Abstract
The aims of high-throughput (HTP) protein production systems are to obtain well-expressed and highly soluble proteins, which are preferred candidates for use in structure–function studies. Here, we describe the development of an efficient and inexpensive method for parallel cloning, induction, and cell lysis to produce multiple fusion proteins in Escherichia coli using a 96-well format. Molecular cloning procedures, used in this HTP system, require no restriction digestion of the PCR products. All target genes can be directionally cloned into eight different fusion protein expression vectors using two universal restriction sites and with high efficiency (>95%). To screen for well-expressed soluble fusion protein, total cell lysates of bacteria culture (∼1.5 mL) were subjected to high-speed centrifugation in a 96-tube format and analyzed by multiwell denaturing SDS-PAGE. Our results thus far show that 80% of the genes screened show high levels of expression of soluble products in at least one of the eight fusion protein constructs. The method is well suited for automation and is applicable for the production of large numbers of proteins for genome-wide analysis.
Keywords: Structural genomics, functional genomics, proteomics, protein expression
The function of a gene is manifested by the protein it encodes. Genome sequencing of many organisms (see http://www.ncbi.nlm.nih.gov/) has led to the concept of analyzing protein function on a genome-wide scale. Structural genomics and proteomics (Christendat et al. 2000; Skolnick et al. 2000; Fields 2001), therefore, have become major research foci. The challenge of studying proteins in a global scale is driving the development of high-throughput (HTP) and parallel approaches in protein expression, purification, biochemical analysis, and structure determination.
Several prototypes of HTP protein expression and purification systems have been initiated (Christendat et al. 2000; Edwards et al. 2000; Lesley 2001; Zhu et al. 2001). Cloning and expression in Escherichia coli are favored in many instances because E. coli has relatively simple genetics, is well characterized, has a relatively rapid growth rate, and has few post-translational protein modifications. One disadvantage, however, of expressing heterologous proteins in E. coli is that proteins are frequently expressed as insoluble aggregated folding intermediates, known as inclusion bodies (Paul et al. 1983). Although it may be possible to increase protein solubility by optimizing expression condition or by refolding the recombinant proteins, in the interests of throughput, only a single set of growth or folding conditions can be used.
Gene fusion is another approach that has been successfully used for producing soluble heterologous proteins in E. coli (Uhl'en and Moks 1990). Several carrier proteins are widely used in gene fusion, including thioredoxin (Trx), maltose-binding protein (MBP), glutathione S-transferase (GST), intein, calmodulin-binding protein (CBP), NusA, and cellulose-associated protein (CAP). Although the use of these carrier proteins has resulted in the successful overexpression of many heterologous proteins, each was tested empirically and certainly may not possess maximal solubilizing characteristics. Moreover, each expression scenario requires a specific vector. Recloning cDNA into each of these specific vectors is extremely labor intensive.
Recombinational cloning (RC) methodology was recently developed to minimize the effort required for alternative expression. It uses either cre-lox (Liu et al. 1999) or Int/Xis/IHF (Hartley et al. 2000) recombination to introduce the gene of interest into a recipient vector. In these systems, aberrant recombination or cointegrant products may result from faulty gene transfer to the expression vector. Another limitation is that translation fusions of the recombination att or lox sites and a few extra nucleotide sequences are required to ensure successful gene transfer. In some cases, such as protein crystallography, in which longer translation fusions are potentially more detrimental to the proteins, a conventional cloning approach with shorter translation fusions is more appropriate. In the present study, we established a new procedure for the parallel cloning of genes into multiple fusion expression vectors without restriction digestion. The main objective here was to rapidly screen for well-expressed soluble proteins that can be used in structural and functional genomics.
Results
Parallel cloning of target genes into multiple fusion protein expression vectors
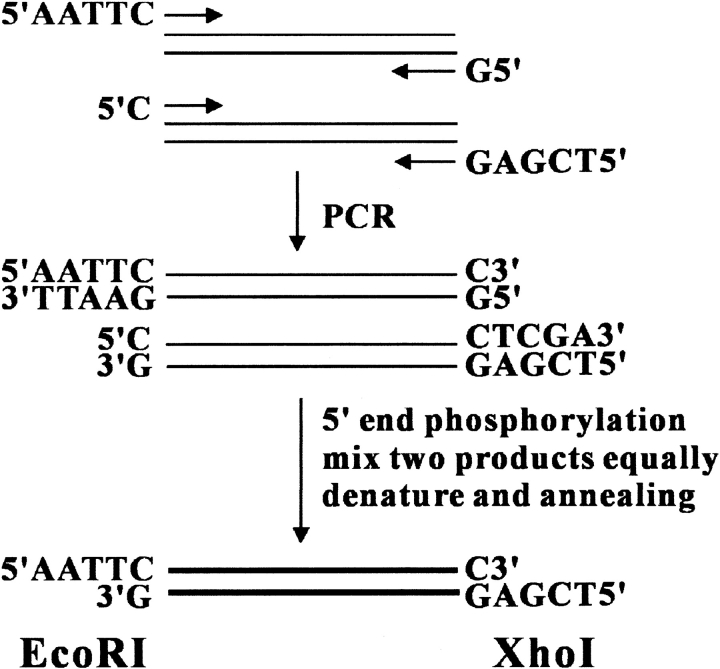
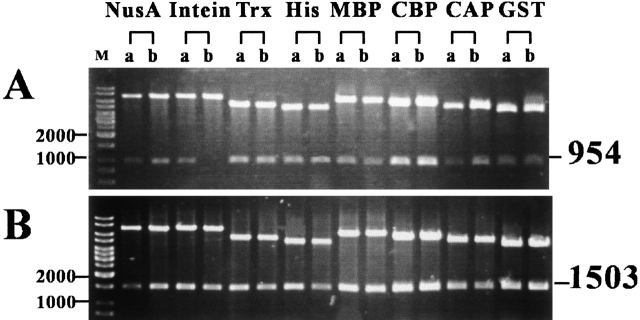
We applied the "sticky end PCR method" (Zeng 1998) to generate DNA products with 5` EcoRI and 3` XhoI sticky ends. As illustrated in Figure 1 ▶, the method requires four PCR primers and reactions in two separate tubes. Both PCR products were purified and mixed equally. After denaturation and renaturation, ∼25% of the final product carries two cohesive ends and is ready for ligation even without restriction digestion. Therefore, this method is suitable for cloning any gene, even genes with internal EcoRI or XhoI restriction sites. To optimize cloning efficiency, sticky-end PCR products were 5` phosphorylated with T4 polynucleotide kinase and the vectors were dephosphorylated by calf intestine alkaline phosphatase. Together, these procedures increase the efficiency of PCR products into multiple expression vectors. As shown in Figure 2 ▶, two independent clones of each ligation reaction were analyzed by restriction digestion. Among the 16 clones of the eight different fusion protein expression vectors, 15 (Fig. 2A ▶) and 16 (Fig. 2B ▶) were identified as successful clones. We applied this method to clone ∼40 genes into these eight expression vectors (>300 cloning reactions) with a >95% success rate.
Fig. 1.
Moleclular cloning strategy. Four PCR primers and reactions were used in two separate tubes. An equal amount of the two PCR products were mixed, and then the 5` ends were phosphorylated with T4 polynucleotide kinase. After denaturing (95°C for 5 min) and renaturing (65°C for 10 min), ∼25% of the final products carry EcoRI (5`) and XhoI (3`) cohesive ends and are ready for ligation with the vectors.
Fig. 2.
Recombinant DNA plasmids purified from JM109(DE3). Eight different fusion protein expression vectors are indicated above. Two independent clones from each construct were isolated for characterization (lanes A and B). Plasmid DNAs were purified in a 96-well format using Millpore's Motage plasmid mini-prep kit; 3–5 μL mini-prep DNA was restriction digested with EcoRI and XhoI and separated in 0.8% agarose gel. A 1-kb DNA ladder (from MBI Fermentas, USA) was used as marker (M) and shown in the far left lane. The expected sizes (in base pair) of the desirable restriction fragments of two different target genes are indicated on the right of the figure.
Induction and screening of soluble fusion proteins
Because bacteria host strain JM109(DE3) is suitable for both plasmid DNA preparation and high level protein expression, it was used initially in this investigation. If the digested vectors were tested as efficacious (∼100% in a single cloning reaction), bacterial colonies were directly induced with isopropyl β-D-thiogalactoside (IPTG) to produce proteins even without examining each individual clone by restriction mapping or colony PCR.
To identify well-expressed and highly soluble fusion proteins, 2 mL of culture was used for small-scale induction. Briefly, bacterial cultures in log phase (OD600∼0.6) were induced with IPTG at 20°C for 24 hr. We found that low temperature and long induction time facilitate correct protein folding; for instance, the fusion protein of yeast Hop2 (encoded by open reading frame YGL033W; Table 1) and Trx is soluble at 20°C but not at 37°C.
Table 1.
Soluble target proteins with high expression levels
| Organism | Gene | Protein size (kD) |
| Yeast | YHL024W | 80.1 |
| YOR351C | 56.9 | |
| YLR394W | 53.9 | |
| YBR233W | 45.8 | |
| YDR065W | 42.9 | |
| YPL018W | 42.8 | |
| YOL104C | 40.9 | |
| YER106W | 35.8 | |
| YHR014W | 33.3 | |
| YIL144Wa | 28.2 | |
| YPL199C | 26.8 | |
| YGL033W | 25.0 | |
| YCR086W | 21.7 | |
| YIL132C | 25.0 | |
| YMR048W | 36.3 | |
| YPL200W | 18.3 | |
| Mammalian | U47110b | 100.0 |
| U47110b | 35.2 | |
| U47110b | 24.2 | |
| U47110b | 8.8 | |
| P97801 | 32.3 | |
| AAC25954 | 31.0 | |
| NP_064587 | 30.5 | |
| AJ404613 | 27.5 | |
| NP_036520 | 19.7 | |
| XP_043137 | 16.3 | |
| Plant | AAF75761 | 27.9 |
| CAC17699c | 28.4 | |
| CAC17699c | 28.4 | |
| O04701 | 26.8 | |
| Insect | BAB17671 | 69.6 |
| AAF58245 | 44.0 |
a Only the N-terminal 256 aa was expressed.
b Full-length and three truncated proteins were expressed.
c Wild type and mutant protein with mutation in the amino acid residue 128.
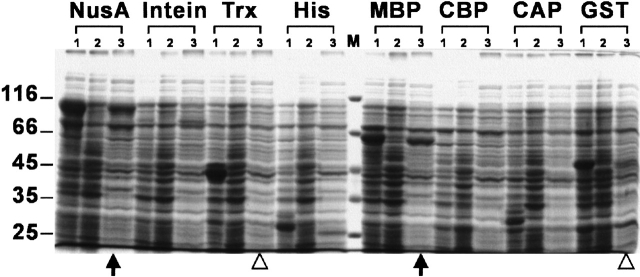
In a parallel analysis of protein solubility, host cells were harvested and lysed in 96-well plates as described under Materials and Methods. Insoluble materials in total cell lysates were removed by centrifugation using a Ti25 rotor, which allows parallel processing of 96 samples; therefore, this system is suitable for automation. To increase the accuracy of protein solubility testing, an ultracentrifugal force (90,000g) was applied to eliminate partially folded protein aggregates. As illustrated in Figure 3 ▶, we applied this HTP system to the expression of yeast Csm2 protein (encoded by open reading frame YIL132C; Table 1). SDS-PAGE was used to separate proteins from total cell lysates induced with or without IPTG induction (Fig. 3 ▶, lanes 1 and 2) and from the soluble protein fraction induced with IPTG induction (Fig. 3 ▶, lane 3). NusA and MBP fusion proteins were found to be well induced and soluble; on the other hand, GST and Trx fusion proteins were expressed in insoluble forms (Fig. 3 ▶).
Fig. 3.
Analysis of budding yeast Csm2 (YIL132C) fusion proteins. Samples of total proteins and soluble protein fractions were separated on a 10% SDS-PAGE under reducing conditions and stained with Coomassie Blue. Lane 1, whole cell lysates of induced cells; lane 2, whole cell lysates of uninduced cells; lane 3, soluble proteins with induction. Eight different fusion proteins are indicated above. The molecular weight standards are shown in the center and labeled on the left (×1,000). NusA and MBP fusion proteins show high solubility (indicated by arrows below the lanes of soluble protein fractions); on the other hand, GST and Trx fusion proteins are well induced but not soluble (indicated by open triangles).
If the proteins were poorly expressed, the DNA clones were retransformed into other host strains, for example, BL21-Gold(DE3) or BL21-CondonPlus(DE3), in an attempt to alleviate problems related to codon bias or protein toxicity. For example, none of the eight fusion proteins of Drosophila Phyl protein (accession number AAF58245; Table 1) were induced in JM109(DE3); on the contrary, NusA-Phyl and GST-Phyl fusion proteins were highly expressed and soluble in BL21-CondonPlus(DE3) (data not shown).
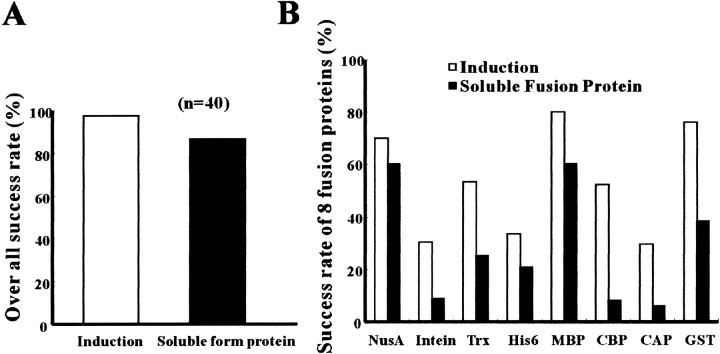
We have cloned and expressed more than 40 proteins from various organisms. The overall successful rate of obtaining soluble proteins, at least in one of the eight expression constructs tested, is >80% (Fig. 4A ▶). The soluble ratio of individual fusion protein is shown in Figure 4B ▶. Often, the larger fusion tags are superior for enhancing protein solubility; for example, the success ratio of soluble NusA (54 kD), MBP (42 kD), and GST (24 kD) fusion proteins are 60%, 60%, and 38%, respectively. Target fusion proteins that have been successfully expressed by this method are listed in Table 1. These target proteins alone range from 9 kD to 100 kD, and the largest soluble fusion protein expressed in this study was ∼150 kD.
Fig. 4.
(A) Statistical analysis of soluble protein ratio obtained in at least one of the eight expression constructs. (B) Eight different gene fusions and their effects were also compared. A total of 40 different genes were tested in this study. Well-induced and highly soluble fusion proteins were identified visually by comparing the relative density of protein bands in SDS-PAGE as shown in Figure 3 ▶.
Generalized protein purification strategy
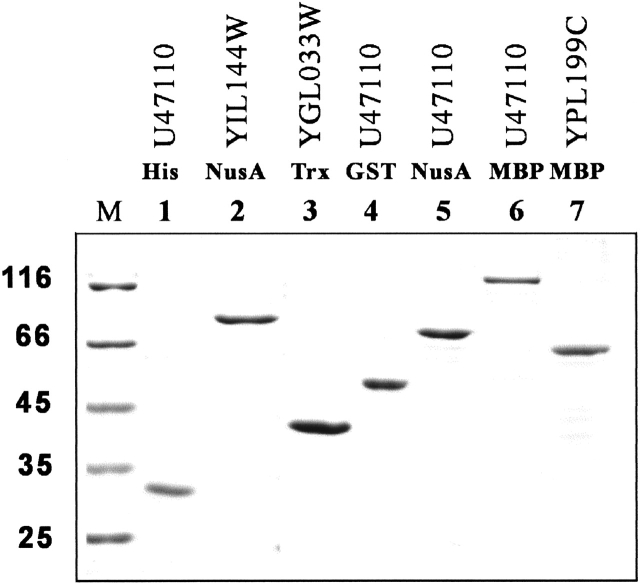
In an HTP process, it is absolutely essential that purification does not depend on the tedious optimization of conditions that exploit subtle differences in protein size, charge, or hydrophobicity. Therefore, it is advantageous to use expression vectors with multiple tagging for affinity purification. Almost all expression vectors used in this study were engineered with an NH2-terminal affinity tag, a cleavage site of protease (e.g., thrombin or factor Xa), and a COOH-terminal His-tag. Recombinant fusion proteins were first isolated by various affinity chromatography columns (glutathione agarose, amylose resin, etc) and then further purified by Ni2+-resin. Routinely, fusion proteins with typical yields (5–20 mg per liter of Luria-Bertani [LB] culture) and purity (>90%) have been obtained (Fig. 5 ▶). Because all these fusion constructs can be proteolytically cleaved to remove the NH2-terminal fusion partners, it is of interest to examine if the cleaved target proteins are still soluble. Thus far, we have tested three yeast target proteins (Trx-YGL033W, MBP-YPL199C, and Nus-YIL144W; Table 1) with thrombin, and all yielded soluble products (data not shown).
Fig. 5.
SDS-PAGE analysis of purified proteins. The gel shows typical yields (5–20 mg per liter of Luria-Bertani [LB] culture) and purity (∼90%) obtained from two steps of affinity purification. The database accession or open reading frame number of the expressed proteins and their fusion tags are indicated. The molecular weight standards are labeled on the left.
Discussion
In the postgenomic era, HTP protein expression technologies are essential tools. Conceivably, the most important characteristic of a protein that determines its feasibility for functional analysis is its solubility. High solubility is also strongly correlated with the success of structural studies using either NMR or X-ray crystallography. The method described here allows one to clone and express multiple fusion proteins in E. coli efficiently. The success ratio for obtaining highly expressed and soluble products in one of the eight fusion constructs is >80%, which is superior to the results of other structural or functional genomic studies (Christendat et al. 2000; Edwards et al. 2000).
Sticky-end PCR and directional cloning methods allow one to obtain multiple expression plasmids without restriction digestion. This is a somewhat conventional cloning approach, but it has several advantages compared with other methods, such as the RC approach. First of all, it is simpler. It allows direct cloning of PCR products into multiple expression vectors. RC methods require at least two cloning steps. Second, it is more accurate in theory and also in practice. With an RC approach, faulty gene transfer might occur because of aberrant recombination or cointegrant vector products. Third, it is less detrimental to proteins. This method introduces only two new amino acids encoded by the restriction sites, whereas RC methods include att or lox sites, as well as other extra sequences to achieve a precise gene transfer. Longer translation fusions introduced by a cloning procedure are usually more harmful to proteins.
All procedures in this study, including DNA cloning, plasmid preparation, protein induction, and cell lysis are based on using a standard 96-well format in an efficient and reproducible manner, making these procedures suitable for automation. All 96 samples could be subjected to ultracentrifugation fractionation in the Beckman Ti25 rotor. Therefore, it is possible to integrate the process of sample transfer from 96-well plate to 96 centrifugal tubes using a robotic protocol, although manual operation is still required afterward.
High speed centrifugation (90,000g) ensures the separation of highly soluble and properly folded proteins from insoluble or partially aggregated materials. Denaturing SDS-PAGE was applied to identify these soluble fusion proteins. Consequently, there is little chance of finding false positives in this screening procedure. After identifying clones expressing soluble fusion proteins, the rest of the cell lysates can be used for other purposes such as small-scale affinity purification.
With our protocols for rapid subcloning, solubility screening, and parallel protein purification, we will be able to provide a large number of high purity fusion proteins for structure–function studies. For protein crystallography, carrier fusion protein domains can be proteolytically cleaved during or after the first step of affinity purification. The resulting His-tagged target proteins can be isolated by Ni2+-resin or other conventional chromatography methods. These proteins can also be used to make protein microarrays, allowing for the parallel characterization of diverse biochemical activities, such as enzymatic assays, protein–protein, protein–nucleic acid, and receptor–ligand interactions. The protein chips may also be applied to screen for new drugs. We have succeeded in the biochemical characterization of at least three fusion proteins expressed in this study (data not shown), indicating that these fusion proteins retain a part of or even the full biochemical activity of the target proteins.
In summary, we have developed an HTP molecular cloning and protein expression system using E. coli. It allows us to screen effectively for well-expressed and highly soluble proteins. The same approach can be applied for alternate cloning of all potential target genes into vectors of different expression systems, including yeast, insect, and mammalian cells, as well as cell-free in vitro systems. Last, but not least, this method is well suited for automation and will be a useful tool for the production of proteins for use in structural and functional genomic studies.
Materials and methods
Molecular cloning
A PCR cloning strategy, referred to as the sticky-end PCR method (Zeng 1998), was applied to generate PCR products bearing cohesive ends compatible with 5` EcoRI and 3` XhoI sites (Fig. 1 ▶). The method requires four PCR primers and reactions in two separate tubes. Both PCR products were purified and mixed equally and then treated with T4 polynucleotide kinase (New England Biolabs) and ATP (Sigma). After denaturing (95°C for 5 min) and renaturing (65°C for 10 min), ∼25% of the final products carried cohesive ends and were ready for ligation.
Fusion protein expression vectors used in these studies were purchased from Novagen, New England Biolabs, or Amershan Pharmacia. We engineered two new universal cloning sites (EcoRI and XhoI) into those vectors. Briefly, the original vectors were cut with restriction enzymes as close to the 3` end of the N-terminal fusion genes. The appropriate DNA cassette was chosen to retain the reading frame of the fusion over EcoRI and XhoI restriction sites and to introduce 6 histidine amino acid residues between XhoI and stop codon. A specific cleavage sequence of protease (e.g., thrombin or factor Xa) was introduced immediately after the EcoRI site and before the coding sequence of target protein; this was achieved by stringent design of the sticky-end PCR primers. To prepare vectors for ligation reactions, the vectors were restriction digested with EcoRI and XhoI and then dephosphorylated with calf intestinal alkaline phosphotase (New England Biolabs).
Plasmid DNA purification was performed in a 96-well format using Millpore's Motage plasmid miniprep kit. Eight different expression vectors were used here for parallel cloning. Two independent clones were isolated and characterized from every cloning reaction. Therefore, soluble protein products of six different genes (48 cloning/96 protein induction) were screened simultaneously.
Small-scale protein induction
Host E. coli strain JM109(DE3) (Novagen) was chosen for plasmid preparation as well as protein induction. Host strains, BL21-Gold(DE3) or BL21-CondonPlus(DE3) (Stratagene), were also used for expression in the case of low-level protein induction in JM109(DE3). Single colonies were grown overnight in LB medium with ampicillin (50 μg/mL) or kanamycin (30 μg/mL) at 37°C. Two 18-μL overnight cultures were inoculated in 2 mL LB (with 1% glucose) and grown at 37°C for 3 hr (OD600∼ 0.6). The cells were cooled in 20°C incubators, induced with or without 0.4 mM IPTG, and subsequently grown for an additional 20 hr. To harvest the cells, 500-μL cultures from each well of the 96-wells were transferred to a new 96-well plate. Culture medium was placed onto a Sorvall RTH750 microplate carrier and centrifuged for 10 min at 4000 rpm. Cell pellets were suspended in 1.5X SDS-PAGE sample buffer and boiled for 5 min.
Protein solubility test
For protein solubility assays, cell pellets from 1.5 mL of culture with IPTG induction were resuspended in 40 μL of ice-cold buffer B (250 mM sucrose, 25 mM Tris-HCl at pH 7.0, 1 mM EGTA, lysozyme 0.3 mg/mL) and incubated on ice for 20 min. The suspensions were mixed with 160 μL of ice-cold lysis buffer (0.1% Triton X-100, 150 mM NaCl, 0.1 unit Benzonase, 1 mM EGTA, 25 mM Tris-HCl at pH 7.0) followed by incubation at 4°C for another 20 min. Benzonase (Novagen, USA) was used here to digest bacteria genomic DNA and RNA. Insoluble materials were removed by centrifugation at 90,000g for 45 min in the Ti25 rotor (Beckman, USA). Soluble fractions (∼100 μL) were then mixed with an equal volume of 3X SDS-sample buffer and boiled immediately for 5 min. Both total cell extracts and soluble fractions were analyzed on 8% to 12% denaturing SDS-PAGE. The proteins (gels) were visualized by Coomassie Blue staining.
Successful expression of soluble fusion protein was scored as follows: Eight different fusion constructs for each target protein were examined. At least one of these constructs must yield a high level of expression and also remain soluble after an ultracentrifugation fractionation procedure, as described previously. Successfully expressed soluble proteins were analyzed by SDS-PAGE and visually identified by Coomassie Blue staining.
Acknowledgments
We gratefully thank Dr. Yi-Ping Hsueh, Su-Ming Hu, and Dr. Chih-Hsiang Leng for helpful discussions; Yu-Jing Hsiao and Shu-Chun Chang for assistance in this study; and Dr. Chung Wang for providing comments on the manuscript. This work was supported by Academia Sinica and National Science Council (NSC90–2321-B-001–015 to A.H.-J. Wang and NSC90–2321-B-001–014 to T.-F. Wang).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
HTP, high throughput
IPTG, isopropyl β-D-thiogalactoside
LB, Luria-Bertani
RC, recombinational cloning
PCR, polymerase chain reaction
Trx, thioredoxin
MBP, maltose-binding protein
GST, glutathione S-transferase
CBP, calmodulin-binding protein
CAP, cellulose-associated protein
SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0205202.
References
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Savchenko, A., Cort, J.R., Booth, V., Mackereth, C.D., Saridakis, V., Ekiel, I., et al. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7 903–909. [DOI] [PubMed] [Google Scholar]
- Edwards, A.M. Arrowsmith, C.H. Christendat, D. Dharamsi, A., Friesen, J.D. Greenblatt, J.F., and Vedadi, M. 2000. Protein production: Feeding the crystallographers, and NMR spectroscopies. Nat. Struct. Biol. [Suppl.] 7 970–972. [DOI] [PubMed] [Google Scholar]
- Fields, S. 2001. Proteomics: Proteomics in genomeland. Science 291 1221–1224. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley, S.A. 2001. High-throughput proteomics: Protein expression and purification in the postgenomic world. Protein Expr. Purif. 22 159–164. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. 1999. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 81300–1309. [DOI] [PubMed] [Google Scholar]
- Paul, D.C., Van Frank, R.M., Muth, W.L., Ross J.W., and Williams, D.C. 1983. Immunocytochemical demonstration of human proinsulin chimeric polypeptide within cytoplasmic inclusion bodies of Escherichia coli. Eur. J. Cell Biol. 31 171–174. [PubMed] [Google Scholar]
- Skolnick, J., Fetrow, J.S., and Kolinski, A. 2000. Structural genomics and its importance for gene function analysis. Nat. Biotechnol. 18283–287. [DOI] [PubMed] [Google Scholar]
- Uhl'en, M. and Moks, T. 1990. Gene fusion for purposes of expression: An introduction. Methods Enzymol. 185129–143. [DOI] [PubMed] [Google Scholar]
- Zeng, G. 1998. Sticky-end PCR: New method for subcloning. Biotechniques 25 206–208. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamzyor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293 2101–2105. [DOI] [PubMed] [Google Scholar]