Abstract
Asp residues are significantly under represented in β-sheet regions of proteins, especially in the middle of β-strands, as found by a number of studies using statistical, modeling, or experimental methods. To further understand the reasons for this under representation of Asp, we prepared and analyzed mutants of a β-domain. Two Gln residues of the immunoglobulin light-chain variable domain (VL) of protein Len were replaced with Asp, and then the effects of these changes on protein stability and protein structure were studied. The replacement of Q38D, located at the end of a β-strand, and that of Q89D, located in the middle of a β-strand, reduced the stability of the parent immunoglobulin VL domain by 2.0 kcal/mol and 5.3 kcal/mol, respectively. Because the Q89D mutant of the wild-type VL-Len domain was too unstable to be expressed as a soluble protein, we prepared the Q89D mutant in a triple mutant background, VL-Len M4L/Y27dD/T94H, which was 4.2 kcal/mol more stable than the wild-type VL-Len domain. The structures of mutants VL-Len Q38D and VL-Len Q89D/M4L/Y27dD/T94H were determined by X-ray diffraction at 1.6 Å resolution. We found no major perturbances in the structures of these Q→D mutant proteins relative to structures of the parent proteins. The observed stability changes have to be accounted for by cumulative effects of the following several factors: (1) by changes in main-chain dihedral angles and in side-chain rotomers, (2) by close contacts between some atoms, and, most significantly, (3) by the unfavorable electrostatic interactions between the Asp side chain and the carbonyls of the main chain. We show that the Asn side chain, which is of similar size but neutral, is less destabilizing. The detrimental effect of Asp within a β-sheet of an immunoglobulin-type domain can have very serious consequences. A somatic mutation of a β-strand residue to Asp could prevent the expression of the domain both in vitro and in vivo, or it could contribute to the pathogenic potential of the protein in vivo.
Keywords: Protein structure, X-ray diffraction, protein stability, β-sheet, aspartic acid
Aspartic acid generally would appear to be acceptable anywhere on a protein surface. However, statistical analysis of the distribution of amino acids in protein structures has shown that Asp residues are under represented substantially in β-chain regions (McGregor et al. 1987; Muñoz and Serrano 1994). Experimental studies of Minor and Kim (1994) and of Smith et al. (1994), though slightly different from each other, showed that the amino acid Asp has the least propensity to be found in β-sheets, when Gly and Pro residues are not considered. Similar results were obtained by theoretical calculations in a study that took only the effect of a residue's side chain steric clashes with its own backbone into consideration (Street and Mayo 1999) and when β-sheet propensities were modeled by a complex energy function (Finkelstein 1995). To explore in more detail the reasons for this biased distribution of Asp residues, we constructed and studied Q→D mutants of a β-domain, namely, an immunoglobulin light-chain variable domain (VL) of human κ4 protein Len (Huang et al. 1997). In agreement with the above observations, the stability of the VL-Len domain decreased significantly when Gln residues Q38 or Q89, located in a β-sheet, were replaced by an Asp residue (Fig. 1 ▶). To understand the reasons for the decline of stability upon introduction of the above Asp residues into a β-sheet of the VL domain, we determined the structures of mutants that contained Asp38 or Asp89 and compared them to known structures of parent molecules. As a control, we also determined the influence on stability of aspargine substitution at positions 38 and 89.
Fig. 1.
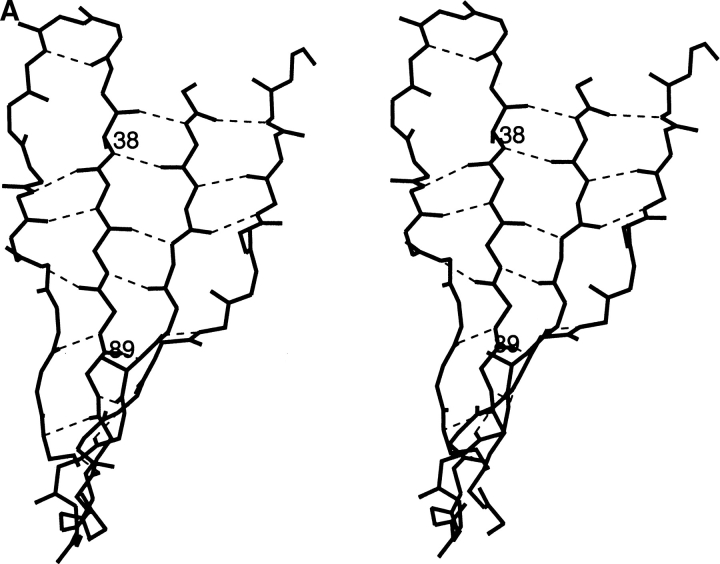
Two orientations of the β-sheet from VL-domain Len that contains residues 38 and 89 are shown in stereo. Only the main-chain atoms of the polypeptide are pictured along with the hydrogen bonds between them. The residues 38 and 89 are shown as alanines for clarity. Panel A focuses on residue 38 and panel B focuses on residue 89.
Results
Protein expression
Glutamine residues 38 and 89 in human κ4 VL Len (Huang et al. 1997) were mutated to aspartic acid and aspargine residues in both the wild-type VL-Len background (mutants Q38D, Q38N and Q89D, Q89N) and in a stabilized VL-Len variant (Raffen et al. 1999), in which residue 27d had been changed from tyrosine to aspartic acid (mutants Q38D/Y27dD, Q38N/Y27dD and Q89D/Y27dD, Q89N/Y27dD). The Q89D and Q89N mutations also were introduced into a highly stabilized triple mutant VL-Len, M4L/Y27dD/T94H (Pokkuluri et al. 2002), yielding quad mutants Q89D/M4L/ Y27dD/T94H and Q89N/M4L/Y27dD/T94H. While soluble protein was obtained from the Q38D, Q38N and Q89N single-mutant clones, no soluble protein expression could be observed from the Q89D single-mutant clone. This observation suggested that protein Q89D was too unstable to be expressed in a soluble form. Diminished expression or lack of expression of soluble proteins has been observed previously for unstable proteins (Schiffer et al. unpubl.). Expression of soluble VL-Len-derived domains that contain the Q89D mutation was possible in the Y27dD and the M4L/Y27dD/T94H backgrounds.
Protein stability
The thermodynamic stability values of the wild type, Y27dD, M4L/Y27dD/T94H, Q38D, Q38N, Q38D/Y27dD, Q38N/Y27dD, Q89D/Y27dD, Q89N/Y27dD, Q89D/M4L/Y27dD/T94H, and Q89N/M4L/Y27dD/T94H proteins were determined by the guanidine hydrochloride (GdnHCl) denaturation method; the results are listed in Table 1. The stability value for the VL-Len Q89D mutant, which was not possible to measure directly, as this mutant clone did not express soluble protein, was estimated based on how much the Q89D mutation reduced the stability values of the Y27dD and M4L/Y27dD/T94H variants. To estimate the stability value for the Q89D mutant, we assumed that the components of the ΔG and Cm values that are contributed by specific mutations are largely additive, a reasonable assumption because these substitutions are not at interacting sites of the molecule.
Table 1.
Stability of the mutants
| Cm [GdnHCl] | m (kcal/mol/M) | ΔGunf (kcal/mol) | ΔΔGaunf (kcal/mol) | |
| Wild-type Lenb | 1.76 | −4.4 ± 0.4 | 7.7 ± 0.6 | 0 |
| Y27dDb | 2.37 | −4.0 ± 0.2 | 9.4 ± 0.5 | −2.7 |
| M4L/Y27dD/T94Hc | 2.71 | −4.5 ± 0.1 | 12.1 ± 0.2 | −4.2 |
| Q38Dd | 1.30 | −3.0 ± 0.3 | 3.8 ± 0.3 | 2.0 |
| Q38N | 1.51 | −3.7 ± 0.1 | 5.6 ± 0.1 | 1.1 |
| Q38D/Y27dD | 2.10 | −3.5 ± 0.2 | 7.4 ± 0.4 | −1.5 |
| Q38N/Y27dD | 2.20 | −3.5 ± 0.1 | 7.6 ± 0.1 | −1.9 |
| Q89D/Y27dD | 1.17 | −3.9 ± 0.2 | 4.5 ± 0.3 | 2.6 |
| Q89N/Y27dD | 1.98 | −4.0 ± 0.1 | 7.9 ± 0.1 | −1.0 |
| Q89D/M4L/ Y27dD/T94H | 1.48 | −4.4 ± 0.1 | 6.5 ± 0.2 | 1.2 |
| Q89N/M4L/ Y27dD/T94H | 2.52 | −4.6 ± 0.2 | 11.5 ± 0.5 | −3.3 |
| Q89De | (0.55) | — | — | (5.3) |
| Q89N | 1.39 | −4.9 ± 0.2 | 6.8 ± 0.3 | 1.6 |
a ΔΔGunf = mLen × Cmmut − mLen × CmLen.
b From Raffen et al. 1999.
c From Pokkuluri et al. 2002.
d From Raffen et al. 1998.
e No protein expression: Cm and ΔΔGunf are calculated values, as explained in the text.
The Q38D mutation reduces the stability of both the wild type and Y27dD mutant proteins (Table 1). The Cm value for the VL-Len Q38D and VL-Len Q38N mutants are 1.3 M and 1.51, respectively. They are 0.46 M and 0.25 M less than the Cm value of the wild type. The introduction of the Q38D and Q38N mutations into the Y27dD construct (Cm = 2.37 M) reduces its Cm by 0.27 M and 0.17 M, respectively as is reflected in the Cm values of 2.10 M and 2.20 M for the double mutants Q38D/Y27dD and Q38N/Y27dD. The stability is affected more dramatically by the introduction of the Q89D mutation than by introduction of the Q38D mutation. The Cm value of the Q89D/Y27dD mutant is 1.17 M; therefore the introduction of Q89D into the Y27dD mutant reduces its Cm by 1.20 M. The introduction of Q89D mutation into the triple mutant (Cm = 2.71 M) reduces its Cm value by a similar amount, 1.23 M; Cm of the quad mutant is 1.48 M. If the Cm of the wild type were to be reduced by the same amount, the calculated Cm for the Q89D mutant protein would be 0.53–0.56 M, a value reflective of an unstable protein. The Cm values of Q89N/Y27dD and Q89N/M4L/Y27dD/T94H are 1.98 M and 2.52 M, respectively. Therefore, the introduction of Q89N reduces the Cm values of the parent molecule by 0.39 M and 0.19 M, respectively. This compares with the introduction of Q89N mutation into the wild-type protein, which reduces the wild-type value by 0.37 M to a Cm of 1.39 M.
Protein structures
The crystal structure of VL-Len Q38D was determined in the presence of UO22+ ions. No crystals were obtained without UO22+ ions. Q38D was crystallized in the P63 (Table 2) space group with one monomer in the asymmetric unit. The protein also has a low association constant in solution (Raffen et al. 1998). In the Q38D structure, the "40 loop" (consisting of residues 38–43) is wider than it is in the native VL-Len domain. The UO22+ interacts with the side-chain carboxyl group of Asp38 and the carbonyl oxygen of residue 42. The closest distance between Asp38 and the carbonyl oxygen of residue 42 is 2.84 Å. The χ1 and χ2 values for the Asp38 side chain in the Q38D mutant are −170° and −166°. With these χ values used for Asp38 modeled in the native Len structure (PDB code 1lve), the above closest distance is reduced by ∼0.3 Å, from 2.84 Å to 2.51 Å. Therefore it appears that the widening of the 38–43 loop in protein Len is required to accommodate an Asp residue at position 38.
Table 2.
Crystallographic data
| Q89D/M4L/Y27dD/T94H | Q38D | |
| Space group | P212121 | P63 |
| a (Å) | 62.9 | 65.8 |
| b (Å) | 104.8 | 65.8 |
| c (Å) | 42.4 | 48.1 |
| VM (Å3/dalton) | 2.8 | 2.4 |
| R merge (last shell) % | 6.1 (26.3) | 7.4 (20.4) |
| Highest resolution (Å) | 1.6 | 1.6 |
| No. of reflections used with │Fo│>3σ│Fo│ | 34586 | 14410 |
| Final R-factor | 0.195 | 0.230 |
| Free R-factor (10% data) | 0.222 | 0.289 |
| No. of refined protein atoms | 1729 | 858 |
| No. of solvent molecules | 336a | 105b |
| Rmsd bond length (Å) | 0.005 | 0.009 |
| Rmsd bond angle (°) | 1.3 | 1.6 |
| Mean temperature factor | ||
| For protein atoms (Å2) | 17.6 | 14.3 |
| For solvent molecules (Å2) | 32.7 | 23.3 |
a Includes two 2-propanol molecules.
b Includes one Zn2+ ion and one UO22+ ion.
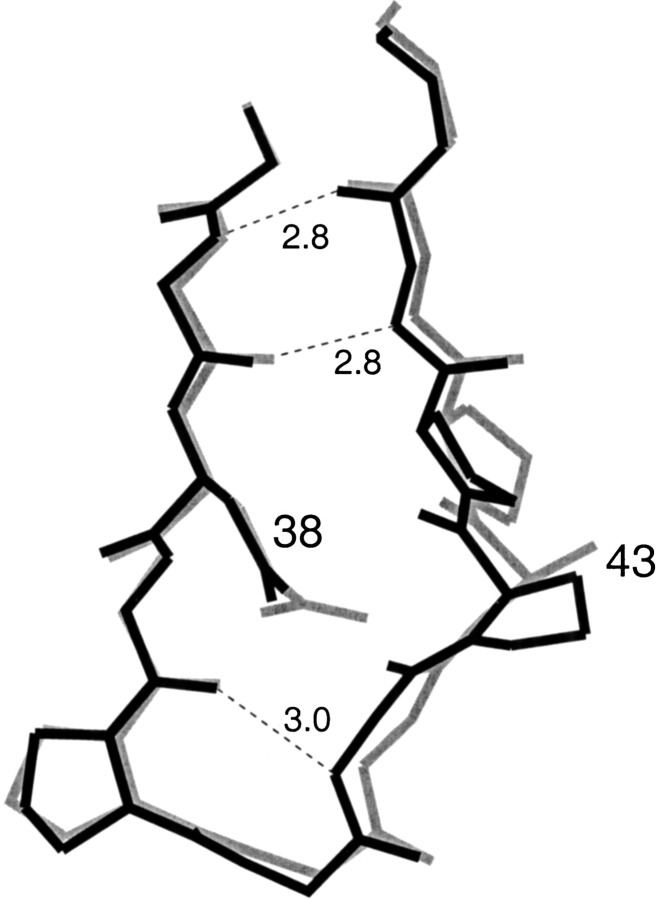
The decrease in stability was less when Asp was introduced at the equivalent residue 38 in κ1 Rei VL domain (Chan et al. 1996); ΔΔG was 1.2 kcal/mol in Rei compared to 2 kcal/mol in the Len protein. A possible reason for the difference is that in protein Rei, residue 43 is Ala while in Len it is Pro. With the more flexible Ala residue instead of Pro, a wider loop is possible, that is, the wider loop can better accommodate the Asp side chain. Actually, the conformations of the "40 loop" in Q38D crystallized with UO22+ and the "40 loop" in wild type VL-Rei are similar, as shown in Figure 2 ▶.
Fig. 2.
The conformations of the "40 loop" in Len Q38D (black) and in κ1 protein Rei (gray) from PDB code 1rei are shown. The hydrogen bonds are indicated by dashed lines.
The VL-Len Q89D/M4L/Y27dD/T94H quad mutant is isomorphous with its parent triple mutant (Pokkuluri et al. 2001). The crystallographic data are shown in Table 2. In both structures, a noncovalent dimer is in the asymmetric unit. Both molecules are also dimers in solution. For the individual domains, the root mean square deviation of α carbons between the two mutants is 0.14 Å and 0.11 Å, respectively. Since modeling of Asp89 in the wild-type Len protein suggested some close contacts, we expected that the segment with Asp89 might not be well defined. Contrary to our expectations, the overall temperature factor of the Q89D/M4L/Y27dD/T94H structure is slightly lower than that of the parent triple mutant, and the segment that includes residues 86 to 103 is well defined.
To understand the details of the structural changes introduced by the Q89D mutation, we examined the φ, ξ, and χ angles in both monomers of the dimer for both the Q89D/M4L/Y27dD/T94H structure and the parent triple-mutant structure. Table 3 lists the significant differences in φ, ξ, and χ angles between the two structures near the mutation site. Changes occur in both monomers of the quad mutant compared to the triple mutant in the ξ values of residues Tyr96, and Ser97 and in the φ values of Phe98 and residue 89. Differences in χ2 values of both monomers occur for Gln90 and Tyr91 and for χ1 values of Phe98. However, there are no significant changes in the φ, ξ, or χ angles for residues in the other neighboring β-strand segment, namely residues 32, 33, and 34. The main-chain hydrogen bonds involving the above segments are not altered.
Table 3.
Differences in dihedral anglesadue to the introduction of Asp at residue 89
| Dihedral angle | Corrb | Residue | mon1c | mon2c | diffd | diffe |
| φ | ✓ | 89 | 6° | 10° | 3° | 1° |
| ξ | 89 | 8° | (4°) | 1° | 3° | |
| φ | Tyr91 | (1°) | 6° | 2° | 3° | |
| φ | Tyr92 | 8° | (3°) | 2° | 3° | |
| ξ | Ser93 | 11° | (3°) | 15° | 7° | |
| φ | Ser93 | (4°) | 6° | 4° | 2° | |
| φ | Thr94 | 8° | (2°) | 11° | 5 | |
| ξ | ✓ | Tyr96 | 8° | 7° | 5° | 4° |
| ξ | ✓ | Ser97 | 12° | 9° | 3° | 0° |
| φ | ✓ | Phe98 | 19° | 15° | 2° | 2° |
| χ2 | ✓ | Gln90 | 8° | 8° | 1° | 0° |
| χ2 | ✓ | Tyr91 | 8° | 11° | 5° | 2° |
| χ2 | Tyr92 | 10° | (1°) | 8° | 3° | |
| χ1 | Ser97 | (1°) | 6° | 2° | 7° | |
| χ1 | ✓ | Phe98 | 16° | 6° | 1° | 9° |
| χ1 | Gln100 | 9° | (4°) | 2° | 3° | |
| χ3 | Gln100 | 13° | (2°) | 2° | 13° |
a The average difference between individual monomers within one structure is 3° for residues 86 to 103.
b Significant changes observed in both monomers are marked by "✓"; values in brackets are not considered significant.
c Difference between Q89D/M4L/Y27dD/T94H and triple mutant.
d Difference between monomers in Q89D/M4L/Y27dD/T94H.
e Difference between monomers in M4L/Y27dD/T94H.
Discussion
As expected from the previously reported statistical, experimental, and modeling studies, both of the Gln to Asp mutations at residues 38 and 89 located at the end and middle of β-strands respectively of VL-Len reduced the stability of the mutant proteins relative to their parent molecules. Clearly the effects on stability of the Gln to Asp mutations at positions 38 and 89 of the VL domain are different. Although both Q→D mutations reduce the stability of the VL, the Q89D mutation in the middle of a β-strand reduces the stability by significantly more (ΔΔG = 5.3 kcal/mol) than does the Q38D mutation at the end of a β-strand (ΔΔG = ∼2.0 kcal/mol). The influence of Asn substitution is less; the Q38N and Q89N mutations reduce the stability of the wild-type protein by 1.1 kcal/mol and 1.6 kcal/mol, respectively. The above values exceed the ΔΔG value of 1.2 kcal/mol for Gln to Asp, and <0.3 kcal/mol for Gln to Asn substituted protein G variants reported by Minor and Kim (1994) and Smith et al. (1994). The differences on mutating residues at the 38 and 89 positions also are evident when the influences on stability of Asp and Asn substitutions are compared. The average reduction in stability is ∼0.6 kcal/mol at residue 38 and ∼4 kcal/mol at residue 89 (see ΔΔG values on Table 1). Our crystallographic investigation of the very destabilizing mutation Q89D of VL-Len was only possible because the mutation could be introduced into a very highly stabilized triple mutant background, a construct previously designed.
Statistical studies on residue locations by McGregor et al. (1987) on 61 proteins found only 67 Asp residues in β-strands. Of these 67 Asp residues, 16 were located in the central region of a β-strand and 51 were located at the ends, where end is defined as the last two residues of a β-strand. Swindells et al. (1995) examined residues in 85 proteins and found only 112 Asp residues in β-sheet regions. Whereas McGregor et al. (1987) found that the rotomers of the Asp residues in β-sheet regions were about equally divided between t and g+ with a small proportion in g− conformation, the t rotomer conformation was the most frequent (58%) in the data analyzed by Swindells et al. (1995). In our studies of VL-Len, we found that both Asp38 and Asp89 are in the t rotomer conformation. With the t rotomer conformation, in general, the closest main-chain atoms are within the same chain segment as the carbonyl oxygen of the residue following the aspartic acid; and are on the adjacent chain segment from the carbonyl oxygen of the hydrogen bonded partner of the Asp+1 residue.
Residues 38 and 89 are both located in the two neighboring extended central strands of a four-strand, antiparallel β-sheet (Fig. 1 ▶). Residue 38 is at the end of a β-strand, residue 89 is in the central region of its β-strand; they are on opposite sides of the domain. Asp89 is relatively confined compared to Asp38. The peptide nitrogen of residue 90 is hydrogen bonded to the carbonyl oxygen of residue 97 on the neighboring β-strand, the peptide nitrogen of residue 39 is not hydrogen bonded because this residue is part of a turn. As shown below, the side chain of Asp38 does not get close to the protein backbone of the neighboring chain, while the geometry of the β-sheet forces the side chain of Asp89 to close proximity.
The closest approach of the Asp38 side chain is to the carbonyl oxygen of residues 39 and 42; but because this loop is not a tight β-turn, the modeled distances are relatively large: 2.5 Å and 2.9 Å, respectively. In the VL-Len Q38D structure, the above distances measured 2.84 and 3.19 Å. Because of the size of the loop, there are no close contacts with neighboring side chains. The φ and ξ values of residue 38 differ in the various mutant Len protein structures we previously determined; the φ and ξ angles range from −118°, 93° in wild-type VL-Len (Pokkuluri et al. 1998) to −145°, 127° in the triple mutant of VL-Len (Pokkuluri et al. 2001). In the VL-Len Q38D mutant, φ and ξ angles of −123°, 103° were found, which fall into the above range. The most likely χ1 and χ2 angles found in the Backbone-Dependent Rotomer Library (Dunbrack et al. 1997) at φ and ξ angles of −120°, 100° are −178° and −166°, close to the χ1 and χ2 values of −170° and −166°, respectively, observed in the Q38D mutant. It has to be pointed out that the conformation of Asp38 as determined in the Q38D structure is influenced by its coordination to the UO22+ ion.
The unfavorable steric interactions of Asp89 in the triple mutant background were modeled to occur with carbonyl oxygens of residues 90 and 97 and the side chain of Phe98. Without the change in φ and ξ angles as a result of the Asp89 substitution, the close contact in the triple mutant would be 2.5 Å for the distance from Asp89 OD1 to the carbonyl of residue 97, given the same χ angles for the Asp side chain. The distance between Asp89 OD1 and the carbonyl of residue 90 is 3.3Å in the modeled structure. Examination of the structures of the mutant and parent molecules showed that with the Asp89 mutation, the structure adjusted to accommodate this residue, with acceptable φ and ξ angles and nonbonded contacts with no obvious strain visible in the molecule. Table 3 shows that there are correlated changes in both monomers in the φ angles of residues 89 and 98 and in the ξ angles of residues 96 and 97. Correlated changes also were observed in the χ angles of residues Gln90, Tyr91, and Phe98, summarized in Table 3.
The φ, ξ values of residue Gln89 varied within restricted ranges (−149°, 129° to −143°, 142°) in previously determined structures of variants of the VL-Len domain. The average values found for Asp89 (−150°, 133°) for the two independent monomers are close to the above ranges found for Gln89. The average observed χ1 and χ2 values are 170° and 49° respectively (Table 4). These χ values compare with −171.7°, 61.2° observed at φ, ξ values of −150°, 130° in the Backbone-Dependent Rotomer Library (Dunbrack et al. 1997) where they represent 20% of the conformations in the above φ, ξ interval.
Table 4.
Average dihedral angles of the quad and triple mutants near residue 89
| Quad | Triple | Quad | Triple | |||||||
| φ | ξ | φ | ξ | χ1 | χ2 | χ3 | χ1 | χ2 | χ3 | |
| Asp/Gln89 | −150 | 133 | 142 | 140 | 170 | 49 | — | 170 | −162 | — |
| Gln90 | −109 | 136 | −113 | 138 | 71 | 83 | 22 | 70 | 75 | 29 |
| Tyr91 | −128 | 36 | −131 | 35 | 48 | −77 | — | 48 | −87 | — |
| Tyr96 | −65 | 153 | −66 | 160 | −66 | −13 | — | −66 | −14 | — |
| Ser97 | −152 | 148 | −152 | 158 | 53 | — | — | 57 | — | — |
| Phe98 | −99 | 155 | −116 | 156 | −88 | 67 | — | −77 | 70 | — |
In the quad mutant, the average φ and ξ values for residue Phe98 are −99°, 155°; these values are comparable to values observed in other Len structures. The χ1 and χ2 values for residue Phe98 are −88°, 67°, which are not favorable; these values compare to previously observed χ1 and χ2 values −77°, 70 in the triple mutant. In the Backbone-Dependent Rotomer Library at φ and ξ values of −100°, 150°, 74% of the residues have χ values of −66°, 98°.
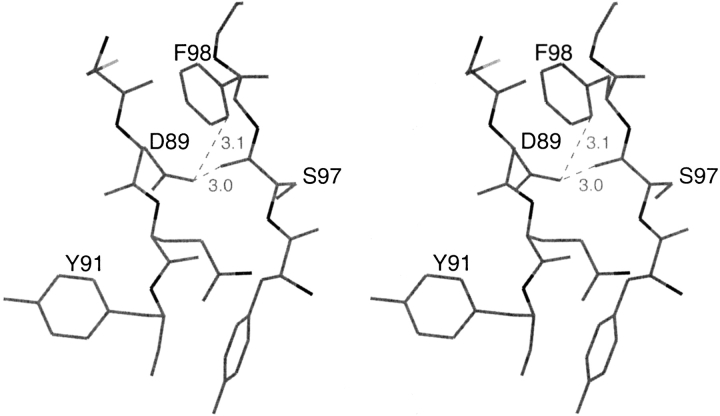
The close contacts made by Asp89 in both monomers are with atom OD1 of Asp89, which is 3.05 Å, 2.94 Å (distances for first and second monomers) from residue 97 carbonyl oxygen and 3.10 Å, 3.12 Å from Phe98 side-chain atom CD1. The ∼3 Å distance between two oxygen atoms and the 3.1 Å distance between an oxygen and a carbon atom are close, but within reasonable limits of nonbonded interactions. However, the interaction of the negative charge on the carboxyl oxygen of Asp89 with the partial negative charge on the carbonyl oxygen of residue 97 is unfavorable. An unfavorable interaction of the carboxyl oxygen of Asp89 with the partial negative charge of the carbonyl oxygen of residue 90 also occurs; the distance between Asp89 OD1 and the carbonyl oxygen of residue 90 is 3.67 and 3.59 in the two monomers.
Although the introduction of Asp at position 89, in the middle of a β-strand of an immunoglobulin variable domain had a major effect on the stability of the protein, it did not cause a major structural perturbance. We do not know the effect of these substitutions on the stability of the unfolded state, therefore we have to discuss the lowering of stability based on the folded state. We find that the observed change in stability is influenced by several factors that include changes in dihedral angles, rotomers, close contacts between atoms, and mainly by the unfavorable charge-charge interactions between the negative charge of the Asp and the partial negative charge of the carbonyl oxygens. The importance of the charge-charge interaction is shown by comparison of the Asp and Asn mutants, the Asp residues are clearly more destabilizing (see Table 1). The neutral Asn residues have similar steric constraints resulting in similar rotomer preferences as the Asp residues, but do not carry a negative charge. Depending on the distance to a neighboring backbone carbonyl the negative charge on the Asp residue could result in varying degrees of destabilization.
A negative charge can alter stability by having both local and global effects. Though Glu affects the global charge distribution of the protein, its longer side chain places the negative charge further from the backbone carbonyls. The Q89D mutation appears to introduce a local repulsion with the protein backbone carbonyl oxygen because the introduction of Glu, a negatively charged residue with a longer side chain, at position 89 has only a relatively minor effect on stability (for the Q89E mutant ΔG = 6.3 kcal/mol; X. Cai, unpubl.). In a structurally related protein, myelin protein zero (MPZ), negatively charged Asp99 residue appears to be required at an equivalent position to Asp89 in Len. Asn substitution at this position leads to a disease state (Marques et al. 1999). In MPZ, positively charged residues Lys101 and Arg38 are in hydrogen bonding distances from Asp99 (Shapiro et al. 1996) and therefore compensate for the negative charge.
Our data are in agreement with the statistical work of McGregor et al. (1987) in which fewer Asp and Asn residues were found in the middle than in the ends of β-strands. We demonstrated that the introduction of Asp in the middle of a β-sheet (at residue 89) is greater than twofold more destabilizing than the substitution of Asp at the end of the β-sheet (at residue 38), the difference in the destabilizing effect of Asn at residues 89 and 38 is less, about a factor of 1.5.
The detrimental effect of Asp within a β-sheet on the stability of an immunoglobulin-type domain could be very serious. A somatic mutation to Asp could prevent the expression of the domain both in vitro and in vivo, or could contribute to the fibril or amorphous precipitate formation of the expressed mutant protein leading to a disease state (Stevens 2000).
Materials and methods
VL mutagenesis, expression and purification
Construction of the vectors producing recombinant VL protein Len and mutants derived from it has been described (Wilkins Stevens et al. 1995). Site-specific mutations of these VL domains were produced with the MORPH Site-Specific Plasmid DNA Mutagenesis Kit (5 Prime to 3 Prime, Inc.) by recombinant PCR (Higuchi et al. 1988) or by the method of Kunkel et al. (1987). All mutations were verified by dideoxy sequencing (Sanger and Coulson 1975). Expression and purification of VLs were performed as described (Wilkins Stevens et al. 1995) except that the Escherichia coli host strain JM83 (Vieira and Messing 1982) was used to improve expression of certain VLs. Extinction coefficients for purified VLs were calculated from the amino-acid sequence with the Wisconsin Sequence Analysis Package (Version 8, September 1994; Genetics Computer Group). Light-chain residues are numbered and the complementarity determining regions (CDRs) are defined according to Kabat et al. (1991).
VL stability measurements
Equilibrium guanidine hydrochloride (GdnHCl) denaturation of VLs was measured by following the increase in tryptophan fluorescence that occurs upon exposure of Trp35, which is highly quenched in the native form. Samples containing 1.5 μM VL, 10 mM sodium phosphate, pH 7.5, and various concentrations of GdnHCl were incubated overnight at 25°C. Fluorescence was measured at 25°C with an SLM Aminco SPF-500C spectrofluorometer (SLM Instruments) with excitation at 295 nm and emission at 350 nm. Raw data were corrected for buffer fluorescence and the denaturation curves were analyzed by the linear extrapolation method using the equation derived by Santoro and Bolen (1988). Nonlinear least-squares fitting, performed with the program KaleidaGraph (Synergy Software), yielded values for ΔGunf, the free energy of unfolding in the absence of denaturant; m, the free energy change per mole of denaturant added; and the statistical error associated with these parameters. The concentration of GdnHCl at the midpoint of the denaturation curve (Cm) was given by ΔGunf/m. The decrease in stability, ΔΔG°unf, was calculated relative to the wild-type protein by multiplying ΔCm, the difference between the mutant and wild-type Cm, by the m value for the wild-type protein (Cupo and Pace 1983).
Structure determination
The Q38D mutant of Len was crystallized, by the hanging drop method, from 25% PEG monomethyl ether 550, 0.01 M zinc sulfate, and 0.1 M MES, pH 6.5 in presence of 1 mM uranyl acetate. Protein concentration was 10 mg/mL. Initially, diffraction data to 2.6 Å were collected using R-Axis IIc and later to 1.6 Å at Argonne National Laboratory Structural Biology Center's beamline 19ID at the Advanced Photon Source. The diffraction data were processed with Denzo (Otwinowski and Minor 1997) and merged and scaled using Scalepack (Otwinowski and Minor 1997).
The unit cell of Q38D contains one monomer per asymmetric unit (VM = 2.4). The structure of Q38D was solved by molecular replacement with the program AMoRe (Navaza 1994) using the model of wild-type Len (PDB code: 1lve; atoms beyond CB removed for residue 38). A solution was obtained with a correlation coefficient of 42% and an R-factor of 44.2%. Rigid body refinement of the resulting model improved the correlation coefficient to 55% and R-factor to 38.7% for 10.0–4.0 Å data. This model was refined further with the program X-PLOR (Brünger 1992) using simulated annealing and positional and restricted individual B-factor refinement. The program CHAIN (Sack 1988) was used for manual rebuilding between cycles. The structure solution and initial refinement were performed using the R-Axis data. Synchrotron data were used in later stages of refinement. Data to 1.6 Å resolution were introduced in small steps. Data collection and final refinement statistics are presented in Table 3.
The Q89D/M4L/Y27dD/T94H mutant of protein Len was crystallized by the hanging drop method from 20% PEG 4000, 20% 2-propanol, 0.1 M sodium citrate, pH 5.6. The protein concentration was 10 mg/mL. Diffraction data to 1.6 Å were collected at Argonne National Laboratory Structural Biology Center's beamline 19ID at the Advanced Photon Source. Before flash cooling for data collection, crystals were soaked briefly in a cryoprotectant solution containing 30% PEG 4000 in addition to all other constituents (except the protein) of the mother liquor in which the crystals were grown. The diffraction data were processed with Denzo (Otwinowski and Minor 1997) then merged and scaled using Scalepack (Otwinowski and Minor 1997).
The unit cell of Q89D/M4L/Y27dD/T94H was found to be isomorphous to that of M4L/Y27dD/T94H (Pokkuluri et al. 2001). The model of M4L/Y27dD/T94H (PDB code:1eeq) was subjected to rigid body refinement against Q89D/M4L/Y27dD/T94H data; atoms beyond CB were removed for residues 89 in both monomers. The R-factor and R-free were 25.9% and 27.3%, respectively for 8.0–3.0 Å data. This model was refined with the program CNS (Brünger et al. 1998) using a maximum likelihood target for refinement; B-factor and bulk solvent corrections were applied. Manual rebuilding was performed using the program CHAIN (Sack 1988). Resolution of the data used in refinement was increased in several small steps to 1.6 Å. Data collection and refinement statistics are shown in Table 3. We checked if the refinement program forced good stereochemistry on the Q89D/M4L/Y27dD/T94H structure and found that the refined structure agreed with the electron density calculated with 3Fo−Fc coefficients after rigid body refinement but before refinement.
Ramachandran plot of the two structures contained 90% of residues in most favored regions with only Ala51 of both monomers in the disallowed region as observed previously (Steipe et al. 1992). Both the structures were deposited in the Protein Data Bank (codes: Q38D, 1efq; Q89D/M4L/Y27dD/T94M, 1eeu).
Fig. 3.
Stereo figure of the conformation of Asp89 showing the short contacts of its carboxyl group with the carbonyl oxygen of residue 97 and side chain of Phe98; these might contribute to the loss of stability.
Acknowledgments
This work was supported by the U.S. Department of Energy, the Office of Biological and Environmental Research under contract No. W-31–109-Eng-38 and by the U.S. Public Health Service Grant DK43757. Use of the Argonne National Laboratory Structural Biology Center beam line at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research, under contract No. W-31–109-Eng-38. We appreciate the help of SBC staff in data collection, and discussions and insights of Dr. W.C. Hanly.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.4920102.
References
- Brünger, A.T. 1992. X-PLOR, version 3.1. A system for X-ray crystallography and NMR. Yale University Press, New Haven, Connecticut.
- Brünger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J-S., Kuszewski, J., Nigles, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T. and Warren, G.L. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta. Cryst. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Chan, W., Helms, L.R., Brook, I., Lee, G., Ngola, S., McNulty, D., Maleeff, B., Hensley, P., and Wetzel, R. 1996. Mutational effects on inclusion body formation in the periplasmic expression of the immunoglobulin VL domain REI. Folding and Design 1 77–89. [DOI] [PubMed] [Google Scholar]
- Cupo, J.F. and Pace, C.N. 1983. Conformational stability of mixed disulfide derivatives of β-lactoglobulin B. Biochemistry 22 2654–2658. [DOI] [PubMed] [Google Scholar]
- Dunbrack, Jr., R.L. and Cohen, F.E. 1997. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 6 1661–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, A.V. 1995. Predicted β-structure stability parameters under experimental test. Protein Eng. 8 207–209. [DOI] [PubMed] [Google Scholar]
- Higuchi, R., Krummel, B., and Saiki, R.K. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 16 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D-B., Chang, C-H., Ainsworth, C., Johnson, G., Solomon, A., Stevens, F.J., and Schiffer, M. 1997. Variable domain structure of κIV human light chain Len: High homology of the murine light chain McPC603. Mol. Immunol. 18 1291–1301. [DOI] [PubMed] [Google Scholar]
- Kabat, E.A., Wu, T.T., Perry, H.M., Gottesman, K.S., and Foeller, C. 1991. Sequences of proteins of immunological interest. 5th ed. NIH publication No. 91–3242, U.S. Department of Health and Human Services, U.S. Government Printing Office, Washington, D.C.
- Kunkel, T.A., Roberts, J.D., and Zakour, R.A. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154 367–382. [DOI] [PubMed] [Google Scholar]
- Marques Jr., W., Hanna, M.G., Marques, S.R., Sweeney, M.G., Thomas, P.K., and Wood, N.W. 1999. Phenotypic variation of new PO mutation in genetically identical twins. J. Neurol. 246 596–599. [DOI] [PubMed] [Google Scholar]
- McGregor, M.J., Islam, S.A., and Sternberg M.J.E. 1987. Analysis of the relationship between side-chain conformation and secondary structure in globular proteins. J. Mol. Biol. 198 295–310. [DOI] [PubMed] [Google Scholar]
- Minor Jr., D.L. and Kim, P.S. 1994. Measurement of the β-sheet-forming propensities of amino acids. Nature 367 660–663. [DOI] [PubMed] [Google Scholar]
- Muñoz, V. and Serrano, L. 1994. Intrinsic secondary structure propensities of the amino acids, using statistical φ-ξ matrices: Comparison with experimental scales. Proteins: Structure, Function, and Genetics 20 301–311. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. In: Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pokkuluri, P.R., Huang, D.-B., Raffen, R., Cai, X., Johnson, G., Wilkins Stevens, P., Stevens, F.J., and Schiffer, M. 1998. A domain flip as a result of a single amino-acid substitution. Structure 6 1067–1073. [DOI] [PubMed] [Google Scholar]
- Pokkuluri, P.R., Raffen, R., Dieckman, L., Boogaard, C., Stevens, F.J., and Schiffer, M. 2002. Increasing protein stability by polar surface residues: Domain-wide consequences of interactions within a loop. Biophys. J. 8 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffen, R., Wilkins Stevens, P., Boogaard, C., Schiffer, M., and Stevens, F.J. 1998. Reengineering immunoglobulin domain interactions by introduction of charged residues. Protein Eng. 11 303–309. [DOI] [PubMed] [Google Scholar]
- Raffen, R., Dieckman, L.J., Szpunar, M., Wunschl, C., Pokkuluri, P.R., Dave, P., Wilkins Stevens, P., Cai, X., Schiffer, M., and Stevens, F.J. 1999. Physicochemical consequences of amino acid variations that contribute to fibril formation by immunoglobulin light chains. Protein Sci. 8 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack, J.S. 1988. CHAIN—A crystallographic modeling program. J. Mol. Graphics 6 224–225. [DOI] [PubMed] [Google Scholar]
- Sanger, F. and Coulson, A.R. 1975. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94 441–448. [DOI] [PubMed] [Google Scholar]
- Santoro, M.M. and Bolen, D.W. 1988. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27 8063–8074. [DOI] [PubMed] [Google Scholar]
- Shapiro, L., Doyle, J.P., Hensley, P., Colman, D.R., and Hendrickson, W.A. 1996. Crystal structure of the extracellular domain from Po, the major structural protein of peripheral nerve myelin. Neuron 17 435–449. [DOI] [PubMed] [Google Scholar]
- Smith, C.K., Withka, J.M., and Regan, L. 1994. A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry 33 5510–5517. [DOI] [PubMed] [Google Scholar]
- Steipe, B., Plückthun, A., and Huber, R. 1992. Refined crystal structure of a recombinant immunoglobulin domain and a complementarity-determining region 1-grafted mutant. J. Mol. Biol. 225 739–753. [DOI] [PubMed] [Google Scholar]
- Street, A.G. and Mayo, S.L. 1999. Intrinsic β-sheet propensities result from van der Waals interactions between side chains and local backbone. Proc. Natl. Acad. Sci. 96 9074–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, F.J. 2000. Four structural risk factors identify most fibril-forming kappa light chains. Amyloid: Int. J. Exp. Clin. Invest. 7 200–211. [DOI] [PubMed] [Google Scholar]
- Swindells, M.B., MacArthur, M.W., and Thornton, J.M. 1995. Intrinsic φ, ξ propensities of amino acids, derived from the coil regions of known structures. Nat. Struct. Biol. 2 596–603. [DOI] [PubMed] [Google Scholar]
- Vieira J. and Messing, J. 1982. The Puc plasmids, an M13Mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19 259–268. [DOI] [PubMed] [Google Scholar]
- Wilkins Stevens, P., Raffen, R., Hanson, D.K., Deng, Y.L., Berrios-Hammond, M. Westholm, F.C. Murphy, C., Eulitz, M., Wetzel, M.R., Solomon, A., Schiffer, M., and Stevens, F.J. 1995. Recombinant immunoglobulin variable domains generated from synthetic genes provide a system for in vitro characterization of light chain amyloid proteins. Protein Sci. 4 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]