Abstract
The Ejl amidase is coded by Ej-1, a temperate phage isolated from the atypical pneumococcus strain 101/87. Like all the pneumococcal cell-wall lysins, Ejl has a bimodular organization; the catalytic region is located in the N-terminal module, and the C-terminal module attaches the enzyme to the choline residues of the pneumococcal cell wall. The structural features of the Ejl amidase, its interaction with choline, and the structural changes accompanying the ligand binding have been characterized by CD and IR spectroscopies, differential scanning calorimetry, analytical ultracentrifugation, and FPLC. According to prediction and spectroscopic (CD and IR) results, Ejl would be composed of short β-strands (ca. 36%) connected by long loops (ca. 17%), presenting only two well-predicted α-helices (ca. 12%) in the catalytic module. Its polypeptide chain folds into two cooperative domains, corresponding to the N- and C-terminal modules, and exhibits a monomer ↔ dimer self-association equilibrium. Choline binding induces small rearrangements in Ejl secondary structure but enhances the amidase self-association by preferential binding to Ejl dimers and tetramers. Comparison of LytA, the major pneumococcal amidase, with Ejl shows that the sequence differences (15% divergence) strongly influence the amidase stability, the organization of the catalytic module in cooperative domains, and the self-association state induced by choline. Moreover, the ligand affinity for the choline-binding locus involved in regulation of the amidase dimerization is reduced by a factor of 10 in Ejl. Present results evidence that sequence differences resulting from the natural variability found in the cell wall amidases coded by pneumococcus and its bacteriophages may significantly alter the protein structure and its attachment to the cell wall.
Keywords: Pneumococcal cell-wall amidases, pneumococcal bacteriophages, Ejl, Ej-1, choline binding proteins, DSC, CD, IR
The cell-wall lytic enzymes encoded by pneumococcus are involved in a variety of physiological functions associated with cell-wall growth, wall turnover, and cell separation (Tomasz 1984). Their polypeptide chains have evolved from the fusion of two independent modules, which are responsible for either the catalytic activity or the attachment to the choline residues of pneumococcal cell wall, which act as an antenna where lysins dock by the so-called choline-binding module, ChBM (García et al. 1990; López et al. 1995, 2000). The cell-wall lytic enzymes coded by many bacteriophages infecting pneumococcus show high similarity to either or both domains of the host major autolysin, the LytA amidase (López et al. 1995). Indeed, pneumococcal cell-wall lytic enzymes provide one of the clearest examples among prokaryotic proteins of a two-module structure whereby similarity between bacteriophage and bacterial DNA allows shuffling of modules by recombination, restructuring both viral and bacterial genomes (López et al. 1995, 2000; García et al. 1998).
The LytA amidase is involved in the separation of daughter cells, and it has been demonstrated to be a virulent factor (Berry et al. 1989). Interestingly, strains that exhibit alterations in their lytic systems appear to contribute to the higher morbidity and mortality of the pneumococcal meningitis (Tuomanen et al. 1988). Therefore, the presence of phage encoded cell-wall lysins in clinical isolates has been considered as an additional factor that could contribute to pneumococcal pathogenicity. Sequence variations in the lytA gene are known to occur naturally (Guillespie et al. 1997; Whatmore and Dowson 1999); however, there is no data about their effects on the autolysin structure or its affinity for the cell wall. Analogously, very little is known about the structural features of the amidases coded by pneumococcal bacteriophages. The pneumococcal clinical strain 101/87 contains the temperate phage Ej-1 coding for the Ejl cell-wall lytic amidase (Díaz et al. 1992). The catalytic function is located in the Ejl N-terminal module and the C-terminal module, composed of six repeats of about 20 amino acids (p1–p6) and a short tail, constitutes the choline-binding module (Díaz et al. 1992). Ejl and the pneumococcal amidase from the R6 strain, LytA, share this type of organization but differ in 15% of their amino acid sequences (Díaz et al. 1992), a percentage far beyond the divergence value (about 5%) found among the LytA amidases of typical pneumococcal strains (Whatmore et al. 2000). Such differences render Ejl activity 13% that of LytA (Díaz et al. 1992). The structural and thermodynamic characterization of LytA has shown that its polypeptide chain seems to be folded into three cooperative domains, two of them involving the N-terminal catalytic module (Varea et al. 2000), and displays a monomer ↔ dimer equilibrium mediated by the ChBM (Usobiaga et al. 1996). This equilibrium shifts to dimer formation upon choline binding to LytA's high-affinity sites (Medrano et al. 1996; Usobiaga et al. 1996). The final region of the ChBM plays a key role on the choline recognition (Varea et al. 2000). Interestingly, the p6 repeat and the tail bear 46% of all the sequence differences between the ChBMs of LytA and Ejl.
This work presents the structural and thermodynamic characterization of Ejl, the first choline-dependent amidase coded by a pneumococcal phage to be studied at this level, using several biophysical approaches (CD and IR spectroscopies, DSC, and analytical ultracentrifugation). The interaction of Ejl with choline and the conformational changes promoted by the ligand are also characterized. Present results provide valuable information regarding the effect of sequence variations on the amidase structure and their influence on the protein attachment to the bacterial surface.
Results and Discussion
Choline–Ejl interactions
The hydrolytic action of pneumococcal cell-wall lysins requires the anchoring of the enzyme to the bacterial envelope by recognition of the choline residues present in pneumococcal teichoic acids. The interaction of the Ejl amidase with choline has been characterized at 25°C by monitoring the variations in the CD spectra and the self-association state of the enzyme induced by the ligand.
CD choline titration curves
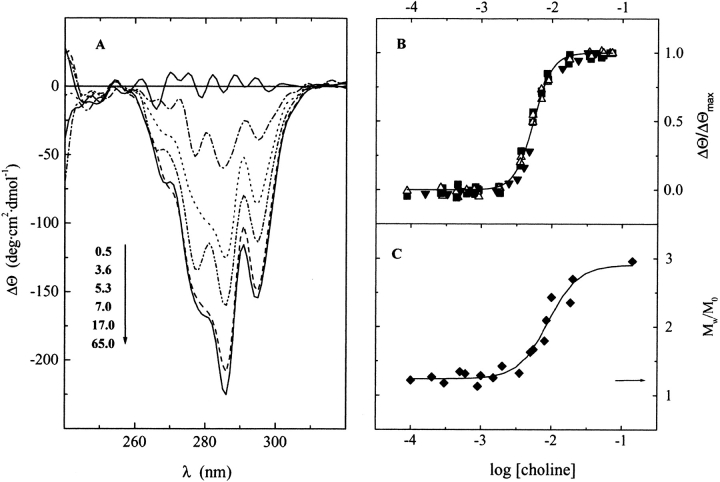
Choline binding to Ejl modifies its far and near CD spectra, inducing a positive maximum at 224 nm and two minima at 295 and 286 nm. Figure 1A ▶ illustrates the differences in the near-UV region of the Ejl spectra produced by increasing concentrations of ligand. The relative ellipticity changes (ΔΘ/ΔΘmax), at the wavelengths yielding the larger variations in Ejl spectra, are shown in Figure 1B ▶ as a function of ligand concentration. The choline titration curves are monophasic, and highly cooperative at all the wavelengths tested, suggesting the presence of a single set of binding sites. Their analysis in terms of the Hill equation (solid line in Fig. 1B ▶) yields a value for the Hill coefficient of 3 and an apparent equilibrium constant, Kapp, of (1.83 ± 0.02) × 102 M−1.
Fig. 1.
Choline titration curves of Ejl amidase. (A) CD differential spectra in the near region of the UV-spectra at increasing concentrations of choline in 20 mM phosphate buffer (pH 8.0, 25°C). The ligand concentrations, in mM units, are indicated in the figure labels. (B) Relative variations in the ellipticity at 286 (filled squares), 295 (open triangles), and 224 (filled triangles) nm; the solid trace is the best-fitting curve calculated for the Hill equation, using a Kapp of 1.8 × 102 M−1 and a Hill coefficient of 3. (C) Dependence of the apparent weight-average molecular mass of Ejl (5.5 μM) on choline concentration. The arrow indicates the Mw/M0 ratio of unbound Ejl (M0 is the monomer molecular weight).
Choline-dependent self-association equilibria of Ejl amidase
The self-association state of the Ejl amidase and its dependence on choline concentration was investigated by sedimentation equilibrium and exclusion chromatography. Figure 1C ▶ shows the relative apparent average molecular weight (Mw/M0) of Ejl, at 5.5 μM, as a function of choline concentration. In the absence of the ligand, Ejl sediments with an apparent molecular weight, Mw, slightly higher than that of the monomer (Mw/M0 = 1.2), indicative of partial self-association. The average molecular weight increases with choline concentration, reaching a value that corresponds to approximately three times that of the monomer under saturating conditions. The choline concentration providing the half variation in Mw/M0 is 9 ± 1 mM. These results suggest that the Ejl amidase could exist either as monomer/dimer/trimer or monomer/dimer/tetramer equilibria that shift towards the self-associated forms of the amidase by choline binding. A better description of the sedimentation profiles was obtained assuming formation of dimers and tetramers (data not shown).
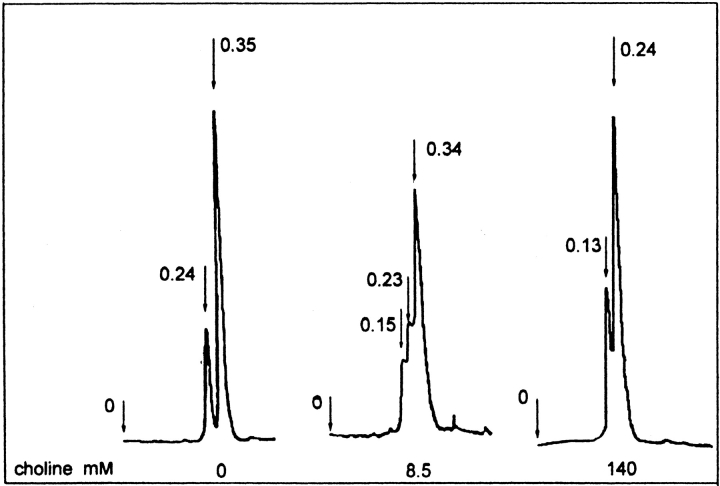
To distinguish between both possible models, Ejl self-association was further characterized by analytical gel filtration chromatography (Fig. 2 ▶), using an initial protein concentration of 5.5 μM. In the absence of choline, the elution profile of Ejl shows two peaks characterized by Kav values of 0.24 and 0.35, and the slowest eluted form is the major component. At 8.5 mM choline a third peak with a Kav value of 0.15 is also observed. At saturating choline concentrations, the peak at 0.35 disappears and the intermediate specie (Kav = 0.24) becomes the most populated form at this protein concentration. The anomalous migration exhibited by the cell wall lysins coded by pneumococcus and its bacteriophages (Varea et al. 2000) does not allow to estimate the molecular masses of the Ejl self-associated forms by exclusion chromatography using the calibration curve. However, the value of 3 found for the Mw/M0 ratio at 140 mM choline would mean the presence of a single Ejl oligomer at saturating choline concentrations, if trimers were the higher molecular weight state stabilized by choline. In contrast, the observation of two oligomers under these conditions (Fig. 2 ▶) are consistent with formation of Ejl dimers (Kav = 0.24), the most populated form, and tetramers (Kav = 0.15). Therefore, present results allow inferring that the Ejl amidase actually exists as monomer ↔ dimer ↔ tetramer equilibria regulated through choline–amidase interactions, being Ejl monomers (Kav = 0.35) and dimers the populated states in the choline-free amidase.
Fig. 2.
Analytical size-exclusion chromatography of Ejl amidase at increasing choline concentrations. Variation of the self-association state of the Ejl amidase on choline concentration (20 mM phosphate buffer; pH 8.0). The arrows indicate the exclusion volume and the elution positions of the different Ejl species expressed as Kav values: 0.35 (monomer), 0.24 (dimer), and 0.13 (tetramer). The initial Ejl concentration was 5.5 μM.
Conformational characterization of the Ejl amidase
Secondary structure
The content in secondary structure of Ejl was estimated by analyzing its far-UV CD and IR spectra in combination with the data provided by sequence-based prediction methods.
CD and IR spectroscopies
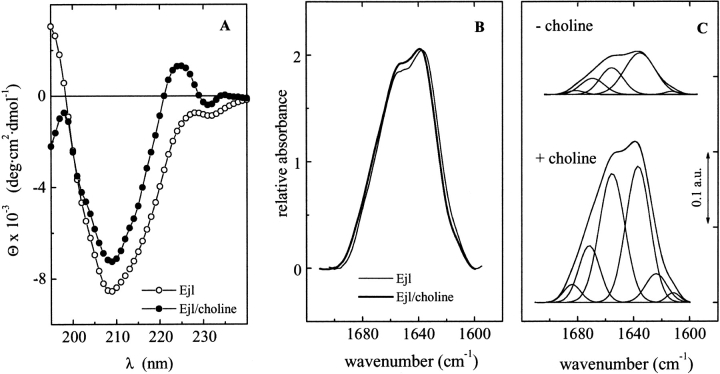
Figure 3A and B ▶ shows the CD and IR spectra of the free and choline-bound Ejl amidase at 25°C. The far-UV CD spectrum of Ejl is characterized by a large minimum centered at 209 nm and a minor one at around 232 nm. The analysis of the experimental curve (see Materials and Methods) yielded the following percentages for the secondary structure elements: 31 ± 3% antiparallel β-sheet, 5 ± 1% parallel β-sheet, 20 ± 2% β-turns, 12 ± 1% α-helix, and 32 ± 3% of unordered structure. In the absence of the ligand, the amide I band of Ejl in D2O can be resolved into three major components at 1634, 1656, and 1670 cm−1 and two minor ones at around 1680 and 1611 cm−1 (Fig. 3C ▶), whose relative area and proposed assignments are listed in Table 1. The band at 1634 cm−1 indicates the presence of β-sheets (Arrondo et al. 1993; Goormaghtight et al. 1994), although it can also contain contributions from nonstructured conformations (Martínez et al. 1996; Chehin et al. 1998). The band at 1656 cm−1 in D2O is usually produced by α-helix (Arrondo et al. 1993; Goormaghtight et al. 1994). However, bands originated from structures with dihedral angles close to α-helix (Arrondo and Goñi 1999) or even bands arising from large loops (Petrelski et al. 1991; Wilder et al. 1992) have also been described at this frequency. Finally, the component at 1670 cm−1 has been associated to β-turns, whereas that at 1680 cm−1 could arise from either β-turns or the high-frequency component of antiparallel β-sheet (Surewicz and Mantsch 1988; Arrondo et al. 1993). The intensity of this component would be less than 1/10 of the low-frequency band around 1630–1638 cm−1 (Fraser and Macrae 1973; Castresana et al. 1988); therefore, the presence of the weak band at 1680 cm−1 in conjunction with the strong band at 1634 cm−1 could denote the presence of antiparallel β-sheet conformations.
Fig. 3.
Far-UV CD spectra and Amide I band (D2O) of Ejl in the absence and presence of choline. (A) CD spectra of Ejl in the absence (open circles) and presence of 20 mM choline (filled circles) (buffer: 20 mM phosphate pH 8.0; T = 25°C). (B) Comparison of the experimental amide I band of Ejl in the absence (thin trace) and presence of choline (thick trace), in arbitrary units. (C) Amide I band decompositions of the Ejl amidase; the parameters derived for the components are shown in Table 1.
Table 1.
Band frequencies, fractional areas, and proposed structure assignments of the components derived from curve fitting analysis of the amide I band of Ejl, in D2O, under native conditions
| Choline | ||||
| − | + | |||
| Assignment | Band (cm−1) | Area % | Band (cm−1) | Area % |
| High-frequency β-sheet | 1681 | 4 | 1685 | 4 |
| Turns | 1670 | 14 | 1672 | 12 |
| α-Helix and loops | 1656 | 27 | 1656 | 40 |
| β-Sheet and unordered | 1634 | 53 | 1636 | 35 |
| β-Strands | — | — | 1624 | 8 |
| 1611a | 2 | 1611 | 1 | |
a Bands at around 1611 cm−1 in native protein arise from amino acid side chains.
Prediction methods
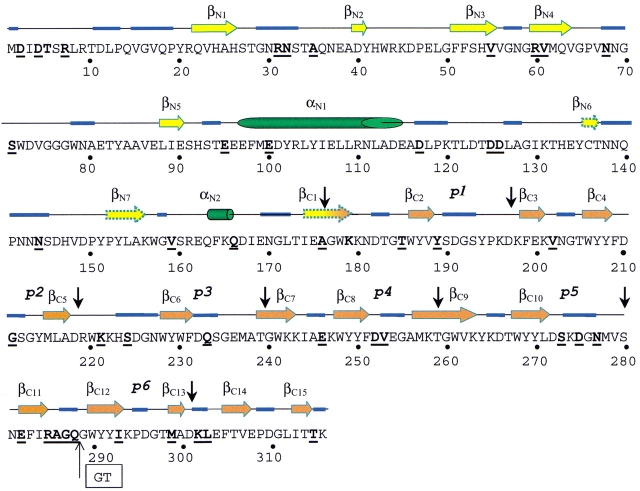
The secondary structure of Ejl was predicted by means of three different methods, i.e., PHD, Jpred, and PSIpred. The three approaches yield similar results, although the percentages of β-sheet predicted by Jpred and PSIpred are higher than that of PHD. The predicted structure corresponds to a mixed protein with a secondary structure composition of 9–14% α-helix, 27–38 % β-sheet, and 53–59 % random conformations. Figure 4 ▶ shows the distribution of secondary structure elements along the Ejl sequence for those residues whose conformation is predicted with an accuracy higher than 85.7% (reliability index ≥6) by at least two of the three methods. Segments involving residues 135–137, 152–155, and 174–179 are also predicted to be in an extended conformation but with lower reliability (Fig. 4 ▶). The N-terminal module is predicted to be composed of short β-strands connected by long loops plus the long α-helix formed by residues 97 to 114 and a shorter one at the end of the module. The ChBM, an all β-region, shows a repetitive pattern as expected from its sequence repeats. The aromatic clusters of the ChBM, with a consensus sequence WYYX, would be forming short β-strands as it also does the region around the aromatic residue placed 7–8 amino acid upstream of each aromatic cluster. It is worth noting that positive bands in CD spectra around 220 nm (Fig. 3A ▶) have been associated to tryptophane residues in β-strand (Freskgard et al. 1994). The Gly residues located at both sides of the aromatic clusters are highly conserved in ChBMs (García et al. 1998; López et al. 2000). Their position in the loops suggests an antiparallel disposal of the β-strands, because glycine is a conserved residue in β-hairpins that contributes to the tight turn between β-strands. The presence of many Pro and Gly residues in the regions predicted to be loops supports this assignment. Moreover, these regions contain 75% of the sequence differences between LytA and Ejl amidases. Despite the differences found between repeats, inherent to limitations of prediction methods, present results suggest that the amino acids forming the choline-binding repeats would probably fold into a β-hairpin, involving the aromatic residues, followed by a rather long loop.
Fig. 4.
Predicted secondary structure of Ejl. The arrows (β-strand), cylinders (α-helix), and blue traces (loops) correspond to those amino acid residues whose conformation is predicted with accuracy higher than 85.7% by at least two of the methods employed (PHD, PSIpred, and the Jpred servers); discontinuous arrows indicate three β-strands predicted with lower reliability. Vertical black arrows indicate the sequence region comprising the sequence repeats (p1–p6) of the ChBM. Bold letters indicate the Ejl amino acid muted in relation to LytA amidase. The "GT" inset indicates the position of the double deletion in the p6 repeat of Ejl.
Table 2 summarizes the experimental and theoretical results derived for the Ejl secondary structure composition. It is worth noting the excellent agreement of the CD estimates for α-helical and β-sheet conformations with those derived from the prediction methods. Analogously, the percentages of β-turns derived from CD and IR analyses are in reasonable agreement, given the limits inherent to both methodologies. In contrast, the contributions of the bands at 1656 and 1634 cm−1 (usually ascribed to α-helix and β-sheet) to the total area of the amide I band are significantly higher than the values derived by the other two methods for these conformations. Furthermore, the band characteristic of unordered structures (1648–1640 cm−1) does not appear in the deconvolved amide I band of the native amidase, although the Ejl CD data were consistent with a content in random structure of about 32%. Taking into account the high sensitivity of far-UV CD spectra to α-helical and unordered conformations, the present results suggest that the IR bands due to the Ejl non periodical structures would overlap those at 1656 and/or 1635 cm−1, as has been reported in other systems (Petrelski et al. 1991; Wilder et al. 1992; Arrondo and Goñi 1999).
Table 2.
Secondary structure composition of Ejl amidase derived from prediction methods CD and IR spectroscopies
| Secondary structure prediction | CD | IR | |||||
| %a | %c | ||||||
| Component | % | Component | − | + | Component | − | + |
| β-Strand | 35 ± 4 | β-sheetb | 36 | 42 | β-sheet and unordered | 58 | 48 |
| Turns, loops, others | 52 ± 2 | turns | 20 | 19 | turns | 14 | 12 |
| Non regular structures | 32 | 31 | |||||
| α-Helix | 10 ± 2 | α-helix | 12 | 8 | α-helix and loops | 28 | 40 |
The signs (−) and (+) indicate the absence or presence of choline.
a Errors are within ±10% of the estimated value.
b Antiparallel and parallel β-sheet are, respectively, 31 and 5% without choline, and 37 and 5% with choline.
c Errors are within ±5% of the estimated value.
Influence of choline binding on Ejl secondary structure
The analysis of the CD spectrum indicates that the modification of Ejl secondary structure induced by choline binding is rather small to be soundly considered, and the spectroscopic changes probably reflect variations in the chiral contribution of the aromatic side chains. On the other hand and according to IR data, choline binding increases the contribution of the 1656 cm−1 component by about 13%, and reduces the total percentage of extended structures by a similar amount (Table 1). In addition, a new component at 1624 cm−1 is also observed (Table 1). Bands at this frequency are not common in native structures, and have been assigned to protein segments in an extended configuration with their peptide residues nonforming intramolecular β-sheet, but hydrogen bonded to other molecular structures (Arrondo et al. 1994; Castresana et al. 1988). The apparent invariance in the β-sheet content of Ejl upon ligand binding shown by CD data together with the good agreement of the IR and CD estimates (Table 2) could indicate that the IR band due to nonperiodical segments would move to higher wavenumber positions, overlapping that from α-helix under choline saturating concentrations. These results indicate that choline binding to Ejl promotes small local rearrangements (Susi 1969) that seem to mainly affect flexible segments of the ChBM.
Thermal stability: Cooperative domain organization
Differential scanning calorimetry
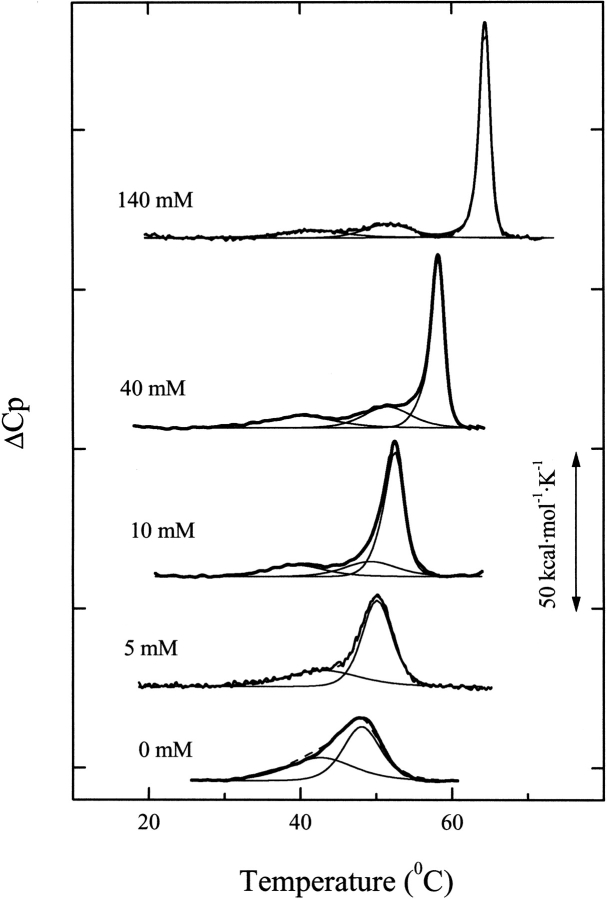
Figure 5 ▶ shows the heat capacity profiles of Ejl in the absence and presence of increasing concentrations of choline. In the absence of the ligand, denaturation takes place with an enthalpy change, ΔHd, of 192 kcal per mole of monomer, and the calorimetric scan is characterized by a broad endotherm with a shoulder at 41.5°C and a maximum at 48.0°C. The deconvolution analysis of the experimental profile shows that the endotherm can be described in terms of two transitions with Tm values of 42.9 (ΔH1 = 84 kcal mol−1) and 48.0°C (ΔH2 = 109 kcal mol−1), which slightly deviate from the two-state model (ΔHvH1/ΔH1 = 0.81; ΔHvH2 /ΔH2 = 1.14) (Table 3). These results would be consistent with a polypeptide chain organized into two cooperative domains that would probably correspond to the catalytic and choline binding modules.
Fig. 5.
Effect of choline concentration on the DSC transitions of Ejl amidase. Calorimetric profiles of the Ejl amidase (27.4 μM) at the choline concentrations indicated in the curve labels (20 mM phosphate buffer, pH 8.0). Bold traces represent the experimental curves, the thin traces depict the results of deconvolution analysis, and the dashed line is the fitted profile. ΔCp values are in kcal•(mole of monomer)−1•K−1.
Table 3.
Thermodynamic parameters for thermal denaturation of Ejl amidase in 20 mM phosphate buffer, pH 8.0a
| [Choline] (mM) | Tm1b (°C) | ΔH1c (kcal mol−1) | ΔH1vHc (kcal mol−1) | Tm2b (°C) | ΔH2c (kcal mol−1) | ΔH2vHc (kcal mol−1) | Tm3b (°C) | ΔH3 (kcal mol−1) | ΔH3vHc (kcal mol−1) | ΔHd (kcal mol−1) |
| 0 | 42.9 | 84 | 72 | — | — | — | 48.0 | 109 | 130 | 192 |
| 3 | 42.8 | 59 | 66 | — | — | — | 49.5 | 133 | 152 | 188 |
| 5 | 43.2 | 63 | 65 | — | — | — | 50.3 | 144 | 159 | 206 |
| 10 | 39.6 | 36 | 78 | 49.4 | 44 | 93 | 52.3 | 140 | 231 | 223 |
| 20 | 40.4 | 53 | 73 | 50.2 | 47 | 88 | 55.2 | 130 | 258 | 228 |
| 40 | 40.2 | 46 | 63 | 51.5 | 54 | 103 | 58.1 | 141 | 317 | 239 |
| 80 | 39.7 | 37 | 73 | 52.3 | 55 | 80 | 62.1 | 158 | 373 | 255 |
| 140 | 41.7 | 34 | 72 | 51.7 | 54 | 100 | 65.0 | 155 | 404 | 238 |
| 280 | 39.4 | 29 | 84 | 49.5 | 58 | 72 | 68.2 | 193 | 386 | 280 |
a Ejl concentration was 27.4 μM. ΔHi is the calorimetric enthalpy change of transition i; ΔHivH is the van't Hoff enthalpy of transition i, calculated as 4RTmiΔCpmi / ΔHi, where ΔCpmi is the maximum of the Cp function, Tmi is the temperature of the maximum, and R is the gas constant. ΔHd is the total denaturation enthalpy change. Calorimetric ΔH values are in kcal per mole of Ejl monomer.
b Estimated error ±0.3°C.
c Estimated error ±12%.
Ejl denaturation displays a complex dependence on choline concentration. At subsaturating concentrations of the ligand the transition centered at 48°C is shifted toward higher temperatures, suggesting its relation with the choline binding module, while the lower temperature transition remains basically unchanged. Thus, at 5 mM choline the endotherm can be resolved into two apparently independent transitions with van't Hoff to calorimetric enthalpy ratios very close to 1, the value expected for the two-state denaturation model. However, Ejl denaturation cannot be further described in terms of two steps at ligand concentrations higher than that providing half-saturation (Fig. 5 ▶). The deconvolution of the heat capacity curve at 10 mM choline showed that the well-resolved transition that takes place at lower temperature proceeds with a calorimetric enthalpy change of 36 kcal per mole of monomer, which is significantly lower than its van't Hoff enthalpy change (Table 3). Furthermore, the highest temperature peak cannot be explained in terms of a single step, and its analysis suggests the presence of two overlapping transitions under the envelope (ΔH>ΔHvH). Further increase in choline concentration gives rise to a progressive sharpening and upward shift of the major peak, and the new transition, centered around 51°C, appears clearly resolved at 40 mM choline and above. The peak near 40°C is always detected, although its calorimetric enthalpy change is significantly reduced as the ligand concentration increases. It is worth noting that the denaturation profiles of Ejl at subsaturating choline concentrations will not be affected by ligand dissociation from denatured complexes, because the free initial concentration of choline is more than two orders of magnitude higher than that of Ejl and, therefore, the saturation fraction remains practically unchanged as denaturation proceeds. Also, the appearance of Ejl oligomers as resolved species in exclusion chromatography (Fig. 2 ▶) indicates that reequilibration among them is slow.
Table 3 summarizes the parameters derived from deconvolution of the DSC curves. No measurable variations in ΔCp have been observed upon Ejl denaturation. This may be due to the high content in the secondary structure that thermally denatured Ejl has according to its CD and IR spectra (data not shown). The rather small influence of choline binding on the Tm1 value suggests that the lowest temperature transition corresponds to the N-terminal catalytic module. Also, the apparently saturating effect of choline on the Tm2 values indicates that the transition centered around 51°C involves either a different conformation or a specific region of the N-terminal module stabilized by means of interdomain interactions upon choline binding to the ChBM (Brants et al. 1989). Data reported in Table 3 shows that the calorimetric ΔH values accompanying the two first transitions of the choline-bound Ejl are significantly lower than their ΔHvH values, which remain within the experimental uncertainty choline concentration independent. These results would be, therefore, consistent with the existence of two choline-bound Ejl conformations with different N-terminal module stability and in slow equilibrium. Taking into account that the first transition is detected even at 140 mM choline, where Ejl dimers and tetramers coexist, the simplest explanation of our DSC results would be that tetramerization increased the stability of the N-terminal module, thus inducing the appearance of the peak at 51.5°C. The finding that a single transition accounts for denaturation of the N-terminal module in the unbound amidase, despite the presence of Ejl monomers and dimers, also supports the hypothesis that tetramerization stabilizes the N-terminal module, increasing its Tm value by about 12°C. The denaturation of the ChBM under saturating choline concentrations does not conform to a two-state process (ΔHvH3/ΔH3 > 2) but the possible presence of two distinct Ejl conformations that may differ in their thermodynamic properties hampers a further analysis of the highest temperature peak. Nevertheless, the increase of the ΔHvH3/ΔH3 ratio with choline concentration up to values around 2.5, due to the increment of ΔHvH3, indicates the existence of tight cooperative interactions among the ChBMs in the Ejl n-mers (dimers or tetramers) stabilized by choline.
CD thermal denaturation studies
Figure 6 ▶ shows the temperature-induced changes in the ellipticity values at 203 and 224 nm, the regions of maximum variability, with and without choline. The variations starting around 65°C occur well above the end of the calorimetric transitions and denote additional events undergone by the denatured amidase; therefore, the comparison of the calorimetric and dichroic transitions was not extended beyond 65°C. Simulation of the CD transition curves by means of equation 1 reveals that the ellipticity changes at 203 nm can be well explained in terms of the lowest temperature transition observed by DSC (Fig. 6A ▶). On the other hand, the ellipticity changes at 224 nm, sensitive to choline concentration, take place within the same temperature interval as the highest temperature DSC peak, and can be basically ascribed to denaturation of the ChBM (Fig. 6B ▶). A somewhat better fit is achieved by assuming a contribution of around 10% from the N-terminal transitions centered at 42.9°C (0 mM choline) and 50.2°C (20 mM choline). These results further support the previous assignment of the DSC transition centered at 48.0°C to denaturation of the ChBM in the absence of choline. Table 4 summarizes the contributions of the different transitions to the ellipticity changes derived from the analysis of CD transition curves using equation 1 at both wavelengths.
Fig. 6.
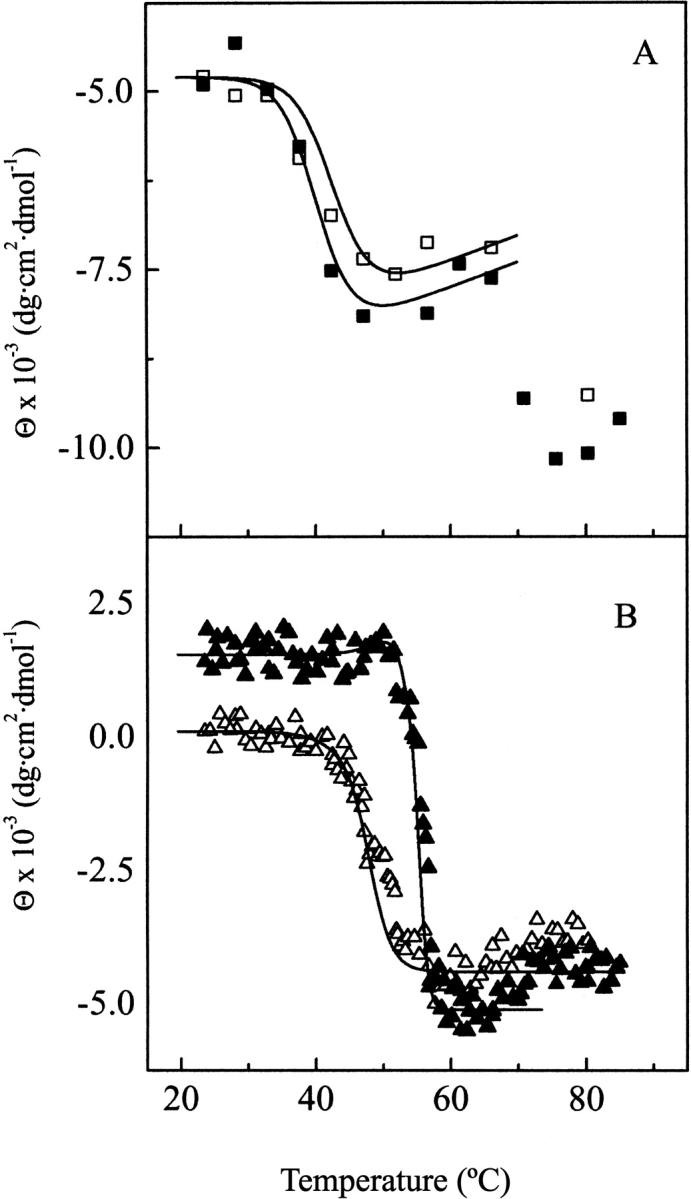
Temperature CD titration curves of Ejl. (A) and (B) depict the elliptiticity changes at 203 (squares) and 224 nm (triangles) as a function of temperature both in the absence (open symbols) and in the presence of 20 mM choline (full symbols), (5.5 μM Ejl; 20 mM phosphate buffer pH 8.0). Solid lines are the theoretical curves calculated according to equation 1 with the parameters reported in Table 4.
Table 4.
Contribution of thermal transitions to the thermally induced variations in Ejl CD spectrum at 203 and 224 nm derived by equation (1)
| ΔΘi • 10−3 (dg dmol−1 cm2) | |||
| Choline (mM) | Tm °C | λ203 | λ224 |
| 0 | 42.9 | −5.07 + 0.035 • ta | −0.6 |
| 48.0 | — | −3.9 | |
| 20 | 40.4 | −4.7 + 0.035 • t | — |
| 50.2 | — | 0.6 | |
| 55.2 | — | −7.3 | |
a The ellipticity change at 203 nm is a function of temperature due to the temperature dependence of Θ203 after the thermal transition.
Comparison of Ejl and LytA amidases
Ejl and LytA have similar secondary structures according to their CD and IR spectra (Varea et al. 2000). However, it is worth noting that the lack of two residues (G289–T290 in LytA) in the p6 repeat of Ejl will short the small predicted loop connecting β-strands C11 and C12, a key region for the choline–amidase interaction (Varea et al. 2000). In contrast with the little effect that the sequence differences have on the secondary structure, they strongly modify the amidase stability and its organization into cooperative domains. The ChBM of Ejl is significantly destabilized relative to that of LytA, and the 11°C decrease of the Tm value is even higher than that resulting from deletion of the last 37 amino acids in LytA (Varea et al. 2000). On the other hand, the catalytic module of LytA seems to be organized into two cooperative blocks, whose transition temperatures are 40.9 and 49.6°C (Varea et al. 2000), whereas that of Ejl forms a single cooperative domain that denatures at 42.9°C in the absence of choline. These differences denote alterations in the native tertiary contacts of the catalytic module.
The choline recognition and the regulation of the amidase self-association by the ligand binding are also dependent on the amidase sequence. Ejl seems to have a single set of binding sites, whose apparent affinities are similar to those of the LytA lower affinity binding sites (Medrano et al. 1996). However, Ejl dimerization reveals that the high-affinity loci involved in LytA dimerization are still present in Ejl, although their affinity for choline is reduced by one order of magnitude. The structural changes derived from the absence of G289–T290 in Ejl could account for the lower affinity of these Ejl loci for choline, because the spatial orientation of the region comprising the 11 final residues is essential for the nonequivalence of LytA choline binding sites (Varea et al. 2000).
The preferential binding of choline to Ejl dimers and tetramers shows that the formerly observed choline-regulated self-association of LytA is a feature shared by other pneumococcal cell-wall amidases. Ejl dimerization would probably involve, as in the LytA amidase (Varea et al. 2000), the C-terminal tail, given the high percentage of identity in their sequences. However, the interdimeric interactions providing tetramerization should involve specific mutations of Ejl, because none of the homologous amidases encoded by pneumococcus or its bacteriophages tetramerize upon choline binding (unpublished results). Tetramerization might constitute a mechanism for compensating the low affinity of Ejl for choline (Medrano et al. 1996; Usobiaga et al. 1996). However, the inhibition of the phage amidase by free choline requires lower ligand concentration than LytA, indicating a less efficient anchoring of Ejl to the cell wall (Díaz et al. 1992). It seems, therefore, that the higher affinity of LytA amidase for choline, due to the nonequivalence of choline binding sites, allows a more efficient attachment to the bacterial surface.
Materials and methods
Protein purification and chemicals
Ejl amidase (36.535 kDa) was purified from Escherichia coli HB101 (pNE103) cells, by affinity chromatography on DEAE-cellulose (Sanz et al. 1988), and the purity of the isolated protein samples was routinely analyzed by SDS-PAGE (Laemmli 1970). Before use, protein samples were extensively dialyzed at 4°C against the appropriate buffer (20 mM phosphate pH 8.0, with or without choline) and then centrifuged at 13,000 rpm for 5 min. Ejl concentration was determined spectrophotometrically using a molar absorption coefficient at 280 nm of 113,630 M−1 cm−1 for the amidase monomer. Choline concentration was measured by differential refractometry (Medrano et al. 1996). All reagents (Sigma Chemical Co.) were of analytical grade.
Circular dichroism spectra
Circular dichroism spectra were recorded in a JASCO J-720 spectropolarimeter (Jasco Corp.), as reported elsewhere (Varea et al. 2000), using Ejl concentrations of 27.4 and 5.5 μM, respectively, for the near- and far-UV regions. The observed ellipticities were converted to mean residue ellipticities, Θ, using a mean molecular mass per residue of 116. Thermal denaturation experiments were carried out by increasing the temperature at a scanning rate of 20°C/h, allowing the temperature to equilibrate for 5 min before recording the spectrum. The analysis of secondary structure composition was performed using the CDNN software package that uses precompiled neural networks for deconvolution of CD spectra, using an spectra data base of 33 proteins (Bohm et al. 1992). The set includes proteins with significant chiral aromatic side-chain contributions on their far-UV CD spectra to get reliable predictions for Ejl structure. Deconvolution of CD spectra using the Lincomb method (Perczel et al. 1992), which explicitly include the aromatic side-chain contributions, yielded secondary structure contents for Ejl consistent with those of CDNN. However, the Lincomb method only allows a global estimation for β-structure (turns plus β-sheet) (Varea et al. 2000), making the CDNN results more suitable for comparing with IR and prediction data estimates.
The CD thermal denaturation curves at a fixed wavelength were theoretically simulated as a linear combination of the variations contributed by the different elementary transitions characterized by DSC by using equation 1 (Varea et al. 2000), where fi(T)
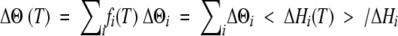 |
(1) |
is the fractional concentration of the denatured form of domain i at temperature T and ΔΘi is the maximum ellipticity change contributed by this transition. The fi(T) values were calculated as fi(T) = 〈ΔHi(T)〉/ΔHi, using the thermodynamic data determined from DSC experiments. The values of ΔΘi were varied until the best fit to the experimental CD denaturation curves was obtained.
Fourier-transform infrared spectra
Infrared spectra were recorded in a Nicolet Magna II 550 FTIR spectrometer (Nicolet Corp.) equipped with a MCT detector and following the procedure described in Varea et al. (2000). Aliquots containing about 300 μg of protein in 10 mM phosphate (pH 8.0), with or without choline, were dried and rehydrated by adding 25 μL of D2O, and allowed to undergo proton–deuterium exchange, until the amide band was invariant with time. Data treatment and band decomposition of the original amide I have been described elsewhere (Arrondo et al. 1993; Arrondo and Goñi 1999).
Prediction methods
Secondary structure predictions were performed by three different approaches using the PHD (Rost and Sander 1994), the PSIpred (Jones 1999), and the Jpred (Cuff and Barton 1999) methods. The PHD and PSIpred are based on the use of the evolutionary information contained in multiple sequence alignments as input to neural networks, while Jpred is a consensus method that incorporates four different approaches to calculate its consensus prediction.
Sedimentation equilibrium
Sedimentation equilibrium experiments were performed by centrifugation of 80-μL samples at 15,000 rpm and 25°C in an Optima XL-A analytical ultracentrifuge (Beckman Instruments, Inc.), using 12 mm double-sector six-channel epon-charcoal centerpieces (Usobiaga et al. 1996). To obtain the weight apparent average molecular weights, Mw, a sedimentation equilibrium model for single species was fitted to individual data sets, using the conservation of a signal algorithm (Minton 1994) from EQASSOC and XLAEQ programs as reported by Usobiaga et al. (1996). The partial specific volume of Ejl calculated from amino acid composition was 0.72 mL g−1 (Laue et al. 1992).
Analytical size-exclusion chromatography
The molecular weight markers and samples (usually 50 μL at 5.5 μM) were eluted at room temperature in 20 mM phosphate buffer, pH 8.0 (with or without choline) at a rate of 0.5 mL min−1, using a calibrated Superose 12 FPLC column (Pharmacia Biotech). The solute behavior was characterized as kav = (Ve − V0)/(Vt − V0), where Ve, V0, and Vt correspond, respectively, to the elution volume of the solute, the void volume, and the total volume of the bed.
Differential scanning calorimetry
DSC measurements were performed using a Microcal MC-2 instrument (Microcal, Inc.) at a heating rate of 0.33 K min−1, unless otherwise stated and under an extra constant pressure of 2 atm. The standard DA-2 and Microcal Origin software were used for data acquisition and analysis. The excess heat capacity functions were obtained after baseline subtraction of the buffer baseline. The thermal transitions involving the N-terminal module of Ejl amidase were found to be irreversible, while denaturation of the ChBM was partially reversible, particularly in the presence of choline. The influence of the irreversible steps on the thermograms was checked by running samples at 0.2, 0.75, and 1.0 K min−1. The heat capacity profiles of Ejl recorded at different scanning rates did not show significant variations in the form or in the enthalpy of the whole endotherm; therefore, the kinetic control of the denaturation processes can be discarded and the thermodynamic analysis of the DSC curves is allowed (Freire et al. 1990; Sánchez-Ruiz 1995).
Conclusions
The analysis of the Ejl secondary structure using prediction methods and experimental data from CD and IR spectroscopies suggests that this amidase is mainly composed of β-strands connected by long loops, and that the α-helical conformation seems to be restricted to the catalytic module, located at the N-terminal position. According to prediction results, the aromatic clusters of the ChBM would form short β-strands, each constituting repeat folds into a β-hairpin followed by a rather long loop. A similar pattern, without the final loop, is also predicted for the terminal C-tail. According to DSC data, the polypeptide chain of choline-free Ejl folds into two cooperative domains, corresponding to the catalytic and choline binding modules, and Ejl monomers exists in equilibrium with dimers.
The binding of choline induces small rearrangements in the Ejl secondary structure but enhances Ejl self-association by preferential binding to dimeric and tetrameric species. The interaction of Ejl with choline stabilizes, by direct binding, the ChBM. Besides, the structural changes induced by the ligand binding seem to stabilize, in a fraction of Ejl molecules, the N-terminal module whose transition temperature increases by about 12°C. This stabilization is mediated by interdomain interactions, and might be related to Ejl tetramerization. DSC results also reveal the presence of tight intersubunit interactions among the ChBMs in the choline–Ejl complex, probably due to formation of dimeric and/or tetrameric cooperative domains upon the choline induced self-association of Ejl.
The sequence differences between Ejl and LytA yield a phage amidase that significantly differs from that of the host in its tertiary and quaternary structure. These structural changes, together with those derived from punctual amino acid substitutions, should account for the affinity decrease towards the choline residues acting as amidase receptors on the pneumococcal cell wall. In addition, the conformational differences between the N-terminal catalytic modules of both amidases revealed by thermal denaturation experiments might also contribute to the lower efficiency of Ejl in hydrolyzing the bacterial cell wall (Ejl specific activity is 13% of LytA; Díaz et al. 1992). These results provide the first direct evidence on the structural changes that may result from the sequence variation found among pneumococcal cell-wall amidases coded by either different strains (Guillespie et al. 1997; Whatmore and Dowson 1999) or its bacteriophages (López et al. 1995).
Acknowledgments
This work was supported by DGICYT Grants PB96-0850, PB96-0809, BIO2000-1307, and BMC2000-1002, and by CAM Grant of the Program for Strategic Groups. We thank M.V. López Moyano for excellent technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ChBM, choline-binding module
Ejl, amidase encoded by the Ejl gene from Ej-1 phage
LytA, major autolysin from S. pneumoniae
IR, Fourier transform infrared spectroscopy
X, aromatic residue
DSC, differential scanning calorimetry
ΔH, calorimetric enthalpy change
ΔHvH, van't Hoff enthalpy change
〈ΔHi(T)〉, excess enthalpy change of transition i at temperature T
Tm, temperature of the maximum in the heat capacity function. HWHH is the half-width at half-height of IR bands
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.4680102.
References
- Arrondo, J.L.R. and Goñi, F.M. 1999. Structure and dynamics of membrane proteins as studies by infrared spectroscopy. Prog. Biophys. Mol. Biol. 72 367–405. [DOI] [PubMed] [Google Scholar]
- Arrondo, J.L.R., Castresana, J., Valpuesta, J.M., and Goñi, F.M. 1994. Structure and thermal denaturation of crystalline and noncrystalline cytochrome oxidase as studied by infrared spectroscopy. Biochemistry 33 11650–11655. [DOI] [PubMed] [Google Scholar]
- Arrondo, J.L.R., Muga, A., Castresana, J., and Goñi, F.M. 1993. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 59 23–56. [DOI] [PubMed] [Google Scholar]
- Berry, A.M., Lock, R.A., Hansman, D., and Paton, J.C. 1989. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm, G., Murh, R., and Jaenicke, R. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5 191–195. [DOI] [PubMed] [Google Scholar]
- Brandts, J.F., Hu, C.Q., Lin, L.-N., and Mas, M.T. 1989. A simple model for proteins with interacting domains. Applications to scanning calorimetry data. Biochemistry 28 8588–8596. [DOI] [PubMed] [Google Scholar]
- Castresana, J., Muga, A., and Arrondo, J.L.R. 1988. The structure of proteins in aqueous solutions and assessment of triose phosphate isomerase structure by Fourier-transform infrared spectroscopy. Biochem. Biophys. Res. Commun. 152 69–75. [DOI] [PubMed] [Google Scholar]
- Chehin, R., Thorolfsson, M., Knappskog, P.A., Martínez, A., Flatmark, T., Arrondo, J.L.R., and Muga, A. 1998. Domain structure and stability of human phenylalanine hydroxylase inferred from infrared spectroscopy. FEBS Lett. 422 225–230. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A. and Barton, C.J. 1999. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins Struct. Funct. Genet. 34 508–519. [DOI] [PubMed] [Google Scholar]
- Díaz, E., López, R., and García, J.L. 1992. Ej-1 a temperate bacteriophage of Streptococcus pneumoniae with a Myrividae morphotype. J. Bacteriol. 174 5516–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, R.D.B. and MacRae, T.P. 1973. Infrared spectrometry. In Conformation in fibrous proteins and related synthetic polypeptides, pp. 94–125. Academic Press, New York.
- Freire, E., van Osdoll, W.W., Mayorga, O., and Sánchez-Ruíz, J.M. 1990. Calorimetrically determined dynamics of complex unfolding transitions in proteins. Annu. Rev. Biophys. Chem. 19 159–188. [DOI] [PubMed] [Google Scholar]
- Freskgard, P.-O., Martensson, L.-G., Jonhasson, P., Jonsson, B.-H., and Carlsson, U. 1994. Assignment of the contribution of the tryptophan residues to the circular dichroism spectrum of hyman carbonic anhydrase II. Biochemistry 33 14281–14288. [DOI] [PubMed] [Google Scholar]
- García, P., García, J.L., García, E., Sánchez-Puelles, J.M., and López, R. 1990. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene 86 81–88. [DOI] [PubMed] [Google Scholar]
- García, J.L., Sánchez-Beato, A.R., Medrano, F.J., and Lopez, R. 1998. Versatility of the choline binding domain. Microb. Drug. Res. 4 25–36. [DOI] [PubMed] [Google Scholar]
- Goormaghtight, E., Cabiaux, V., and Rysschaert, J.-M. 1994. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. III Secondary structures. Subcell. Biochem. 23 405–450. [DOI] [PubMed] [Google Scholar]
- Guillespie, S.H., Mchugh, T.D., Ayres, H., Dickens, A., Efstratiou, A., and Whiting, G.C. 1997. Allelic variation in Streptococcus pneumonia autolysin (N-Acetyl-muramoyl-L-alanine amidase). Infect. Immun. 65 3936–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Laue, T.M., Shah, B.D., Ridgeway, T.M., and Pelletier, S.L. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical ultracentrifugation in biochemistry & polymer science (eds. S.E. Harding, H.C. Horton, and A.J. Rowe), pp. 90–125. Royal Society of Chemistry, London.
- López, R., García, E., García, P., and García, J.L. 1995. Architecture and domain interchange of pneumococcal cell wall lytic enzymes. Dev. Biol. Stand. 85 273–281. [PubMed] [Google Scholar]
- López, R., González, M.P., García, E., García, J.L., and García, P. 2000. Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol. 151 437–443. [DOI] [PubMed] [Google Scholar]
- Martínez, A., Haavik, J., Flatmark, T., Arrondo, J.L.R., and Muga, A. 1996. Conformational properties and stability of tyrosine hydroxylase studied by infrared spectroscopy. J. Biol. Chem. 271 19737–19742. [DOI] [PubMed] [Google Scholar]
- Medrano, F.J., Gasset, M., López-Zumel, C., Usobiaga, P., García, J.L., and Menéndez, M. 1996. Structural characterization of the unligated and choline-bound forms of the major pneumococcal autolysin Lyta amidase. J. Biol. Chem. 271 29152–29161. [DOI] [PubMed] [Google Scholar]
- Minton, A.P. 1994. Conservation of signal: A new algorithm for the elimination of the reference concentration as an independently variable parameter in the analysis of sedimentation equilibrium. In Modern analytical ultracentrifugation (eds. T.H. Schuster and T.H. Lave), pp. 81–92. Birkhaüser, Boston.
- Perczel, A., Park, K., and Fasman, G.D. 1992. Analysis of the circular dichroism spectrum of proteins using the convex constraint algorithm: A practical guide. Anal. Biochem. 203 83–93. [DOI] [PubMed] [Google Scholar]
- Petrelski, S.J., Byler, D.M., and Liebman, M.N. 1991. Comparison of various molecular forms of bovine trypsin: Correlation of infrared spectra with X-ray crystal structures. Biochemistry 30 133–143. [DOI] [PubMed] [Google Scholar]
- Rost, B. and Sander, C. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins Struct. Funct. Genet 19 55–72. [DOI] [PubMed] [Google Scholar]
- Sánchez-Ruíz, J.M. 1995. Differential scanning calorimetry of proteins. Subcell. Biochem. 24 133–176. [DOI] [PubMed] [Google Scholar]
- Sanz, J.M., López, R., and García, J.L. 1988. Structural requirements of choline derivatives for "conversion" of pneumococcal amidase. FEBS Lett. 232 308–312. [DOI] [PubMed] [Google Scholar]
- Surewicz, W.K. and Mantsch, H.H. 1988. New inside into protein secondary structure from resolution-enhanced infrared spectra. Biophys. Biochim. Acta 953 225–130. [DOI] [PubMed] [Google Scholar]
- Susi, H. 1969. Infrared spectra of biological macromolecules and related systems. In Structure and stability of biological macromolecules (eds. S.N. Timasheff and L. Stevens), pp. 575–663. Dekker, New York.
- Tomasz, A. 1984. Building and breaking of bonds in the cell wall of bacteria: The role for autolysin. In Microbial cell wall sinthesis and autolysis (ed. C. Nombela), pp. 3–12. Elsevier, Amsterdam.
- Tuomanen, E., Pollack, H., Parkinson, A., Davidson, M., Fackland R., Rich, R., and Zak, O. 1988. Microbiological and clinical significance of a new property of defective lysis in clinical strains of pneumococci. J. Infect. Dis. 158 36–43. [DOI] [PubMed] [Google Scholar]
- Usobiaga, P., Medrano, F.J., Gasset, M., García, J.L., Saíz, J.L., Rivas, G., Laynez, J., and Menéndez, M. 1996. Structural organization of the major autolysin from Streptococcus pneumoniae. J. Biol. Chem. 271 6832–6838. [DOI] [PubMed] [Google Scholar]
- Varea, J., Saiz, J.L., López-Zumel, C., Monterroso, B., Medrano, F.J., Arrondo, J.L., Iloro, I., Laynez, J., García, J.L., and Menéndez, M. 2000. Do sequence repeats play an equivalent role in the choline-binding module of pneumococcal LytA amidase? J. Biol. Chem. 275 26842–26855. [DOI] [PubMed] [Google Scholar]
- Whatmore, A. and Dowson, C. 1999. G. The autolysin-encoding gene (lytA) of Streptococcus pneumonia displays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell-wall lytic enzymes. Infect. Inmun. 67 4551–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore, A.M., Efstratiou A., Pickerill, A.P., Broughton, K., Woodard, G., Sturgeon, D., George, R., and Dowson, C.G. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Inmun. 68 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder, C.L., Friedrich, A.D., Potls, R.O., Daumy, G.O., and Fracoeur, M.L. 1992. Secondary structural analysis of two recombinant murine proteins, interleukins 1 alpha and 1 beta: Is infrared spectroscopy sufficient to assign structure? Biochemistry 31 27–31. [DOI] [PubMed] [Google Scholar]