Abstract
ADP-ribosyltransferases including toxins secreted by Vibrio cholera, Pseudomonas aerurginosa, and other pathogenic bacteria inactivate the function of human target proteins by attaching ADP-ribose onto a critical amino acid residue. Cross-species polymerase chain reaction (PCR) and database mining identified the orthologs of these ADP-ribosylating toxins in humans and the mouse. The human genome contains four functional toxin-related ADP-ribosyltransferase genes (ARTs) and two related intron-containing pseudogenes; the mouse has six functional orthologs. The human and mouse ART genes map to chromosomal regions with conserved linkage synteny. The individual ART genes reveal highly restricted expression patterns, which are largely conserved in humans and the mouse. We confirmed the predicted extracellular location of the ART proteins by expressing recombinant ARTs in insect cells. Two human and four mouse ARTs contain the active site motif (R-S-EXE) typical of arginine-specific ADP-ribosyltransferases and exhibit the predicted enzyme activities. Two other human ARTs and their murine orthologues deviate in the active site motif and lack detectable enzyme activity. Conceivably, these ARTs may have acquired a new specificity or function. The position-sensitive iterative database search program PSI-BLAST connected the mammalian ARTs with most known bacterial ADP-ribosylating toxins. In contrast, no related open reading frames occur in the four completed genomes of lower eucaryotes (yeast, worm, fly, and mustard weed). Interestingly, these organisms also lack genes for ADP-ribosylhydrolases, the enzymes that reverse protein ADP-ribosylation. This suggests that the two enzyme families that catalyze reversible mono-ADP-ribosylation either were lost from the genomes of these nonchordata eucaryotes or were subject to horizontal gene transfer between kingdoms.
Keywords: ADP-ribosylation, recombinant proteins, PSI-BLAST, orthologous genes, paralogous gene, cross-species PCR, database searches
The purpose of this study was to identify all recognizable human and mouse members of the family of toxin-related mono-ADP-ribosyltransferases; to clone, sequence, and chromosomally map their genes and cDNAs; to express the gene products as recombinant proteins and assay their enzyme activities; and to assess structural and sequence similarities between bacterial and mammalian ADP-ribosyltransferases.
ADP-ribosylation is an enzyme-catalyzed post-translational protein modification in which the ADP-ribose moiety is transferred from NAD+ to a specific amino acid in a target protein while the nicotinamide moiety is released (Althaus et al. 1985; Jacobson and Jacobson 1989; Aktories 1991; Haag and Koch-Nolte 1997; Rappuoli and Montecucco 1997; Smith 2001). This stereospecific reaction is catalyzed by mono- and poly-ADP-ribosyltransferases (mARTs and pARTs). mARTs catalyze the transfer of a single ADP-ribose moiety onto a specific amino acid side chain of a target protein; pARTs (also designated poly-ADP-ribose-polymerases or PARPs), additionally can catalyze the elongation and branching of ADP-ribose units on ADP-ribosylated targets. In the absence of target proteins, some ARTs also exhibit NAD-glycohydrolysis (NADase) activity. The mART subfamily includes many well-known bacterial toxins as well as a number of mammalian and avian ecto-enzymes. Members of the pART subfamily have been identified in a broad spectrum of eucaryotes, including man, Drosophila, Arabidopsis, but not in yeast or procaryotes.
Most known mARTs transfer ADP-ribose onto arginine residues. Arginine-specific ARTs include cholera toxin and Escherichia coli heat-labile enterotoxin, which target the alpha-subunit of heterotrimeric G-proteins; C2 toxin of Clostridium botulinum, VIP2 of Bacillus cereus, and SpvB of Salmonella entericae, all of which target actin; ALT of T4-bacteriophage, which targets E. coli RNA-polymerase; DRAT of Rhodospirillum rubrum, which targets dinitrogenase reductase; ExoS of Pseudomonas aeruginosa, which targets ras; and mouse ART1 and ART2, which target integrins and other cell surface proteins (Aktories 1991; Ludden 1994; Haag and Koch-Nolte 1997; Rappuoli and Montecucco 1997; Okazaki and Moss 1998; Han et al. 1999; Otto et al. 2000). Additionally, mARTs have been identified, which specifically target the side chains of other amino acid residues, including diphthamide in elongation factor 2 (diphtheria toxin and pseudomonas exotoxin A); cysteine in inhibitory G-proteins (pertussis toxin); asparagine in rho and related small GTP binding proteins (C3 exoenzymes of Clostridium botulinum and Staphylococcus aureus) (Aktories 1991; Wilson and Collier 1992; Barbieri 2000; Wilde et al. 2001). In contrast, pARTs (e.g., PARP, tankyrase) target glutamic acid in histones, telomerase binding protein, and others (Burkle 2001; Smith 2001).
Like phosphorylation, ADP-ribosylation is a reversible post-translational modification that can be used as a mechanism to regulate endogenous protein functions. A reversible ADP-ribosylation cycle was first corroborated in the photosynthetic bacterium Rhodospirillum rubrum, in which nitrogen fixation is regulated by an mART-mediated ADP-ribosylation of the key enzyme dinitrogenase reductase (Ludden 1994). An ADP-ribosylhydrolase (ARH) reverses ADP-ribosylation of dinitrogenase reductase. (This ARH is designated dinitrogenase reductase ADP-ribosylarginine glycohydrolase or DRAG). A homolog of this enzyme, designated ADP-ribosylarginine hydrolase (ARH), has been identified in humans and mice (Moss et al. 1992).
ARTs show remarkable plasticity in amino acid sequences, a feature that hampers the in silico identification of distant gene family members (Domenighini and Rappuoli 1996; Koch-Nolte et al. 1996a; Bazan and Koch-Nolte 1997; Okazaki and Moss 1998). The crystal structures of eight bacterial toxins and of chicken PARP uncovered common features of the NAD-binding pocket: a unique topology of six conserved β-strands and an α-helix form the upper and lower jaws of a Pacman-like active site crevice (Choe et al. 1992; Ruf et al. 1996; Han et al. 1999, 2001). Only a single amino acid residue, the catalytic glutamic acid residue in the fifth β-strand, is strictly conserved in all structures (Carrol and Collier 1984). Two other conserved residues in the first and second β-strand (arginine and serine in most mARTs, histidine and tyrosine in the two diphthamide-specific mARTs, and in all known pARTs have been used to divide the ART family into two subgroups, i.e., the R-S-E motif group and the H-Y-E motif group) (Domenighini and Rappuoli 1996; Koch-Nolte et al. 1996a; Bazan and Koch-Nolte 1997). A second glutamic acid, two residues upstream of the catalytic E in the loop connecting the fourth and fifth β-strand, is found in all arginine-specific ARTs, by far the largest subfamily of known ARTs (R-S-EXE).
Recent improvements in database search tools, in particular the development of the iterative, position-sensitive PSI-BLAST program, have made it possible to identify distant protein family members even when they share only very limited amino acid sequence identities (Altschul and Koonin 1998). PSI-BLAST generates a search matrix on the basis of a multiple sequence alignment in which conserved residues are given a higher weight than nonconserved residues. Here, we describe the use of this tool to provide evidence that mammalian mARTs indeed are related to ADP-ribosylating toxins. Moreover, the results uncover remarkable "holes" in the phylogeny of mARTs in the completed genomes of yeast, worm, fly, and mustard weed. Remarkably, this distribution pattern is mirrored by similar "holes" in the phylogeny of the ARH family.
Results
Identification, cloning, and sequencing of human and mouse ART/Art genes
To identify and clone the members of the gene family of toxin-related mARTs from the human and the mouse, we used a combination of strategies. First, we derived degenerate PCR primers corresponding to conserved regions of known mammalian ART1–ART5 and used these for cross-species “Zoo”-PCR analyses. A representative result is shown in Figure 1 ▶. PCR products of the expected size were generated from genomic DNAs of most organisms analyzed. These were then subcloned and sequenced. The results revealed that in the vast majority of cases, the PCR products derived from the respective ART ortholog. Thereby, we obtained fragments of hitherto unknown murine Art3 and Art4 starting from the known sequences of human ART3 and ART4 (Koch-Nolte et al. 1997). Similarly, starting from the known sequence of mouse Art5, we obtained a fragment of the hitherto unknown human ART5 gene (Okazaki et al. 1996; Glowacki et al. 2001). Moreover, we thus obtained respective ART sequences from several other mammalian species. Occasionally, cloned Zoo-PCR products contained, in addition to the ART-ortholog, a fragment derived from an evident ART-paralog. These always corresponded to one of the other known ARTs (ART1–5). Full-length cDNA and genomic clones were then obtained for all known human and mouse ART genes by classic cloning techniques (Figs. 2, 3 ▶ ▶).
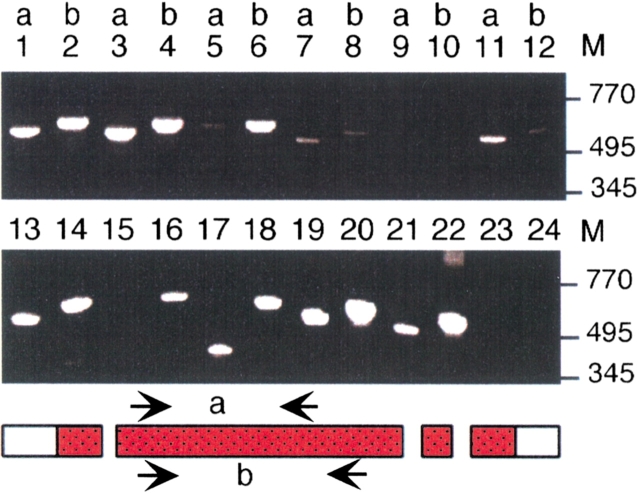
Fig. 1.
Cross-species "Zoo"-PCR analyses of ART5. Genomic DNAs were subjected to PCR-amplification with two pairs of ART5-specific primers (a and b) ending in codons for conserved amino acids. Reaction products were size fractionated by agarose gel electrophoresis and visualized by staining with ethidium bromide. Genomic DNAs were from: rhesus monkey: lanes 1–2, macaque monkey: lanes 3– 4, pig: lanes 5–6, rat: lanes 7–8, control (H2O): lanes 9–10, mouse: lanes 11–12, human: lanes 13–14, tree shrew: lanes 15–16, house shrew: lanes 17–18, cow: lanes 19–20, sheep: lanes 21–22, chicken: lanes 23–24. M = size of marker fragments in base pairs.
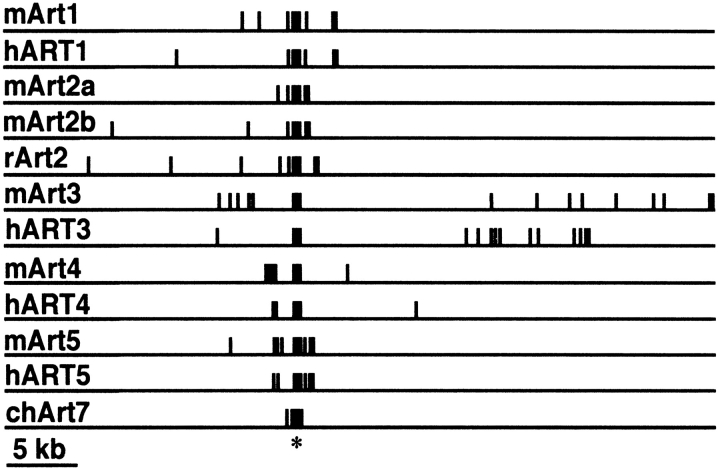
Fig. 2.
Schematic diagram of ART gene exon/intron structures. Exons and flanking introns were sequenced from respective P1 and PAC genomic DNA clones. Exons are depicted as closed boxes, introns and flanking regions as lines. Genes were aligned with respect to the major exon encoding the catalytic domain (*). The sizes of exons and introns are shown in Figure 4 ▶.
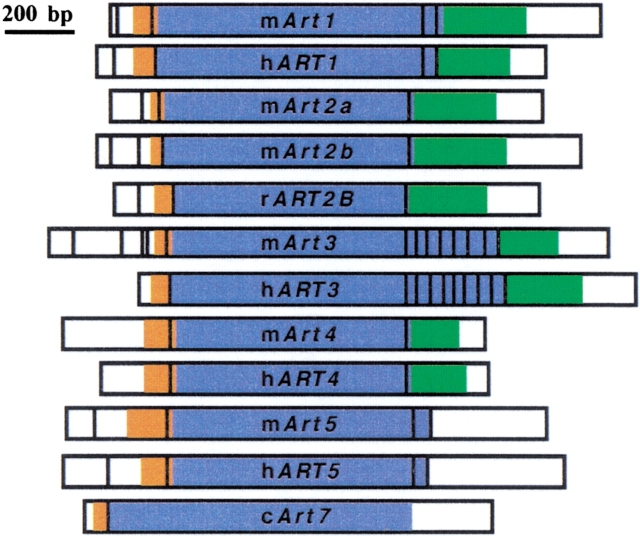
Fig. 3.
Schematic diagrams of the ART cDNA exon compositions. The major splice variant is depicted for each ART gene transcript. Noncoding 5` and 3` untranslated regions are white, coding regions are color coded: leader peptide: orange, native peptide: blue, GPI-anchor signal: green.
To determine whether the human or other mammalian genomes contain additional ART gene family members, we next used the deduced full-length human and mouse ART1-ART5 amino acid sequences as input for database searches (Table 1). The tBLASTn program was used to search nucleotide databases, and PSI-BLAST to search protein databases. tBLASTn can detect significant amino acid sequence homologies (>25% sequence identity) encoded in nucleotide sequences. PSI-BLAST can detect much more distantly related sequences (<5% amino acid sequence identity), but is restricted to the database of protein sequences (Altschul and Koonin 1998). Previous studies (Braren et al. 1997; Koch-Nolte et al. 1997) have demonstrated that tBLASTn can successfully detect paralogous mammalian ART-sequences (e.g., ART3, 4, and 5 using ART1 and ART2 as input). This is facilitated by the fact that a single large exon typically encodes most of the native protein of mammalian ARTs (Fig. 3 ▶).
Table 1.
Chromosomal localization and dbEST representation of the human and mouse ART and ARH gene families
| Human | Mouse | |||||
| gb | ESTs | Band | gb | ESTs | cM | |
| ART1 | S74683 | 1 | 11p15 | X95825 | 49 | 7.49 |
| ART2A | X65060 | 0 | 11q13 | X52991 | 3 | 7.48 |
| ART2B | na | 0 | 11q13 | X87612 | 1 | 7.48 |
| ART3 | X95827 | 40 | 4q13.1–q13.3 | AJ311769 | 82 | 5.51 |
| ART4 | X95826 | 12 | 12p12.2–p13.1 | Y08300 | 3 | 6.70 |
| ART5 | Y16836 | 6 | 11p15 | Y08028 | 12 | 7.49 |
| ARH1 | L13291 | 1 | 3q13.31–33 | L13290 | 145 | na |
| ARH2 | Al333260 | 23 | 13q31 | Al503324 | 24 | na |
| ARH3 | AJ313333 | 135 | 1p34.1–35.3 | AW209835 | 50 | na |
| GAPD | M33197 | 7541 | 12p13 | M32599 | 1854 | 6 |
| HPRT | M31642 | 120 | Xq26.1 | J00432 | 276 | X |
The number of matching expressed sequence tags (ESTs) in the public database (release June 2001) is given for each gene. Chromosomal localizations are shown as cytogenetic bands for the human and as distances from the centromere in cM for the mouse.
tBLASTn searches of the recently completed human genome sequence (Lander et al. 2001) revealed only a single hitherto unknown pseudogene (designated ART2BP) in addition to the already identified ART2 pseudogene (Haag et al. 1994) and four functional human ART gene family members (ART1, ART3, ART4, and ART5) (Okazaki et al. 1994; Koch-Nolte et al. 1997) (Table 1). The mouse has functional orthologs for each of these genes (designated Art1, Art2a, Art2b, Art3, Art4, and Art5). tBLASTn searches of the EST database identified one or more EST derived from each of the functional murine and human ART genes, but did not reveal any other related paralogue (Table 1). Similarly, PSI-BLAST searches did not uncover any other mammalian ART-paralogs in the protein database, but did connected the mammalian ARTs with ADP-ribosylating bacterial toxins (see below).
Chromosomal localization and exon/intron structures of mouse and human Art/ART genes
We determined the chromosomal localization of human ART genes by PCR screening of human–mouse somatic cell hybrids and confirmed the assignments by sequence similarity searches of the completed human genome sequence (Lander et al. 2001). To map the mouse Art genes we first identified an informative allelic polymorphism for each Art gene and then determined its distribution in a panel of genomic DNAs from a backcross of (C57BL/6J × Mus spretus) × Mus spretus (Table 1). All ART orthologs mapped to regions of conserved linkage synteny. Interestingly, ART1 and ART5 are arranged in a “head-to-head” orientation in close proximity in both the mouse (Glowacki et al. 2001) and humans.
The exon/intron structures of all human and mouse ART genes were determined by comparative sequencing of full-length cDNA and genomic DNA clones (Fig. 2 ▶). In each case, a single long exon encodes most of the native protein, whereas separate exons encode the N- and C-terminal signal peptides (Fig. 4 ▶). ART2 and ART4 do not contain any additional coding exons. ART1 and ART5 contain one additional small exon encoding the C-terminal end of the native protein. ART3 contains several small exons in this region. The 5` untranslated region is split into several distinct exons in most ART genes (Fig. 4 ▶). These results confirm and markedly extend previous findings on the gene structures of mouse Art1 (Braren et al. 1998), rat ART2 (Haag et al. 1996), and mouse Art5 (Glowacki et al. 2001).
Fig. 4.
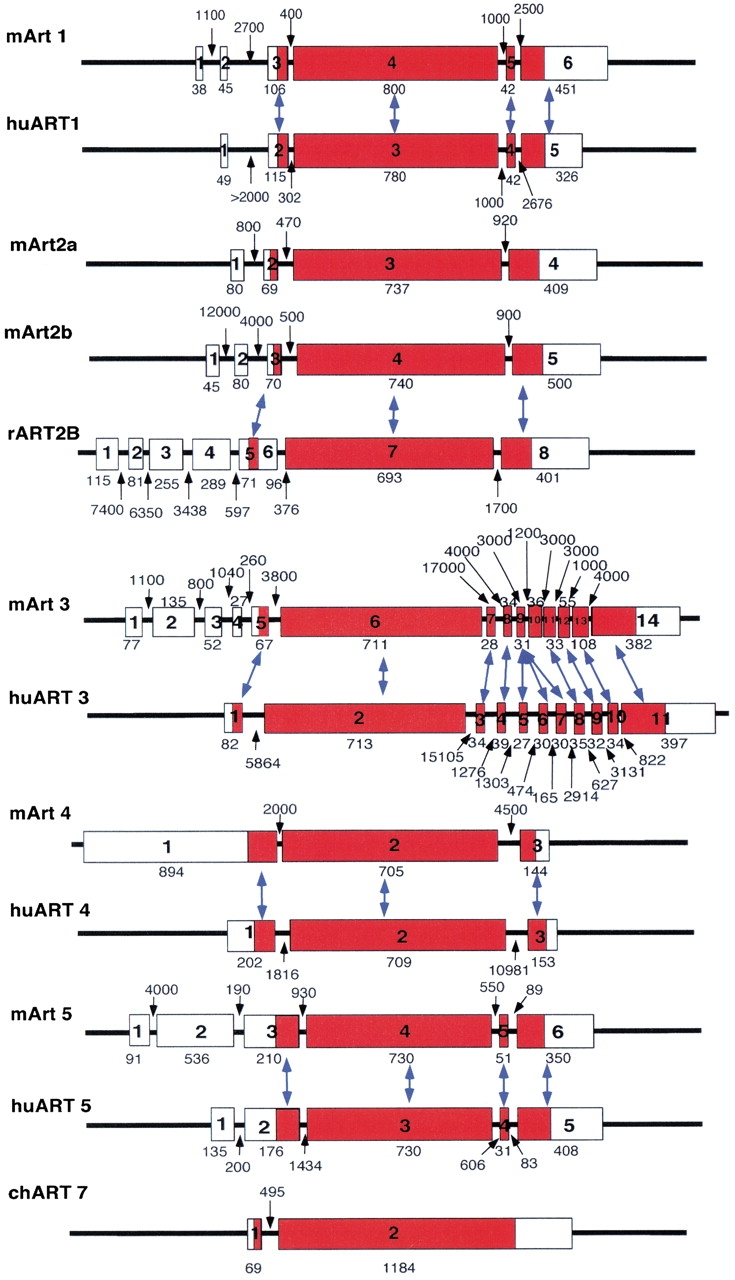
Exon-intron structures of human and mouse ART genes. Exons are depicted as boxes, introns as lines. Numbers indicate lengths of exons and introns in base pairs. Corresponding exons in ART orthologs are connected by arrows. Note that a small exon in the 3` coding region of human ART3 is triplicated with respect to its mouse Art3.
Common and distinctive features of the deduced ART amino acid sequences
All ARTs contain hydrophobic N-terminal peptides that exhibit high signal sequence probabilities (Figs. 5, 6 ▶ ▶). The predicted extracellular localization was confirmed by expression cloning in insect cells (see below). All human and mouse ARTs except for ART5 end in a second prominent stretch of hydrophobic amino acids, a characteristic feature of GPI-anchored membrane proteins. Four cysteine residues are strictly conserved in all mammalian ART proteins. These are also found in the three known avian ecto-ARTs. Structure-based alignments of mammalian ARTs and bacterial ARTs of known structure indicate that the second and third cysteine residues lie in close proximity, and therefore, probably form a disulfide bond. Human and mouse ART1, ART3, and ART5 each contain an extra pair of cysteine residues near the C-terminal end. The positions of these residues are conserved in the respective species orthologs but not among paralogs. Human and mouse ART1, ART5, and mouse ART2.1, ART2.2 contain residues corresponding to the R-S-EXE motif of arginine specific mARTs. ART3 and ART4 of both the human and the mouse show nonconservative amino acid deviations in this motif (Figs. 5, 6 ▶ ▶).
Fig. 5.
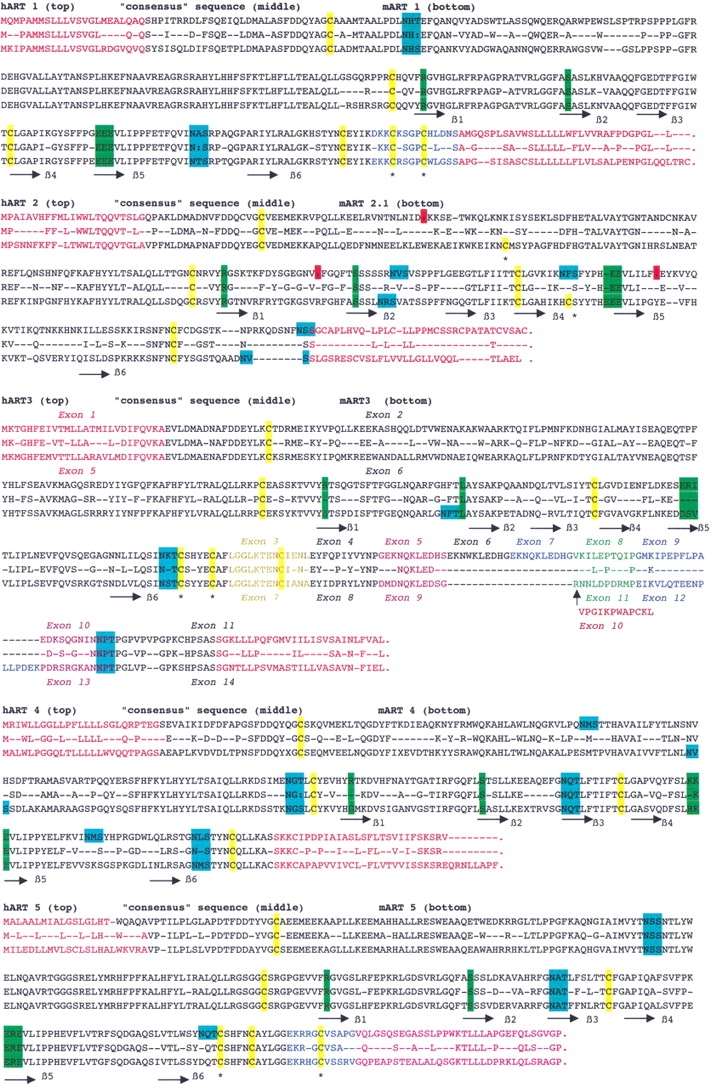
Amino acid sequence alignments of orthologs ARTs. Alignments of the deduced amino acid sequences of each pair of ART-orthologs are shown with the human sequence on top and mouse sequence on bottom. Conserved residues are shown in the middle of each alignment. Predicted secondary structure units lining the six conserved β strands of the active site crevice are marked below the alignment. Different colors are used for amino acids encoded by different exons. The four cysteine residues common to all ARTs are highlighted in yellow, additional ART-specific pairs of cysteine are marked by asterisks. Potential asparagine-linked glycosylation sites are highlighted in blue. Residues corresponding to the R-S-EXE motif of arginine-specific ARTs are marked in green. Three premature stop codons of human ART2AP are marked in red.
Fig. 6.
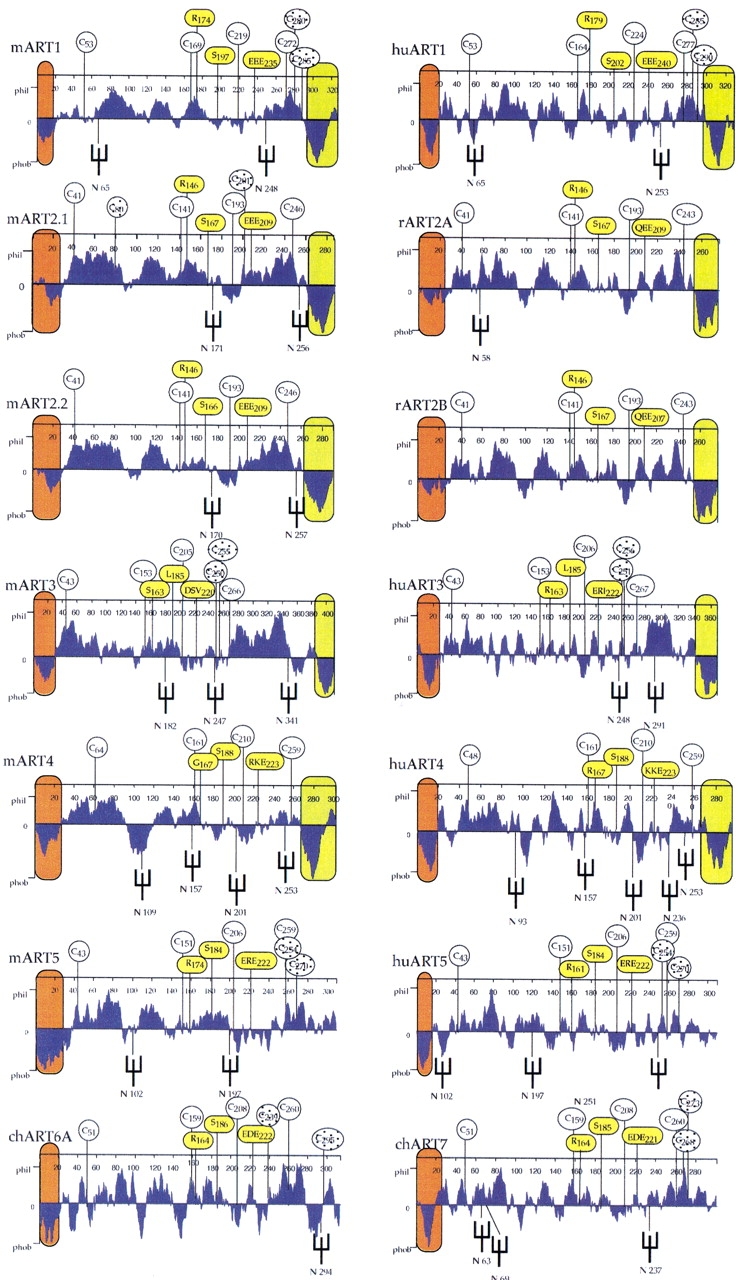
Hydropathy profiles of human and mouse ART proteins. Hydropathy profiles were calculated with a window setting of 19 amino acid residues. Putative hydrophobic amino- and carboxy-terminal signal peptides are colored orange and green, respectively. Positions of potential asparagine-linked oligosaccharide side chains are marked by forks. The four cysteine residues conserved in all mammalian ARTs are marked by circles with numbers indicating their respective positions from the amino terminus. Additional cysteine residues found only in individual ARTs are marked by shaded circles. Residues corresponding to the R-S-EXE motif of arginine specific mARTs are marked by yellow circles.
Tissue-specific expression patterns of ART genes
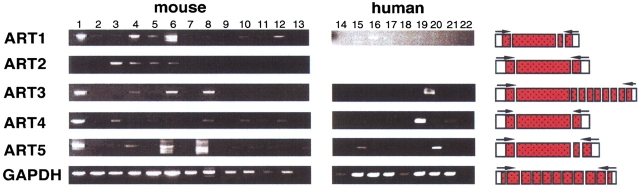
To determine whether human and mouse ART genes are expressed ubiquitously or in a tissue-restricted manner, we performed semiquantitative reverse transcription PCR (RT-PCR) (Fig. 7 ▶) and serial Northern blot analyses (Fig. 8 ▶). The results reveal restricted expression patterns for the individual ART genes. ART1 is most prominently expressed in skeletal muscle, ART2 in T cells, ART3 and ART5 in testis. ART4 shows a broader pattern of expression with prominent signals in heart, lung, liver, and spleen. Tissue specificity is largely conserved in the human and mouse.
Fig. 7.
Reverse transcription PCR analyses of ART gene expression. Panels of mouse and human cDNAs were subjected to reverse transcription PCR analyses using primers from separate exons as illustrated schematically. Products after 31 cycles were size fractionated by agarose gel electrophoresis and visualized by ethidium bromide staining. cDNAs were from: heart: lane 1, brain: lane 2, spleen: lane 3, lung: lane 4, liver: lane 5, skeletal muscle: lane 6, kidney: lane 7, testis: lane 8, 7-day embryo: lane 9, 11-day embryo: lane 10, 15-day embryo: lane 11, 17-day embryo: lane 12, control (H2O): lane 13, colon: lane 14, ovary: lane 15, peripheral blood lymphocytes: lane 16, prostate: lane 17, small intestine: lane 18, spleen: lane 19, testis: lane 20, thymus: lane 21, and control (H2O): lane 22.
Fig. 8.
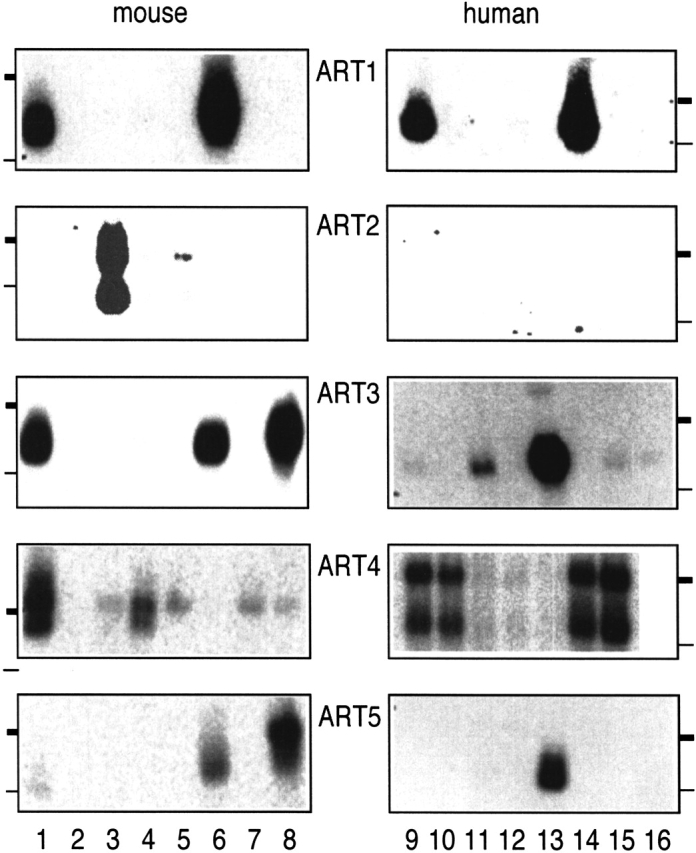
Northern blot analyses of human and mouse ART gene expression. Northern blots with mouse and human RNAs were hybridized with radiolabeled ART-specific probes, and bound activity was visualized by autoradiography. Thick and thin bars indicate the migration positions of 2.4kb and 1.35kb marker fragments, respectively. Mouse RNAs were from heart: lane 1; brain: 2; spleen: 3; lung: 4; liver: 5; skeletal muscle: 6; kidney: 7; and testis: 8. Human RNAs for ART1 and ART2 were from heart: lane 9; brain: 10; placenta: 11; lung: 12; liver: 13; skeletal muscle: 14; kidney: 15; and pancreas: 16; for ART3 and ART5, pancreas: lane 9; adrenal medulla: 10; thyroid: 11; adrenal cortex: 12; testis: 13; thymus: 14; small intestine: 15; and stomach: 16; and for ART4, spleen: lane 9; lymph node: 10; thymus: 11; appendix: 12; peripheral blood leukocytes: 13; bone marrow: 14; and fetal liver: 15.
Recombinant expression and enzymatic analysis of epitope-tagged soluble ARTs
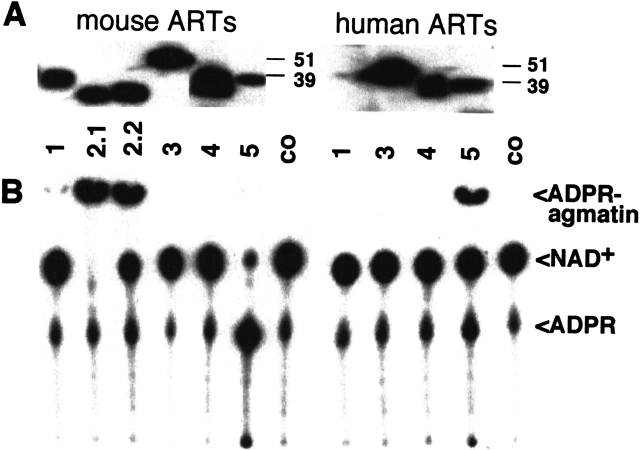
We designed insect cell expression constructs to test the function of the putative N-terminal signal sequences. In case of ART1–ART4, we deleted the GPI anchor signal sequence to obtain soluble proteins. Engineered C-terminal epitope tags allowed their detection and purification. Indeed, the ART proteins were efficiently produced and secreted by transfected insect cells (Fig. 9A ▶). Using established enzyme assays (Haag et al. 1995; Koch-Nolte et al. 1996b; Braren et al. 1998) we demonstrated potent arginine-specific ADP-ribosylation activity for human and mouse ART1, mouse ART2.1 and ART2.2, and human ART5 (Fig. 9B ▶). Mouse ART5 resembles rat ART2 (Haag et al. 1995) in showing potent NAD-glycohydrolase activity. Neither mouse nor human ART3 and ART4 showed any detectable enzyme activities in these assays.
Fig. 9.
Comparative enzyme assays of purified epitope-tagged ARTs. ARTs were immunoprecipitated from the supernatants of respective baculovirus-infected insect cells with immobilized anti-FLAG monoclonal antibody M2. Purified ARTs were incubated with 32P-NAD+ for 60 min at 37°C in the presence of 2 mM agmatine (an arginine analog). Proteins were analyzed by SDS-PAGE and Western blot analyses using the anti-FLAG antibody (A). Note that human ART1 was not produced efficiently in this system (A, lane 7). Enzyme reaction products were analyzed by thin-layer chromatography followed by autoradiography (B). Co: control precipitates from mock infected cells.
The deduced ART amino acid sequences exhibit conserved structural motifs and amino acid sequence similarities with ADP-ribosylating toxins
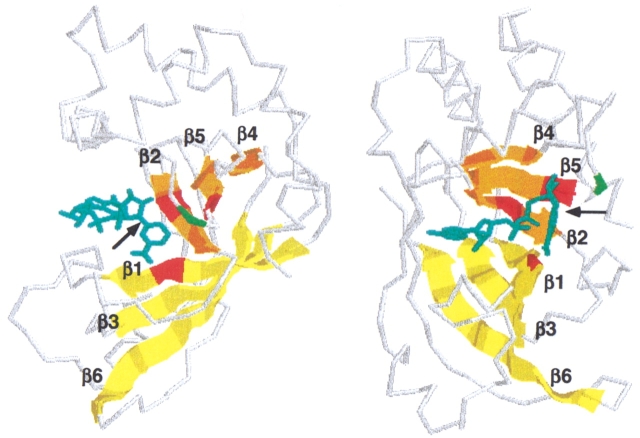
To assess possible structural and sequence similarities between mammalian and bacterial mARTs we used secondary structure prediction and position sensitive database search programs (PHDsec and PSI-BLAST) (Rost and Sander 1993; Altschul and Koonin 1998). The N-terminal portion of mammalian ARTs is predicted to be mainly helical; the C terminal region is dominated by β-sheets. This pattern of secondary structure units is very similar to those of the catalytic domains in the recently crystallized ADP-ribosylating bacterial toxins VIP2 of Bacillus cereus and C3 of Clostridium botulinum (Han et al. 1999, 2001) (Fig. 10 ▶). Moreover, PSI-BLAST connected mammalian mARTs with most known bacterial mARTs (Table 2). The overall sequence identities between vertebrate and bacterial mARTs are <10%, that is, deep in the “twilight zone” of traditional search programs. Significantly, the matches detected by PSI-BLAST cluster in regions corresponding to the active site crevice in VIP2, C3, and other bacterial toxins of known 3D structures (Figs. 10, 11 ▶ ▶).
Fig. 10.
Matches detected by PSI-BLAST cluster in regions corresponding to the active site crevice of VIP2. Regions shown in the multiple sequence alignment (Fig. 11 ▶) are projected onto the backbone of Bacillus cereus VIP2 (Han et al. 1999) with bound NAD (cyan). The two images generated by rasmac using pdb file 1QS2 are rotated by 90° to provide side (left) and front (right) views of the active site crevice. The arrow points to the cleaved bond in NAD. The six conserved β-strands of the lower and upper jaws of the active site crevice are depicted as cartoons and are colored yellow (β4, β5, β2) and orange (β1, β3, β6), respectively. The three residues of the R-S-E motif are in red, the upstream glutamic acid of arginine-specific mARTs is in green.
Table 2.
Tiling paths of PSI-BLAST searches with mARTs and ARHs
| i | *ART1/H.s. | i | VIP2/B.c. | i | *ARH3/H.s. |
| 1 | ART2/M.m. | 1 | C2/C.b. | 1 | *ARH1/H.s. |
| *ART3/H.s. | iota/C.p. | *ORF/A.f | |||
| *ART4/H.s. | C3/C.b.ph | *ORF/M.j. | |||
| ART5/M.m. | *EDIN/S.a. | *ORF/A.a | |||
| ART6/G.g | ToxA/C.d. | *ORF/E.c. | |||
| ART7/G.g | *ORF/B.h. | ORF/S.c. | |||
| 2 | *exoS/P.a. | *C3/S.a. | *ORF/M.l. | ||
| *exoT/P.a. | 2 | *exoS/P.a | 2 | DRAG/R.r. | |
| 3 | C2/C.b. | *exoT/P.a. | DRAG/A.b. | ||
| 4 | *SpvB/S.t.P. | *SpvB/S.t.P. | ORF/Y.e. | ||
| VIP2/B.c. | *ORF/S.p. | ORF/L.m. | |||
| *ORF/B.h. | 3 | ALT/E.c.ph | *ORF/D.r. | ||
| 5 | iota/C.p. | ORF/V.p. | J1/T.c. | ||
| C3/C.b. | 4 | ART7/G..s. | 3 | converged | |
| *C3/S.a. | 5 | *ART1/H.s. | |||
| *ORF/S.p. | ART2/M.m | i | DRAG/R.r. | ||
| 6 | *EDIN/S.a. | ART5/M.m. | 1 | DRAG/A.b. | |
| ORF/V.p. | ART6/G.g. | *ORF/M.j. | |||
| 7 | ALT/E.c.ph | ART7/G.g. | *ORF/A.f | ||
| ToxA/C.d. | 6 | *ART3/H.s. | *ORF/A.a | ||
| 8 | converged | *ORF/Ch.V. | *ORF/E.c. | ||
| 7 | *CT/V.c. | ORF/S.c. | |||
| i | *CT/V.c. | *LT/E.c.P. | *ORF/M.l. | ||
| 1 | *LT/E.c.P. | 8 | DRAT/R.r. | 2 | *ARH1/H.s. |
| 2 | PT/B.p. | DRAT/A.b. | *ARH3/H.s. | ||
| 3 | *MTX/B.s. | 9 | converged | ORF/Y.e. | |
| pier/P.r. | ORF/L.m. | ||||
| ORF/S.c. | i | modA/E.c.ph. | *ORF/D.r. | ||
| 4 | *exoS/P.a | 1 | modB/E.c.ph. | J1/T.c. | |
| *exoT/P.a. | 2 | converged | 3 | converged | |
| 5 | converged | ||||
| i | HvnA/V.f. | ||||
| i | *ETA/P.a. | 1 | HvnB/V.f. | ||
| 1 | DT/C.d.ph | *ORF/P.a. | |||
| 2 | converged | 2 | converged |
PSI-BLAST searches of the nonredundant public database (June 2001) were initiated with the sequences indicated on top. Searches were repeated in successive iterations using a matrix in which conserved residues are given a higher weight. Converged indicates that no more significant matches were detected. A single ortholog is listed for genes that have been cloned from more than one species. Matches are listed with the protein abbreviation (ORF = open reading frame) followed by abbreviations for species designation. Asterisks mark species with completed genome sequences.
Fig. 11.

Multiple sequence alignment of the catalytic cores of vertebrate and bacterial mARTs. Multiple amino sequence alignment of the region corresponding to the six conserved β-strands of the active site crevice (see Fig. 10 ▶). Nonconserved residues in connecting loops are not shown and are indicated by numbers. Known secondary structure units in the crystallized mARTs are underlined (C3, VIP2, CT, LT, PT, DT, ETA, and PARP). Color coding as in (A). Note that the last β strand in VIP2, C3, and related ARTs (marked by asterisks) has been displaced by a strand in opposite orientation relative to that in LT, PARP, and others (i.e., β5 and β6 are parallel in VIP2, whereas they are antiparallel in CT).
PSI-BLAST analyses reveal similar "holes" in the phylogenies of ARTs and ARHs
Remarkably, none of the four other recently elucidated eucaryotic genomes (those of a yeast, a worm, a fly, and a plant) contains any predicted open reading frame as or more closely related to the mammalian ARTs as the bacterial toxins (Table 3). We wondered whether comparable "holes" in phylogeny also occurred in the ARH family, that is, the enyzmes reversing ADP-ribosylation. Indeed, bacterial and human ARHs were connected by PSI-BLAST, whereas no significant matches were detected in the other four completed eucaryotic genomes. Of note, we discovered two hitherto unknown human ARH gene family members (designated ARH2 and ARH3), both of which are represented also in dbEST (see Table 1).
Table 3.
Phylogenetic distribution of ART and ARH genes in completed and unfinished genomes
| Viruses (>500) | Bacteria (33) | Archaea (10) | Eucaria (4) | Vertebr (1) | |
| ARTs in finished genomes | Ch.V./1 | P.a./2 | 0 | 0 | H.s./5 |
| V.c./1 | |||||
| S.a./2 | |||||
| B.h./1 | |||||
| B.s./1 | |||||
| S.p./1 | |||||
| E.c.P./1 | |||||
| S.t.P./1 | |||||
| ARTs in other genomes | E.c.ph/1 | B.c./1 | 0 | P.r./1 | M.m./6 |
| C.b.ph/1 | C.b./1 | G.g./3 | |||
| C.p./1 | |||||
| V.p./1 | |||||
| R.r./1 | |||||
| A.b./1 | |||||
| S.c./1 | |||||
| Unconnected ARTs | C.d.ph/1 | P.a./2 | 0 | 0 | 0 |
| E.c.ph/2 | V.f./2 | ||||
| ARHs in finished genomes | 0 | A.a./1 | M.j./1 | 0 | H.s./3 |
| B.h./1 | A.f./1 | ||||
| D.r./1 | |||||
| E.c./1 | |||||
| ARHs in other genomes | 0 | R.r./1 | 0 | T.c./3 | M.m./3 |
| A.b./1 | |||||
| Y.e./1 | |||||
| L.m./1 | |||||
| S.c./7 | |||||
| pARTs | Ci.V./1 | 0 | 0 | C.e./2 | H.s./14 |
| D.m./3 | M.m./14 | ||||
| A.t./8 | |||||
| pARGs | Ha.V./1 | 0 | 0 | C.e./1 | H.s./1 |
| D.m./1 | M.m./1 | ||||
| A.t./1 |
The number of finished genome sequences for each phylogenetic class is shown on top. Species abbreviations are followed by the number of recognizable respective gene family members. Abbreviations in Tables 2 and 3: Vertebrate: G.g. Gallus gallus, H.s. Homo sapiens, M.m. Mus musculus, Nonvertebrate eucaryotes: A.t. Arabidopsis thaliana, C.e. Caenorhabditis elegans, D.m. Drosophila melanogaster, P.r. Piris rapae, T.c. Tripedalia cystophora, Archaea: A.f. Archaeoglobus fulgidus, M.j. Methanococcus jannaschii, Bacteria: A.b. Azospirillum brasilense, A.a. Aquifex aeolicus, B.c. Bacillus cereus, B.h. Bacillus halodurans, B.s. Bacillus subtilis, B.p.Borditella pertussis, C.b. Clostridium botulinum, C.d. Clostridium difficile, C.p. Clostridium perfringens, D.r. Deinococcus radiodurans, E.c. Escherichia coli, L.m. Listeria monocytogenes, M.l. Mesorhizobium loti, P.a. Pseudomonas aeruginosa, S.a. Staphylococcus aureus, S.c. Streptomyces coelicolor, S.p. Streptococcus pyogenes, R.r. Rhodospirillum rubrum, V.c. Vibrio cholera, V.f. Vibrio fischeri, V.p. Vibrio parahaemolyticus, Y.e. Yersinia enterocolitica, Plasmids: E.c.P. Escherichia coli phage; C.b.ph. Clostridium botulinum phage; C.d.ph. Corynebacterium diphtheria phage. Ch.V. Paramecium bursaria Chlorella virus, Ci.V. Chilo iridescent virus, Ha.V. Helicoverpazea nuclear polyhedrosis virus.
Discussion
In this study, we identified the human and mouse members of the mART and ARH gene families (Table 1). The presence of respective orthologs in many mammalian species indicates that these genes were generated by duplication events at or before the mammalian radiation. Most ART genes exhibit a tightly restricted expression pattern and relatively low transcript levels (Figs. 7, 8 ▶ ▶), possibly reflecting the regulatory function of the ART gene products.
Our enzyme assays with recombinant ARTs confirmed arginine-specific transferase activity for all human and mouse ARTs containing the R-S-EXE motif (Figs. 9, 10 ▶ ▶). ART3 and ART4 lack this motif, and do not display any detectable arginine-specific enzyme activity, indicating that these family members may have acquired a different target specificity or lost enzyme acitivity altogether. In this context, it is interesting to point out the following precedents from procaryotic mARTs:
Some enzymatically inactive ART domains, for example, the N-terminal domains of C2 and VIP2 toxins, have acquired a new, protein-binding function (Han et al. 1999, 2001). Other examples for the acquisition of novel functions by inactivated enzyme family members include caspase-related FLIP (Tschopp et al. 1998) and lens crystallins (Piatigorsky 1998).
Some mARTs, for example, diphtheria toxin, exhibit a narrow target specificity that would not have been detected with the assays applied here (Wilson and Collier 1992).
The enzyme activities of some mARTs, for example, cholera toxin and ExoS, become apparent only after binding to an activator (Rappuoli and Montecucco 1997; Barbieri 2000).
Further studies are required to resolve whether any of these explanations also apply to ART3 and ART4. Note, however, that we did not detect any enzyme activity above background upon adding recombinant ART3 and ART4 proteins to lysates from various cell lines (not shown).
The results of our database searches and structure prediction analyses strongly support an evolutionary relationship of mammalian and bacterial ARTs as well as of mammalian and bacterial ARHs (Fig. 6 ▶; Tables 2, 3). Interestingly, both enzyme families evidently are missing in the four fully sequenced genomes and expressed sequence tags of lower eucaryotes. A similar phylogenetic distribution pattern has been observed for 41 human genes, mostly coding for enzymes (Andersson et al. 2001; Lander et al. 2001). This can be explained either by loss of the respective genes from these nonchordata eucaryotes or by horizontal transfer of the respective genes from bacteria to the vertebrate (or prevertebrate) lineage enzymes (Andersson et al. 2001; Lander et al. 2001). It is of interest to note that many bacterial ARTs are encoded by mobile genetic elements, including bacteriophages (ALT, C3 exoenzyme, diphtheria toxin), pathogenicity island (cholera toxin), and plasmid (SpvB), which could facilitate horizontal gene transfer.
Only four open reading frames of unknown function from bacterial genomes showed significant sequence identity to the mART family (Table 2). The likely significance of these matches is underscored by our recent demonstration that one of these, the SpvB virulence factor of Salmonella, indeed, is an actin-specific mART (Otto et al. 2000; Tezcan-Merdol et al. 2001). By analogy, the other mART-related proteins encoded by these open reading frames are candidate virulence factors (Pallen et al. 2001).
Note that PSI-BLAST failed to connect two small groups of known bacterial mARTs to the R-S-E family (Vibrio fischerii halovibrin alpha and beta, E. coli phage modA and modB; Table 2), whereas structure threading programs (Firestine et al. 1996) reveal that these, indeed, contain an R-S-E motif (Bazan and Koch-Nolte 1997; Wilkens et al. 1997). Evidently the sequences of these mARTs (and the lengths of their connecting loops between conserved β-strands) have diverged too far to be connected with PSI-BLAST. The same holds for the two diphtamidespecific H-Y-E mARTs of Corynebacterium diphtheria and Pseudomonas aeruginosa, and for the large H-Y-E subgroup of pARTs. Hence, it is possible that other orphan open reading frames in the genome databases encode unidentified ARTs. Indeed, a recent study using PSI-BLASTS to search databases of unfinished procaryotic genomes uncovered several additional putative bacterial ARTs (Pallen et al. 2001). It is much less likely that family members were missed among orphan open reading frames in the case of ARHs, for which sequence identities between pro- and eucaryotic family members are much higher than for ARTs.
Several organisms evidently encode both ARTs and ARHs, implicating the presence of endogenous ADP-ribosylation cycles as demonstrated in photosynthetic bacteria (Ludden 1994). Others apparently contain only one of these enzymes (Table 3). In the sole presence of ARTs these often function as virulence factors. Conversely, endogenous ARHs might protect against the deleterious effects of pathogen-encoded ART. For example, it is conceivable that the endogenous ARH of E. coli serves to protect against phage-encoded mARTs (e.g., mod and ALT).
Last, but not least, the results of this study pave the way for future investigations. Art knock-out mice are currently being generated in our laboratories to further elucidate the role of these interesting exoenzymes. Moreover, the recombinant ARTs will facilitate the development of ART inhibitors and of ART-specific antibodies. These hold promise as novel tools for therapeutic interventions.
Materials and methods
Isolation and sequencing of cDNA and genomic DNA
ART-specific primers used in this study are listed in Table 4 (“f” and “rrdquo; in primer names indicate forward or reverse orientation of the primer). Human and mouse cDNAs were subjected to 5` and 3` rapid amplification of cDNA ends (RACE) according to the manufacturer's (Clontech) protocol. Genomic DNAs were prepared as described previously (Koch et al. 1990; Koch-Nolte et al. 1993, 1997). IMAGE consortium ESTs and P1, PAC, and BAC genomic DNA clones were obtained from Genome Systems, and the Resource Center of the German Genome Project (rzpd). Genomic DNA libraries were screened by PCR with ART-specific primers or by hybridization with ART-specific probes. Exon-containing fragments were generated by restriction digestion or PCR amplification, subcloned into plasmids (pBluescript, Stratagene, and pCR2.1, Invitrogen) and sequenced. Sequences obtained from PCR products were confirmed by sequencing clones obtained from two separate PCR reactions.
Table 4.
ART-specific primers used in this study
| Primer | Application | Gene | Location | Nucleotide sequence |
| M14f | 3`RACE | mArt1 | exon 4 | GAGGACACCTTCTTCGGTATC |
| T27f | 3`RACE | mArt3 | exon 9 | CAACCCAGATATGGACAATCAGAAGC |
| T25f | 3`RACE | mArt3 | exon 11 | CAACCTTGACCCTGACAGAATGCC |
| S02f | 3`RACE | mArt4 | exon 2 | GGCCAGTTCCTCTCTGCCTCCCT |
| S05f | 3`RACE | mArt4 | exon 3 | GTGGTTATCGTCTGTCTCTTTTTGG |
| N02f | 3`RACE | mArt5 | exon 4 | ACTCTCTGGAGTTATGATCAGACCTG |
| N03f | 3`RACE | mArt5 | exon 6 | GCTGCAGCTCTCCAGAGCTGGACC |
| T41r | 5`RACE | mArt3 | exon 6 | ACTCCAAAGCAGGTCTGGATGGTTAGCAC |
| M33r | 5`RACE | mArt1 | exon 4 | GGCAGGGCCGCTGTCATG |
| T46r | 5`RACE | mArt3 | exon 5 | CTTCACCTGGAAGATGTCCATTAGAAC |
| S34r | 5`RACE | mArt4 | exon 1 | CCTCCTGGAAGCCACAGCGCCAT |
| S35f | 5`RACE | mArt4 | exon 1 | GCATGCARGGGAATGCCAAGGGT |
| N07r | 5`RACE | mArt5 | exon 2 | CGCTAGTTAAGTCGCCAAATCTGTC |
| N10r | 5`RACE | mArt5 | exon 3 | TGGAGGGGAGTGCTGGCTTGG |
| M18f | MTC | mArt1 | exon 1 | CAGCTTTGCCGCCATGGAGAAGGC |
| M16f | MTC | mArt1 | exon 4 | CATGACAGCGGCCCTGCC |
| M90r | MTC | mArt1 | exon 6 | GCCTGGTACTACCACTCATACC |
| M15f | MTC | hART1 | exon 1 | GGACAAGGCCTAGATGAGG |
| T23f | MTC | mArt3 | exon 5 | ATGAAGATGGGACATTTTGAAATGGTCAC |
| T47r | MTC | mArt3 | exon 14 | TCTGGACTTCCTGTGGGATCCC |
| S03f | MTC | mArt4 | exon 1 | GATGGCGCTGTGGCTTCCAGGAGG |
| S99r | MTC | mArt4 | exon 3 | AGCAGCTCCTTTAAAAAGGAGCCAG |
| L00f | MTC | hART4 | exon 1 | CCTCCTGCAACGATGAGAATCTGGCTCC |
| L99r | MTC | hART4 | exon 3 | GGAGCCACAAGATTTCTTTATACTCTGC |
| N00f | MTC | mArt5 | exon 3 | AGGATGATTCTGGAGGATCTGCTGATG |
| N99r | MTC | mArt5 | exon 6 | CTGCTTCCTGCAGCCGTTCAAAGCCC |
| B11f | Zoo-PCR | mArt1 | exon 3 | CTTTGATGACCAGTACGAGGGCTG |
| B32r | Zoo-PCR | mArt1 | exon 3 | CATAGCCTGGGATCAGCACCTCCTC |
| T21f | Zoo-PCR | mArt3 | exon 6 | GTGCTAACCATCCAGACCTGCTTTGGAGT |
| T42r | Zoo-PCR | mArt3 | exon 6 | CCGAGGAATGCGCACTCGTAGTAGCTGCAGGT |
| L11f | Zoo-PCR | mArt4 | exon 2 | TCTTTTGATGATCAGTACCAAGGCTG |
| L42r | Zoo-PCR | mArt4 | exon 2 | AACTTTAAACAGCTCATAGGGAGG |
| N11f | Zoo-PCR | mArt5 | exon 4 | GATACCTTTGATGATGCCTATGTGGGCTG |
| N41r | Zoo-PCR | mArt5 | exon 4 | GACAACGCCTGGATAGGAGCCCCAAAACA |
| TT01f | chr.mapping | mArt3 | intron | CACGCACTCACATACACAATGTGCC |
| TT31r | chr.mapping | mArt3 | intron | TGATATGATGCCTTCTGGCCTCACC |
| S21f | chr.mapping | mArt4 | intron | CTCCTGAGACAGGGACAGAGAATCAAG |
| S51r | chr.mapping | mArt4 | intron | TTTCTAGGTACTAGGGGGTCAGACTG |
Data deposition requirements: The sequences described here have been deposited in the EMBL database (see Table 1 for accession numbers).
Cross-species “Zoo” PCR
Primers for “Zoordquo; PCR were designed from regions where known ARTs showed strong sequence similarity, for example, primers ended in codons for a conserved cysteine or the catalytic glutamic acid residue. PCR reactions were carried out with genomic DNAs (100–500 ng) from different mammalian species using “hot start” and “touch-down” techniques with Amplitaq Gold polymerase (Stratagene). The initial denaturation was for 8 min at 96°C, followed by 33 cycles for 20 sec at 95°C. The annealing temperature was decreased successively in 5° intervals from 70 to 35°C in the first eight cycles followed by 25 cycles at 65°C. Annealings were performed for 20 sec, extensions for 60 sec (at 72°C), with a final extension of 8 min.
Chromosomal mapping, Southern and Northern blot analyses
Map positions for human ART genes were determined by PCR screening of genomic DNAs from rodent/human somatic cell hybrids as described previously (Koch-Nolte et al. 1993, 1996a). Map positions were confirmed by BLASTn analyses of the recently published human genome sequence (Lander et al. 2001). Mouse Art genes were mapped by PCR screening (Art3, Art4) or by Southern blot analyses (Art1, Art2a, Art2b, Art5) of a panel of 94 genomic DNAs from the (C57BL/6JEi × SPRET/Ei) × SPRET/Ei (BSS) backcross panel available from The Jackson Laboratory Mapping Panel Resource (Rowe et al. 1994). PCR primers flanking informative polymorphic microsatellite repeats in Art3 and Art4 in both cases yielded ∼200 bp C57BL/6J products versus slightly smaller SPRET/Ei products. Informative restriction fragment length variants (RFLVs) used for mapping the other Art genes were: BamH1 RFLVs for Art1, Art2a, and Art3 (6.7, 3.7, and 3.0 kb SPRET/Ei fragments, respectively, versus slight smaller fragments in C57BL/6J), and a Taq1 RFLV for Art5 (C57BL/6J ∼1.7 kb, SPRET/Ei ∼2.0 kb). Southern and Northern blot analyses were performed as described previously (Koch et al. 1990; Koch-Nolte et al. 1997; Braren et al. 1998).
Expression of recombinant ART proteins and enzyme assays
Baculovirus constructs for producing soluble ARTs in which the hydrophobic C-terminal GPI-anchor signal sequence were replaced by tandem His6x and FLAG epitope tags were prepared as described previously (Koch-Nolte et al. 1996b). Recombinant ARTs were purified from insect cell supernatants by affinity chromatography on Talon-columns (Clontech) or Sepharose-immobilized anti-FLAG M2 monoclonal antibody (Sigma), and were incubated in 50 μL enzyme buffer (20 mM Tris pH 8.0, 1 mM ADP-ribose, 5 mM DTT, 2 mM agmatine, 5 μM 32P-NAD+, 0.5 μCi, Amersham-Pharmacia) for 60 min at 37°C. Proteins were analyzed by SDS-PAGE and Western blot analyses; soluble reaction products by thin layer chromatography as described previously (Haag et al. 1995; Braren et al. 1998).
Database searches and sequence deposition
The tBLASTn program was used to screen the human genome sequence (Lander et al. 2001) and the database of expressed sequence tags (dbEST) (Marra et al. 1998). The PSI-BLAST program was used to screen the nonredundant protein database for sequences related to known ARTs and ARHs (blast@ncbi.nlm.nih.gov) (Altschul and Koonin 1998). PSI-BLAST searches were initiated with the BLOSUM62 Matrix; gap penalties were set at 11 (Existence) and 1 (Extension). For the ART family, searches were intitiated with a threshold setting of 0.01; at higher iterations the threshold level was reset manually to a value between 0.1 and 4.0 to include known ARTs and to exclude irrelevant matches. For the ARH family, searches were performed with a threshold setting of 0.01 for all iterations. The sequences described here have been deposited in the EMBL database (see Table 1 for accession numbers).
Amino acid sequence alignment and secondary structure prediction analyses
Multiple sequence alignments were performed with PSI-BLAST, ClustalW, and with a weighted dynamic programming method (HSSP/MaxHom). The generated multiple alignments were used as input for secondary structure predictions that were produced by profile-based neural network systems (PHDsec) (Rost and Sander 1993) (PredictProtein@EMBL-Heidelberg.de). Hydropathy profiles were generated on a Macintosh with the MacMolly software (Softgene) by using the Kyte-Doolittle algorithm and a window setting of 19 amino acid residues. Signal peptide cleavage sites were predicted with the Signal P program (www.cbs.dtu.dk/services/SignalP/).
Acknowledgments
This work was supported by grant No310/3 from the Deutsche Forschungsgemeinschaft to F.K.N., and NIH DK36175 and DK27722 and the Juvenile Diabetes Research Foundation, International to E.H.L. DNAX is fully supported by Schering-Plough. We thank K. Bartels, R. Girisch, and M. Bridget for excellent technical assistance, and H.G. Thiele and S. Rothenburg for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ARH, ADP-ribosylhydrolase
GAPD, glyceraldehyde-3-phosphate dehydrogenase
GPI, glycosylphosphatidylinositol
HPRT, hypoxanthine phosphoribosyltransferase
mART, mono(ADP-ribosyl)transferase
NAD+, nicotine adenine dinucleotide
PBS, phosphate-buffered saline
pART, poly(ADP-ribosyl)transferase
PCR, polymerase chain reaction
RACE, rapid amplification of cDNA ends
RT, reverse transcription
RFLV. restriction fragment length variant
SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis
utr, untranslated region
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0200602.
References
- Aktories, K. 1991. ADP-ribosylating toxins. Springer Verlag, Berlin.
- Althaus, F.R., Hilz, H., and Shall, S. 1985. ADP-ribosylation of proteins. Springer Verlag, Berlin.
- Altschul, S.F. and Koonin, E.V. 1998. Iterated profile searches with PSI-BLAST—A tool for discovery in protein databases. Trends Biochem. Sci. 23 444–447. [DOI] [PubMed] [Google Scholar]
- Andersson, J.O., Doolittle, W.F., and Nesbo, C.L. 2001. Genomics. Are there bugs in our genome? Science 292 1848–1850. [DOI] [PubMed] [Google Scholar]
- Barbieri, J.T. 2000. Pseudomonas aeruginosa exoenzyme S, a bifunctional type-III secreted cytotoxin. Int. J. Med. Microbiol. 290 381–387. [DOI] [PubMed] [Google Scholar]
- Bazan, J.F. and Koch-Nolte, F. 1997. Sequence and structural links between distant ADP-ribosyltransferase families. Adv. Exp. Med. Biol. 419 99–107. [DOI] [PubMed] [Google Scholar]
- Braren, R., Firner, K., Balasubramanian, S., Bazan, F., Thiele, H.G., Haag, F., and Koch-Nolte, F. 1997. Use of the EST database resource to identify and clone novel mono(ADP-ribosyl)transferase gene family members. Adv. Exp. Med. Biol. 419 163–168. [DOI] [PubMed] [Google Scholar]
- Braren, R., Glowacki, G., Nissen, M., Haag, F., and Koch-Nolte, F. 1998. Molecular characterization and expression of the gene for mouse NAD+:arginine ecto-mono(ADP-ribosyl)transferase, Art1. Biochem. J. 336 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle, A. 2001. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 163 1–5. [DOI] [PubMed] [Google Scholar]
- Carrol, S.F. and Collier, R.J. 1984. NAD+ binding site of diphtheria toxin: Identification of a residue within the nicotinamide subsite by photochemical modification with NAD. Proc. Natl. Acad. Sci. 81 3307–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Bennett, M.J., Fujii, G., Curmi, P.M., Kantardjieff, K.A., Collier, R.J., and Eisenberg, D. 1992. The crystal structure of diphtheria toxin. Nature 357 216–222. [DOI] [PubMed] [Google Scholar]
- Domenighini, M. and Rappuoli, R. 1996. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 21 667–674. [DOI] [PubMed] [Google Scholar]
- Firestine, S.M., Nixon, A.E., and Benkovic, S.J. 1996. Threading your way to protein function. Chem. Biol. 3 779–783. [DOI] [PubMed] [Google Scholar]
- Glowacki, G., Braren, R., Cetkovic-Cvrlje, M., Leiter, E.H., Haag, F., and Koch-Nolte, F. 2001. Structure, chromosomal localization, and expression of the gene for mouse ecto-mono(ADP-ribosyl)transferase ART5. Gene 275 267–277. [DOI] [PubMed] [Google Scholar]
- Haag, F., Andresen, V., Karsten, S., Koch-Nolte, F., and Thiele, H. 1995. Both allelic forms of the rat T cell differentiation marker RT6 display nicotinamide adenine dinucleotide (NAD)-glycohydrolase activity, yet only RT6.2 is capable of automodification upon incubation with NAD. Eur. J. Immunol. 25 2355–2361. [DOI] [PubMed] [Google Scholar]
- Haag, F. and Koch-Nolte, F. 1997. ADP-ribosylation in animal tissues: Structure, function and biology of mono(ADP-ribosyl)transferases and related enzymes. Plenum Press, New York. [DOI] [PubMed]
- Haag, F., Koch-Nolte, F., Kühl, M., Lorenzen, S., and Thiele, H.G. 1994. Premature stop codons inactivate the RT6 genes of the human and chimpanzee species. J. Mol. Biol. 243 537–546. [DOI] [PubMed] [Google Scholar]
- Haag, F., Kuhlenbäumer, G., Koch-Nolte, F., Wingender, E., and Thiele, H.G. 1996. Structure of the gene encoding the rat T cell ecto-ADP-ribosyltransferase RT6. J. Immunol. 157 2022–2030. [PubMed] [Google Scholar]
- Han, S., Arvai, A.S., Clancy, S.B., and Tainer, J.A. 2001. Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: Structural insights for recognition specificity and catalysis. J. Mol. Biol. 305 95–107. [DOI] [PubMed] [Google Scholar]
- Han, S., Craig, J.A., Putnam, C.D., Carozzi, N.B., and Tainer, J.A. 1999. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 6 932–936. [DOI] [PubMed] [Google Scholar]
- Jacobson, M.K. and Jacobson, E.L. 1989. ADP-ribose transfer reactions: Mechanisms and biological significance. Springer Verlag, New York.
- Koch, F., Haag, F., Kashan, A., and Thiele, H.G. 1990. Primary structure of rat RT6.2, a nonglycosylated phosphatidylinositol-linked surface marker of postthymic T cells. Proc. Natl. Acad. Sci. 87 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch-Nolte, F., Haag, F., Braren, R., Kuhl, M., Hoovers, J., Balasubramanian, S., Bazan, F., and Thiele, H.G. 1997. Two novel human members of an emerging mammalian gene family related to mono-ADP-ribosylating bacterial toxins. Genomics 39 370–376. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte, F., Haag, F., Kühl, M., van Heyningen, V., Hoovers, J., Grzeschik, K. H., Singh, S., and Thiele, H.G. 1993. Assignment of the human RT6 gene to 11q13 by PCR screening of somatic cell hybrids and in situ hybridization. Genomics 18 404–406. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte, F., Kühl, M., Haag, F., Cetkovic-Cvrlje, M., Leiter, E.H., and Thiele, H.G. 1996a. Assignment of the human and mouse genes for muscle ecto mono (ADPribosyl)transferase to a conserved linkage group on human chromosome 11p15 and mouse chromosome 7. Genomics 36 215–216. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte, F., Petersen, D., Balasubramanian, S., Haag, F., Kahlke, D., Willer, T., Kastelein, R., Bazan, F., and Thiele, H.G. 1996b. Mouse T cell membrane proteins Rt6–1 and Rt6–2 are arginine/protein mono(ADPribosyl)transferases and share secondary structure motifs with ADP-ribosylating bacterial toxins. J. Biol. Chem. 271 7686–7693. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Ludden, P.W. 1994. Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol. Cell. Biochem. 138 123–129. [DOI] [PubMed] [Google Scholar]
- Marra, M.A., Hillier, L., and Waterston, R.H. 1998. Expressed sequence tags—ESTablishing bridges between genomes. Trends. Genet. 14 4–7. [DOI] [PubMed] [Google Scholar]
- Moss, J., Stanley, S.J., Nightingale, M.S., Murtagh, J.J., Monaco, L., Mishima, K., Chen, H.C., Williamson, K.C., and Tsai, S.C. 1992. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J. Biol. Chem. 267 10481–10488. [PubMed] [Google Scholar]
- Okazaki, I.J. and Moss, J. 1998. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J. Biol. Chem. 273 23617–23620. [DOI] [PubMed] [Google Scholar]
- Okazaki, I.J., Kim, H.J., and Moss, J. 1996. Cloning and characterization of a novel membrane-associated lymphocyte NAD:arginine ADP-ribosyltransferase. J. Biol. Chem. 271 22052–22057. [DOI] [PubMed] [Google Scholar]
- Okazaki, I.J., Zolkiewska, A., Nightingale, M.S., and Moss, J. 1994. Immunological and structural conservation of mammalian skeletal muscle glycosylphosphatidylinositol-linked ADP-ribosyltransferases. Biochemistry 33 12828–12836. [DOI] [PubMed] [Google Scholar]
- Otto, H., Tezcan-Merdol, D., Girisch, R., Haag, F., Rhen, M., and Koch-Nolte, F. 2000. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol. Microbiol. 37 1106–1115. [DOI] [PubMed] [Google Scholar]
- Pallen, M.J., Lam, A.C., Loman, N.J., and McBride, A. 2001. An abundance of bacterial ADP-ribosyltransferases—Implications for the origin of exotoxins and their human homologues. Trends Microbiol. 9 302–307. [DOI] [PubMed] [Google Scholar]
- Piatigorsky, J. 1998. Multifunctional lens crystallins and corneal enzymes. More than meets the eye. Ann. NY Acad. Sci. 842 7–15. [DOI] [PubMed] [Google Scholar]
- Rappuoli, R. and Montecucco, C. 1997. Guidebook to protein toxins and their use in cell biology. Oxford University Press, Oxford.
- Rost, B. and Sander, C. 1993. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. 90 7558–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, L., Nadeau, J., Turner, R., Frankel, W., Letts, V., Eppig, J., Ko, M., Thurston, S., and Birkenmeier, E. 1994. Maps from two interspecific backcross DNA panels as a community mapping resource. Mammal. Genome 5 253–274. [DOI] [PubMed] [Google Scholar]
- Ruf, A., Menissier de Murcia, J., De Murcia, G., and Schulz, G.E. 1996. Structure of the catalytic fragment of poly(ADP-ribose)polymerase from chicken. Proc. Natl. Acad. Sci. 93 7481–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. 2001. The world according to PARP. Trends Biochem. Sci. 26 174–179. [DOI] [PubMed] [Google Scholar]
- Tezcan-Merdol, D., Nyman, T., Lindberg, U., Haag, F., Koch-Nolte, F., and Rhen, M. 2001. Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol. Microbiol. 39 606–619. [DOI] [PubMed] [Google Scholar]
- Tschopp, J., Irmler, M., and Thome, M. 1998. Inhibition of fas death signals by FLIPs. Curr. Opin. Immunol. 10 552–558. [DOI] [PubMed] [Google Scholar]
- Wilde, C., Chhatwal, G.S., Schmalzing, G., Aktories, K., and Just, I. 2001. A novel C3-like ADP-ribosyltransferase from Staphylococcus aureus modifying RhoE and Rnd3. J. Biol. Chem. 276 9537–9542. [DOI] [PubMed] [Google Scholar]
- Wilkens, K., Tiemann, B., Bazan, F., and Ruger, W. 1997. ADP-ribosylation and early transcription regulation by bacteriophage T4. Adv. Exp. Med. Biol. 419 71–82. [DOI] [PubMed] [Google Scholar]
- Wilson, B.A. and Collier, R.J. 1992. Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: Active-site structure and enzymic mechanism. Curr. Top. Microbiol. Immunol. 175 27–41. [DOI] [PubMed] [Google Scholar]