Abstract
There is preliminary experimental evidence indicating that the major outer-membrane protein (MOMP) of Chlamydia is a porin. We tested this hypothesis for the MOMP of the mouse pneumonitis serovar of Chlamydia trachomatis using two secondary structure prediction methods. First, an algorithm that calculates the mean hydrophobicity of one side of putative β-strands predicted the positions of 16 transmembrane segments, a structure common to known porins. Second, outer loops typical of porins were assigned using an artificial neural network trained to predict the topology of bacterial outer-membrane proteins with a predominance of β-strands. A topology model based on these results locates the four variable domains (VDs) of the MOMP on the outer loops and the five constant domains on β-strands and the periplasmic turns. This model is consistent with genetic analysis and immunological and biochemical data that indicate the VDs are surface exposed. Furthermore, it shows significant homology with the consensus porin model of the program FORESST, which contrasts a proposed secondary structure against a data set of 349 proteins of known structure. Analysis of the MOMP of other chlamydial species corroborated our predicted model.
Keywords: Chlamydia, major outer-membrane protein (MOMP), porin, topology model
The genus Chlamydia includes three species that infect humans: Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae (Schachter and Dawson 1978; Moulder et al. 1984; Stephens 1999). Using serological techniques, the 15 major C. trachomatis human serovars are divided into two groups, a C complex that includes serovars A, C, H, I, and J, and the B-complex that includes the B, Ba, D, E, L1, and L2 serovars. Serovars F and G are related to the B-complex, while K and L3 are more related to the C complex than to the B-group (Wang and Grayston 1984). In addition, the C. trachomatis mouse pneumonitis (MoPn) serovar, also known as Chlamydia muridarum (Everett et al. 1999), was isolated from a mouse following inoculation with human tissues (Nigg 1942).
Chlamydia are characterized by a unique developmental cycle involving a metabolically inactive infectious form, the elementary body (EB), that after invading the eukaryotic cell differentiates into a metabolically active form called the reticulate body (RB), which is responsible for intracellular replication (Moulder et al. 1984). The MOMP accounts for 60% of the mass of the outer membrane of the EBs (Caldwell et al. 1981; Hatch et al. 1981). Alignment and comparison of MOMP sequences from all the C. trachomatis serovars and C. psittaci have revealed the existence of five constant and four variable domains (VDs) (Stephens et al. 1987; Fitch et al. 1993).
Preliminary evidence suggests that MOMP may have a porin-like function (Bavoil et al. 1984; Wyllie et al. 1998). Bacterial porins are an unorthodox class among integral membrane proteins because their primary sequences are abundant in charged residues, and no hydrophobic stretches long enough to span the membrane as α-helices can be identified (for review, see Schirmer 1998). Structural studies have shown that bacterial porins are folded into a closed barrel-like β-pleated sheet structure delimiting a pore. Consequently, conventional hydropathy methods that predict membrane-spanning segments based on α-helix structures are inadequate for this class of membrane protein (Jeanteur et al. 1991, 1994).
Here, we have predicted the topology of C. trachomatis MOMP using the hydrophobicity algorithm of Schirmer and Cowan (1993) and an artificial neural network (Diederichs et al. 1998). The combination of both methods, the results of which were in excellent agreement, allowed us to propose the topology of MOMP.
Results
Hydrophobicity algorithm
Applying the β-side hydrophobicity algorithm of Schirmer and Cowan (1993) to the sequence of C. trachomatis MoPn MOMP yielded the plot shown in Figure 1A ▶. This method analyzes the sequence for transmembrane β-strands by computing the mean hydrophobicity of each face of a putative β-strand. Therefore, values for even and odd residues have been plotted in separate curves. Peaks with Hs'≥0.6 are likely to indicate membrane-spanning β-strands. As observed with other porins, they occur in pairs as two strands joined by a short loop at the periplasmic end of the barrel, well separated by long loops that are exposed at the cell surface.
Fig. 1.
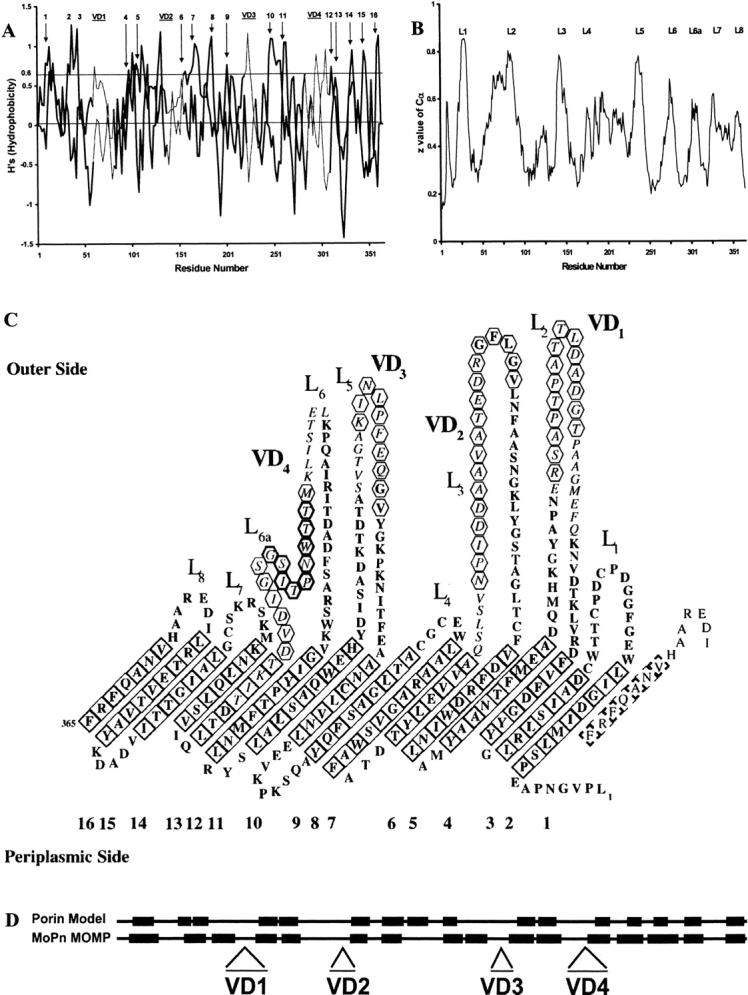
(A) β-side hydrophobicity plot of Chlamydia trachomatis mouse pneumonitis (MoPn) MOMP. Values for even- and odd-numbered residues are plotted as two separate curves. Peaks that have been assigned to transmembrane β-strands in panel C are labeled 1 to 16. The locations of the four variable domains (VD1 through VD4) are indicated by bars at the top of the plot, and the residues corresponding to those regions are joined by thin lines. (B) Topology plot from an artificial neural network program trained on bacterial outer-membrane proteins: predicted z-values are plotted against the C. trachomatis MoPn MOMP amino-acid sequence. The extracellular loops are marked L1 through L8 as assigned in panel C. (C) Predicted topology of the MOMP of C. trachomatis MoPn. The 16-stranded β-barrel has been unrolled and the view is from its outer surface. Italics highlight amino acids in the VDs, while tilted squares indicate residues in transmembrane β-strands, with a bold outline if the side chains are facing away from the pore. Antibodies have been mapped to the segments indicated by hexagons. The conserved peptide in VD4 is indicated by bold hexagons. (The amino-acid sequence is in one-letter code.) (D) Secondary-structure homology using FORESST. The predicted membrane-spanning β-sheets (black boxes) of C. trachomatis MoPn MOMP more closely align with those of the FORESST general porin model after MOMP segments containing the four VDs were deleted.
Very similar plots were obtained for four other chlamydial MOMPs from different species and strains: C. trachomatis, serovars E and L3, C. pneumoniae, strain AR39, and C. psittaci, strain A22/M, which show at least 66% amino-acid identity amongst themselves (data not shown). It can, therefore, be concluded that the predicted topology also is valid for these sequences.
Artificial neural network
To corroborate the above results, we employed an artificial neural network (ANN) that had previously been trained with a set of seven outer-membrane porins with known high-resolution structures (Diederichs et al. 1998). The ANN predicts the z-coordinates of Cα atoms with regard to the outer membrane in the xy-plane. Thus, high z-values indicate long extracellular loops, while low z-values correspond to periplasmic loops. Applying this ANN to MoPn MOMP gave the graph shown in Figure 1B ▶. By design, the ANN will fail to identify short extracellular segments such as loop L4. In MoPn MOMP, the sixth extracellular loop, which corresponds to VD4, contains a nonapeptide conserved among chlamydial species. Because of its proximity to the outer membrane, the ANN identifies this region as a transmembrane segment, nevertheless some immunological studies have shown this conserved region is neither a trans-membrane nor surface exposed. Thus, we propose that this loop is comprised of two well-defined segments protruding outside the cell, termed for clarity L6 and L6a in Figure 1B and 1C ▶, separated by the conserved nonapeptide in close proximity to the outer membrane.
Two-dimensional model
The two-dimensional (2-D) model of Figure 1C ▶ shows the proposed topology of C. trachomatis MoPn MOMP. We used four criteria to assign residues to transmembrane segments: (1) A β-side hydrophobicity index Hs'>0.6; (2) Seven to nine residues are needed to span the core of the membrane with every second residue being hydrophobic and thus, positive and negative Hs' alternate; (3) A z-value Cα between 0.5 and 0.3 given by the ANN; and (4) Segments with high sequence variability (VD) are excluded by negative inference based on immunological, biochemical, and evolutionary evidence, as discussed below.
The model depicts the protein as an unrolled β-barrel viewed from the outside. The 16 β-sheet membrane-spanning segments are represented by tilted squares and account for approximately one-third of all the amino-acid residues. The residues facing away from the pore are represented in bold, while nonbold, tilted squares indicate the residues with side chains that face the pore. All of the residues facing away from the pore are hydrophobic or aromatic, with the exception of D34, D108, H244, D311, and K322. The model also reveals an abundance of aromatic residues along the upper and lower ends of the β-strands, located at the junction between hydrophobic and hydrophilic environments. The barrel edge in contact with the periplasmic space exhibits a pattern of alternating Phe and Tyr. Aliphatic residues in the middle of the β-barrel form a nonpolar ribbon in contact with the membrane lipids. The side of the MOMP facing the outside of the cell predominantly exhibits long loops, while the turns facing the periplasmic space are short. The external loops contain numerous charged side chains, mostly Asp, and to a lesser extent, Glu.
Analysis using the FORESST program
The FORESST program found MOMP to more closely fit its porin model than other possible structural models. The homology between the porin consensus model and our predicted secondary structure (Fig. 1D ▶) was statistically significant (Z = 3.30). The primary difference between the two was that the MOMP contained additional residues in the VDs. These segments are on the large external loops in our model (Fig. 1C ▶). When we shortened the length of the large loops by removing the VDs, the FORESST Z-score increased from 3.30 to 4.04. This supports the hypothesis that MOMP is a porin and that the VDs lie on large external loops.
Discussion
The structural characterization of membrane proteins from microorganisms has provided unique insights into their pathogenesis and has aided in the development of therapeutic and preventive measures (Destenaves and Thomas 2000; Klebe 2000). In the case of the genus Chlamydia, the MOMP is considered to be a major target for the development of vaccines and therapeutic strategies (Caldwell and Perry 1982; Ward 1992; Fitch et al. 1993; de la Maza and de la Maza 1995; Brunham et al. 2000; Stephens, 2000). In contrast to other gram-negative bacteria, peptidoglycan has not been found in the outer membrane of Chlamydia (Moulder et al. 1984). To account for this observation, it has been proposed that the MOMP, along with the 60 kDa and the 12 kDa cysteine-rich proteins, forms a supramolecular structure supported by disulfide bonds that may be responsible for the integrity of the outer membrane (Bavoil et al. 1984; Raulston 1995; Hatch 1996; Wyllie et al. 1998). This supramolecular structure should provide enough plasticity for the structural changes that occur from the time the organism has the compact conformation of the infectious 300 nm EB, to the metabolically active RB that can have a diameter up to 1500 nm. MOMP probably functions as a porin during replication of the RB (Bavoil et al. 1984). However, the MOMP also may act as an adhesin (Su et al. 1990), both at the time the EB infects the host cell and when the RB adheres to the inclusion membrane to interact with the host mitochondria and endoplasmic reticulum (Peterson and de la Maza 1988; Hackstadt et al. 1995).
Crystallization of membrane proteins poses significant technical challenges and, as a result, only a handful of tertiary structures have been solved (Schulz 2000). In the absence of the tertiary structure of the Chlamydia MOMP, we have built a model of the topography of this protein using the structural prediction algorithm of Schirmer and Cowan (1993) and the artificial neural network of Diederichs et al. (1998).
Our model exhibits several structural features that have been identified as common to nonspecific bacterial porins (Schulz 2000). The Chlamydia MOMP has 16 β-sheets spanning the depth of the membrane forming a barrel-like structure with long loops protruding out of the cell, while short turns face the periplasmic space. Its barrel-forming amino acids have hydrophilic and hydrophobic characteristics alternating in a way such that the residues facing the pore are in contact with water, while those facing away from the pore are in contact with the membrane lipids. The model also shows the relative abundance of aromatic residues along the upper and lower ends of the β-strands, located at the interface between the hydrophobic and the hydrophilic environment. This "girdle" of aromatic residues lining the boundaries of the β-barrel has been described in other bacterial porins (Schulz 1994). Schulz (1994) proposed that the function of the Tyr-Phe girdle was to prevent conformational damage of the porin during mechanical movements in the membrane. Like other bacterial porins, the chlamydial MOMP also has a band of middle-sized, nonpolar residues located in the mid-section of the β-barrel (Schulz 1994). Only the first eight N-terminal amino acids were not assigned by our prediction.
An alternative assignment of the transmembrane segments of the MoPn MOMP suggests the presence of 18 β-strands. In this model, the two extra strands would occupy positions five and six, which would be located between loops L2 and L3, and correspond to residues assigned to loop L3 on the 16-strand model of Figure 1c ▶. Even though the topology predicted by the hydrophobicity algorithm does not allow the ruling out of the existence of 18 transmembrane segments, the prediction of the z-coordinates by the ANN algorithm does not support the presence of neither transmembrane segments nor external loops. Therefore, an 18 β-strand model most likely can be excluded based on the combined results of these two prediction methods.
On the MOMP external loops, we found a large number of Asp (18) and Glu (9) residues. The side chains of Asp and Glu have been reported to participate in the binding to the carboxylate groups of lipopolysaccharide (LPS) through divalent cations such as calcium and magnesium (Cowan et al. 1992). Nevertheless, not all the negatively charged residues are located on the external loops. Three Asp residues and two Glu are part of the small turns facing the periplasmic side of the membrane. Negatively charged residues also can be seen in the periplasmic turns of the porins from Rodopseudomonas blastica and Escherichia coli OmpF (Cowan et al. 1992; Kreusch et al. 1994). Furthermore, as expected, residues with the highest bend potential, Gly, Pro, Asp, Tyr, Lys, and Ser, are well represented in the periplasmic turns of the MoPn MOMP (Chou and Fasman 1978).
Chlamydial MOMPs differ from other porins of gram-negative bacteria in that they possess a large number (7 to 10) of Cys residues (Stephens et al. 1987; Stephens, 1999). The ability of MOMP to form intra- and intermolecular disulfide bonds with outer-membrane, cysteine-rich proteins is thought to contribute to the rigidity of the EB, which lacks the peptidoglycan layer found in other gram-negative bacteria (Bavoil et al. 1984; Moulder et al. 1984; Hatch 1996). Our predicted topology of the MoPn MOMP assigns seven of the eight Cys residues to the external loops, while the only Cys crossing the membrane (C204) is poorly conserved among serovars. Interestingly, six of the seven Cys located in the external loops are in L1, L4, and L7, three of the four shorter loops that do not have sequence variation. These three loops are in close proximity to L2, the loop that may act as a latch to stabilize the formation of the porin trimers (see below). Thus, it is possible that loops L1, L4, and L7, by being in or close to the trimer interface, may provide rigidity to the membrane of the EB by way of disulfide bonds, while at the same time allowing the necessary plasticity to the membrane for the RB to form following reduction of the disulfide bridges (Hackstadt et al. 1985). Furthermore, the location of the Cys residues in this model suggests that the MOMP cannot form disulfide bonds with other proteins unless they are surface exposed. In this respect, the N terminus of the 60-kDa, cysteine-rich protein is thought to extend to the surface of the bacterial cell (Everett and Hatch 1995; Mygind et al. 1998).
The functional characterization of the external loops presents a challenge because of their exceptionally high sequence variability (Welte et al. 1991; Schulz 2000). In the E. coli OmpF, of the eight surface exposed loops, the second (L2) connects one subunit with its neighbor by latching into its pore and stabilizing the trimers that are common among bacterial porins (Phale et al. 1998). Residue E71 on L2 forms salt bridges and hydrogen bonds with R100 and R132 on the channel wall in the adjacent subunit. In the case of the chlamydial MOMP, formation of trimers has been suggested although not yet proven (McCafferty et al. 1995; Wyllie et al. 1998). If the second loop (L2), containing VD1, is also the latching loop in chlamydial MOMP, the trimer interface could approximately correspond to the constant domains CD1 and CD5. If a similar latching mechanism is taking place in Chlamydia, a negatively charged residue (E82 in MoPn MOMP) at the end of VD1 conserved in 53 of the 58 MOMP sequences analyzed by Bush and Everett (2001) stands out as a likely candidate. Alternatively, an adjacent, highly conserved (in 55 out of 58 sequences aligned) Arg residue (R81 in MoPn MOMP) also could be involved.
In porins, the third loop (L3) extends inside the pore and is attached to the barrel wall, whereby considerably constricting the lumen of the pore (Cowan et al. 1992). An asymmetrical arrangement of positive and negative charges at the constriction site causes a strong electrostatic field parallel to the plane of the membrane that spreads along the channel governing ion selectivity. In the E. coli OmpF, a positively charged cluster formed by three Arg (R42, R82, and R132) protrudes from the barrel wall to face two negatively charged acidic side chains (D113 and E117) located on L3 (Saint et al. 1996; Phale et al. 2001). We have examined the topology predicted for C. trachomatis MoPn MOMP in search of a similar asymmetrical charge distribution. Loop L3 of the MoPn MOMP is negatively charged with four acidic residues (3 Asp and 1 Glu) and two basic residues (Arg and Lys). Basic residues are, by comparison, more abundant in the transmembrane segments: R39, R50, R109, and R173 located in or close to strands 2, 3, 5, and 7, as it was found in E. coli OmpF (Phale et al. 2001).
As can be seen in Figures 1A and 1B ▶, VD3 and part of VD4 are predicted as transmembrane segments. This assignment has to be rejected on immunological, biochemical, and evolutionary evidence. Already in the original paper by Schirmer and Cowan (1993), it was shown that the Hs' algorithm can produce false positives. Immunological studies and immunoelectronmicroscopy, for example, have shown that mice inoculated with Chlamydia produce antibodies against all four VDs of the MOMP (Kuo and Chi 1987; Baehr et al. 1988; Baghian et al. 1990; Andersen 1991; Peterson et al. 1991; Pal et al. 1993, 1996, 1997a, Pal et al. b, 2000; Qu et al. 1993; Villeneuve et al. 1994). Like with Neisseria gonorrhoeae only the long loops L2, L3, L5, and L6 of C. trachomatis and C. psittaci have VDs probably reflecting the fact that they are more immunoaccessible (van der Ley et al. 1991). Loops L1, L4, and L7 are not only shorter than the loops containing the VDs but, as previously discussed, also have numerous Cys residues probably involved in disulfide bonding and thus, imposing a significant restriction on structural changes (see above). In the case of C. pneumoniae however, the lack of sequence variability suggests that the MOMP is immunorecessive, or it is not surface accessible (Christiansen et al. 1999; Wolf et al. 2001).
Alignment of MOMP sequences from different chlamydial species and serovars identified a conserved nonapeptide within the highly variable VD4: TTWNPTISG (indicated in bold hexagons in Fig. 1C ▶ and corresponding to the sequence TTLNPTIAG in the MOMP of the human C. trachomatis serovars (Stephens et al. 1987; Fitch et al. 1993). The algorithms employed here suggest that this invariant region is a putative transmembrane segment. However, the assignment of loops L6 and L6a immediately before and after this nonapeptide argues otherwise. Su et al. (1990) suggested that this conserved region functions as a cryptic hydrophobic binding site responsible for Chlamydia-host cell interactions. Epitope mapping supports the assumption that this region is not a transmembrane segment. The monoclonal antibody MoPn 13–2 binds to this region of the MoPn MOMP (Pal et al. 1997b). Also, the subspecies-specific monoclonal antibody E4 binds to this invariant region TLNPTIA within the VD4 of all the human chlamydial serovars (Peterson et al. 1988). In vitro, this antibody reacted by immunofluorescence and dot-blot ELISA with native EB of the B- and C-related complexes, but it reacted only with heat-treated EB of the C complex. The fact that a mild treatment exposes this binding site suggests that, even in the members of the C complex, this epitope is not buried in the membrane.
Enzyme digestion, biochemical analysis, and substitution-rate studies also support the location of the VDs outside the membrane. For example, the infectivity of serovar L2 was resistant to treatment with trypsin, while infectivity of the B serovar was significantly reduced following digestion with this enzyme (Su et al. 1988). Comparison of the VD2 and VD4 showed a trypsin-sensitive Lys residue in each of these two sites on the B serovar, while the MOMP of the L2 serovar was cleaved only at the Lys in VD4. Based on these findings, it was concluded that VD2 and VD4 are surface exposed and play a critical role on chlamydial infectivity. Recently, Hughes et al. (2001) showed that a recombinant MOMP in which the VD4 was deleted retained its ability to form pore channels, indicating VD4 is not directly involved in the pore function.
The assignment of VDs to surface-exposed loops also is consistent with the results of Brunham et al. (1994), who found a higher rate of nonsynonymous than synonymous substitutions in the MOMP VDs. This pattern suggests the action of positive selection. Very few genes show evidence of positive selection. The major class of such genes is pathogen surface proteins (van der Ley et al. 1991; Smith et al. 1995; Endo et al. 1996; Bush et al. 1999; Yamaguchi-Kabata and Gojobori 2000; Bush 2001). Here, the hypervariability of the VDs probably helps the pathogen to evade recognition by the immune system of the host.
In summary, we are proposing a topology scheme for the MOMP of Chlamydia consistent with immunological, biochemical, and evolutionary data. The model should help to understand the role of MOMP in the pathogenesis of this microorganism, to develop preventive and therapeutic measures, and to assist in planning further experiments to advance our knowledge of the structure of this protein.
Materials and methods
Prediction of membrane-spanning β-strands
The topology of the MOMP from C. trachomatis, serovars MoPn, E and L3, C. pneumoniae, strain AR39, C. psittaci, and strain A22/M (Accession numbers JT0947, MMCWTE, JE0413, A43587, and A60109, respectively) was predicted using an algorithm developed by Schirmer and Cowan (1993). This algorithm calculates the mean hydrophobicity of one side of a putative β-strand by taking the average of the hydrophobic indices of every second residue within a sliding window of four.
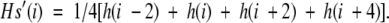 |
To account for the observed preponderance of aromatic residues in flanking positions, the method assigns an arbitrarily increased "hydrophobicity index" of 1.6 to aromatic residues when found in positions i−2 or i+4.
Topology prediction using an artificial neural network
An ANN trained to predict the topology of bacterial outer-membrane proteins with a predominance of β-strands was applied to the MOMP sequences (Diederichs et al. 1998). This ANN, available online (http://strucbio.biologie.uni-konstanz.de), predicts the z-coordinate of Cα atoms in a coordinate frame with the outer membrane in the xy-plane, such that low z-values (<0.3) indicate periplasmic turns, medium z-values indicate transmembrane β-strands, and high z-values (≥0.6) indicate extracellular loops.
Secondary structure comparisons with known porins
We used the computer program FORESST (Fold Recognition from Secondary Structures, Release 1.0, http://absalpha.cit.nih.gov/) to determine the degree of homology between our predicted secondary structure model and a data set of 349 proteins of known structure (Di Francesco et al. 1999; Geetha et al. 1999). Our query sequence consisted of a string of 365 letters, each representing a position in the MoPn MOMP protein sequence. FORESST provides results as Z-scores, which are the number of standard deviations by which the hidden Markov score for the query sequence was above the average score for the family model using a random protein (Cowan et al. 1992; Kreusch et al. 1994). Comparisons with z-scores of 3.0 and higher are considered significantly homologous.
Acknowledgments
The authors gratefully acknowledge the generous assistance of Dr. Sajith Jayasinghe for help with UNIX analyses. This work was supported by Public Health Grants from the National Institute of Allergy and Infectious Diseases to L.M.D. (AI-32248) and R.M.B. (AI-44474) and by a grant of the Swiss National Science Foundation to T.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.3650102.
References
- Andersen, A.A. 1991. Serotyping of Chlamydia psittaci isolates using serovar-specific monoclonal antibodies with the microimmunofluorescence test. J. Clin. Microbiol. 29 707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr, W., Zhang, Y-X., Joseph, T., Su H., Nano, F.E., Everett, K.D.E., and Caldwell, H.D. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. 85 4000–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghian, A,, Shaffer, L., and Storz, J. 1990. Antibody response to epitopes of chlamydial major outer membrane proteins on infectious elementary bodies and of the reduced polyacrylamide gel electrophoresis-separated form. Infect. Immun. 58 1379–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoil, P., Ohlin, A., and Schachter, J. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham, R., Yang, C., Maclean, I., Kimani, J., Maitha, G., and Plummer, F. 1994. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Invest. 94 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham, R.C., Zhang, D.J., Yang, X., and McClarty, G.M. 2000. The potential for vaccine development against chlamydial infection and disease. J. Infect. Dis. 155 749–755. [DOI] [PubMed] [Google Scholar]
- Bush, R.M. 2001. Predicting adaptive evolution. Nature Rev. Genetics 2 387–392. [DOI] [PubMed] [Google Scholar]
- Bush, R.M., and Everett, K.D.E. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Systematic Evol. Microbiol. 51 203–220. [DOI] [PubMed] [Google Scholar]
- Bush, R.M., Fitch, W.M., Bender, C.A., and Cox, N.J. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16 1457–1465. [DOI] [PubMed] [Google Scholar]
- Caldwell, H.D., Kromhout, J., and Schachter, J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, H.D. and Perry, L.J. 1982. Neutralization of Chlamydia trachomatis infectivity with antibodies to the major outer membrane protein. Infect. Immun. 38 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, P.Y. and Fasman, G.D. 1978. Empirical predictions of protein conformation. Ann. Rev. Biochem. 47 215–276. [DOI] [PubMed] [Google Scholar]
- Christiansen, G., Boesen, T., Hjerno, K., Daugaard, L., Mygind, P., Madsen, A.S., Knudsen, K., Falk E., and Birkelund, S. 1999. Molecular biology of Chlamydia pneumoniae surface proteins and their role in immunopathogenesis. Am. Heart J. 138 S491–S495. [DOI] [PubMed] [Google Scholar]
- Cowan, S.W., Schirmer, T., Rummel, G., Steiert, M., Ghosh, R., Pauptit, R.A., Jansonius, J.N., and Rosenbusch, J.P. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358 727–733. [DOI] [PubMed] [Google Scholar]
- de la Maza, L.M. and de la Maza, M.A. 1995. A new computer model for estimating the impact of vaccination protocols and its application to the study of Chlamydia trachomatis genital infections. Vaccine 13 119–127. [DOI] [PubMed] [Google Scholar]
- Diederichs, K., Freigang, J., Umhau, S., Zeth, K., and Breed, J. 1998. Prediction by a neural network of outer membrane β-strand protein topology. Protein Sci. 7 2413–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Francesco, V., Munson, P.J., and Garnier, J. 1999. FORESST: Fold recognition from secondary structure predictions of proteins. Bioinformatics 15 131–140. [DOI] [PubMed] [Google Scholar]
- Endo, T., Ikeo, K., and Gojobori, T. 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13 685–690. [DOI] [PubMed] [Google Scholar]
- Everett, K.D.E., Bush, R.M., and Andersen, A.A. 1999. Emended description of the order Chlamydiales, proposal of two new families and species, revised taxonomy of the family Chlamydiaceae including a new genus and five new species, and standard for the identification of organisms. Intern. J. Syst. Bacteriol. 49 415–440. [DOI] [PubMed] [Google Scholar]
- Everett, K.D.E. and Hatch, T.P. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J. Bacteriol. 177 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, W.M., Peterson, E.M., and de la Maza, L.M. 1993. Phylogenetic analysis of the outer membrane protein genes of Chlamydiae, and its implication for vaccine development. Mol. Biol. Evol. 10 892–913. [DOI] [PubMed] [Google Scholar]
- Geetha, V., di Francesco, V., Garnier, J., and Munson, P.J. 1999. Comparing protein sequence-based and predicted secondary structure-based methods for identification of remote homologs. Prot. Eng. 12 527–534. [DOI] [PubMed] [Google Scholar]
- Hackstadt, T., Scidemore, M.A., and Rockey, D.D. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: Directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusions. Proc. Natl. Acad. Sci. 92 4877–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt, T., Todd, W.J., and Caldwell, H.D. 1985. Disulfide-mediated interaction of the chlamydial major outer membrane protein: Role in the differentiation of chlamydiae. J. Bacteriol. 161 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, T.P. 1996. Disulfide cross-linked envelope proteins: The functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, T.P., Vance, D.W., and Al-Hossainy, E. 1981. Identification of a major envelope protein in Chlamydia spp. J. Bacteriol. 146 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, E.S., Shaw, K.M., and Ashley, R.H. 2001. Mutagenesis and functional reconstitution of chlamydial major outer membrane proteins: VS4 domains are not required for pore formation but modify channel function. Infect. Immun. 69 1671–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur, D., Lakey, J.H., and Pattus, F. 1991. The bacterial porin superfamily: Sequence alignment and structure prediction. Mol. Microbiol. 5 2153–2164. [DOI] [PubMed] [Google Scholar]
- Jeanteur, D., Lakey, J., and Pattus, F. 1994. The porin superfamily: Diversity and common features. In Bacterial cell wall, (eds. J-M. Ghuysen and R. Hankenbeck) pp. 363–380. Elsevier Science B.V., New York.
- Klebe, G. 2000. Recent developments in structure-based drug design. J. Mol. Med. 78 245–246. [DOI] [PubMed] [Google Scholar]
- Kuo, C-C. and Chi, E.Y. 1987. Ultrastructural study of Chlamydia trachomatis surface antigens by immunogold staining with monoclonal antibodies. Infect. Immun. 55 1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreusch, A., Neubüser, A., Schiltz, E., Weckesser, J., and Schulz, G.E. 1994. Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 Å resolution. Protein Sci. 3 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty, M.C., Herring, A.J., Andersen, A.A., and Jones, G.E. 1995. Electrophoretic analysis of the major outer membrane protein of Chlamydia psittaci reveals multimers which are recognized by protective monoclonal antibodies. Infect Immun. 63 2387–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder, J.W., Hatch, T.P., Kuo, C-C., Schachter, J., and Storz, J. 1984. Genus Chlamydia. In Bergey's manual of systemic bacteriology (ed. N.R. Krieg), pp. 729–739. Williams and Wilkins Co., Baltimore.
- Mygind, P., Christiansen, G., and Birkelund, S. 1998. Topological analysis of Chlamydia trachomatis L2 outer membrane protein 2. J. Bacteriol. 180 5784–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, C. 1942. Unidentified virus which produces pneumonia and systemic infection in mice. Science 95 49–59. [DOI] [PubMed] [Google Scholar]
- Pal, S., Cheng, X., Peterson, E.M., and de la Maza, L.M. 1993. Mapping of a surface-exposed B-cell epitope to the variable sequent 3 of the major outer-membrane proteins of Chlamydia trachomatis. J. Gen. Microbiol. 139 1565–1570. [DOI] [PubMed] [Google Scholar]
- Pal, S., Peterson, E.M., and de la Maza, L.M. 1996. Intranasal immunization induces long-term protection in mice against a Chlamydia trachomatis genital challenge. Infect. Immun. 64 5341–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, S., Rangel, J., Peterson, E.M., and de la Maza, L.M. 2000. Immunogenic and protective ability of the two developmental forms of Chlamydiae in a mouse model of infertility. Vaccine 18 752–761. [DOI] [PubMed] [Google Scholar]
- Pal, S., Theodor, I., Peterson, E.M., and de la Maza, L.M. 1997a. Immunization with an acellular vaccine consisting of the outer membrane complex of Chlamydia trachomatis induces protection against a genital challenge. Infect. Immun. 65 3361–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, S., Theodor, I., Peterson, E.M., and de la Maza, L.M. 1997b. Monoclonal immunoglobulin A antibody to the major outer membrane protein of the Chlamydia trachomatis mouse pneumonitis biovar protects mice against a chlamydial genital challenge. Vaccine 15 575–582. [DOI] [PubMed] [Google Scholar]
- Peterson. E.M., Cheng. X., Markoff. B.A., Fielder. T.J., and de la Maza. L.M. 1991. Functional and structural mapping of Chlamydia trachomatis species-specific major outer membrane protein epitopes by use of neutralizing monoclonal antibodies. Infect. Immun. 59 4147–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, E.M. and de la Maza, L.M. 1988. Chlamydia parasitism: Ultrastructural characterization of the interaction between the chlamydial cell envelope and the host cell. J. Bacteriol. 170 1389–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, E.M., Zhong, G., Carlson, E., de la Maza, L.M. 1988. Protective role of magnesium in the neutralization by antibodies of Chlamydia trachomatis infectivity. Infect. Immun. 56 885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phale, P.S., Philippsen, A., Kiefhaber, T., Koebnik, R., Phale, V.P.. Schirmer, T., and Rosenbusch, J.P. 1998. Stability of trimeric OmpF porin: The contributions of the latching loop L2. Biochemistry 37 15663–15670. [DOI] [PubMed] [Google Scholar]
- Phale, P.S., Philippsen, A., Widmer, C., Phale, V.P., Rosenbusch, J.P., and Schirmer, T. 2001. Role of charged residues at the E. coli OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry 40 6319–6325. [DOI] [PubMed] [Google Scholar]
- Qu, Z., Cheng, X., de la Maza, L.M., and Peterson E.M. 1993. Characterization of a neutralizing monoclonal antibody directed at variable domain I of the major outer membrane protein of Chlamydia trachomatis C-complex serovars. Infect. Immun. 64 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulston, J.E.. 1995. Chlamydial envelope components and pathogen-host cell interactions. Mol. Microbiol. 15 607–616. [DOI] [PubMed] [Google Scholar]
- Saint, N., Lou, K-L., Widmer, C., Luckey, M., Schirmer, T., and Rosenbusch, J.P. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. J. Biol. Chem. 271 20676–20680. [PubMed] [Google Scholar]
- Schachter, J. and Dawson, C.R. 1978. Human chlamydial infections. PSG Publishing Co., Littleton, MA.
- Schirmer, T. 1998. General and specific porins from bacterial outer membranes. J. Struct. Biol. 121 101–109. [DOI] [PubMed] [Google Scholar]
- Schirmer, T. and Cowan, S.W. 1993. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 2 1361–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, G.E. 1994. Structure-function relationships in porins as derived from a 1.8 Å resolution crystal structure. In Bacterial cell wall (eds. J.M. Ghuysen and R. Hankenbeck), pp. 343–352. Elsevier Science B.V., New York.
- Schulz, G.E. 2000. β-barrel membrane proteins. Curr. Op. Struct. Biol. 10 443–447. [DOI] [PubMed] [Google Scholar]
- Smith, N.H., Maynard Smith, J., and Spratt, B.G. 1995. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: Evidence of positive Darwinian selection. Mol. Biol. Evol. 12 363–370. [DOI] [PubMed] [Google Scholar]
- Stephens, R.S. 1999. Chlamydia: Intracellular biology, pathogenesis and immunology. ASM Press, Washington.
- Stephens, R.S. 2000. Chlamydial genomics and vaccine antigen discovery. J. Infect. Dis. 181 S521–S523. [DOI] [PubMed] [Google Scholar]
- Stephens, R.S., Sanchez-Pescador, R., Wager, E.A., Inouye, C., and Urdea, M.S. 1987. Diversity of Chlamydia trachomatis major outer membrane protein genes. J. Bacteriol. 169 3879–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Watkins, N.G., Zhang, Y-X., Caldwell, H. 1990. Chlamydia trachomatis-host cell interactions: Role of the chlamydial major outer membrane protein as an adhesin. Infect. Immun. 58 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Zhang Y-X., Barrera, O., Watkins, N.G., and Caldwell, H.D. 1988. Differential effect of trypsin on infectivity of Chlamydia trachomatis: Loss of infectivity requires cleavage of major outer membrane protein variable domains II and IV. Infect. Immun. 56 2094–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ley, P., Heckels, J.E., Virji, M., Hoogerhout, P., and Poolman, J.T. 1991. Topology of outer membrane porins in pathogenic Neisseria spp. Infect. Immun. 59 2963–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A., Brossay, L., Paradis, G., and Hébert J. 1994. Determination of neutralizing epitopes in variable domains I and IV of the major outer membrane protein from Chlamydia trachomatis serovar K. Microbiol. 140 2481–2487. [DOI] [PubMed] [Google Scholar]
- Wang, S-P. and Grayston, J.T. 1984. Microimmunofluorescence serology of Chlamydia trachomatis. In Medical virology III (eds. L.M. de la Maza and E.M. Peterson), pp. 87–118. Elsevier Science Pub., New York.
- Ward, M.E. 1992. Chlamydial vaccines—future trends. J. Infect. 25 11–26. [DOI] [PubMed] [Google Scholar]
- Welte, W., Weiss, M.S., Nestel, U., Weckesser, J., Schiltz, E., and Schulz, G.E. 1991. Prediction of the general structure of E. coli OmpF and PhoE from the sequence and structure of porin from Rhodobacter capsulatus. Orientation of porin in the membrane. Biochim. Biophys. Acta 1080 271–274. [DOI] [PubMed] [Google Scholar]
- Wolf, K., Fischer, E., Mead, D., Zhong, G., Peeling, R., Whitmire, B., and Caldwell, H.D. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69 3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie, S., Ashley, R.H., Longbottom, D., and Herring, A.J. 1998. The major outer membrane protein of Chlamydia psittaci functions as a porin-like ion channel. Infect. Immun. 66 5202–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Kabata, Y. and Gojobori, T. 2000. Reevaluation of amino acid variability of the human immunodeficiency virus type 1 gp120 envelope glycoprotein and prediction of new discontinuous epitopes. J. Virol. 74 4335–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]


