
- Wallis, R., Moore, G.R., James, R., and Kleanthous, C. 1995. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion-controlled association and femtomolar binding for the cognate complex. Biochemistry 34 13743–13750. [DOI] [PubMed] [Google Scholar]
- Whittaker, S.B., Boetzel, R., MacDonald, C., Lian, L.Y., Pommer, A.J., Reilly, A., James, R., Kleanthous, C., and Moore, G.R. 1998. NMR detection of slow conformational dynamics in an endonuclease toxin. J. Biomol. NMR. 12 145–159. [DOI] [PubMed] [Google Scholar]
- Whittaker, S.B., Boetzel, R., MacDonald, C., Lian, L.Y., James, R., Kleanthous, C., and Moore, G.R. 1999. Assignment of 1H, 13C and 15N signals of the DNase domain of colicin E9. J. Biomol. NMR. 14 201–202. [DOI] [PubMed] [Google Scholar]
- Whittaker, S.B., Czisch, M., Wechselberger, R., Kaptein, R., Hemmings, A.M., James, R., Kleanthous, C., and Moore, G.R. 2000. Slow conformational dynamics of an endonuclease persist in its complex with its natural protein inhibitor. Protein Sci. 9 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]